Candida africana taxonomical status is controversial. It was proposed as a separate species within the Candida albicans species complex; however, phylogenetic analyses suggested that it is an unusual variety of C. albicans. The prevalence of C. albicans-related species (Candida dubliniensis and C. africana) as vulvovaginal pathogens is not known in Argentina. Moreover, data on antifungal susceptibility of isolates causing vulvovaginal candidiasis is scarce. The aims of this study were to establish the prevalence of C. dubliniensis and C. africana in vaginal samples and to evaluate the antifungal susceptibilities of vaginal C. albicans species complex strains. We used a molecular-based method coupled with a new pooled DNA extraction methodology to differentiate C. dubliniensis and C. africana in a collection of 287 strains originally identified as C. albicans isolated from an Argentinian hospital during 2013. Antifungal susceptibilities to fluconazole, clotrimazole, itraconazole, voriconazole, nystatin, amphotericin B and terbinafine were evaluated by using the CLSI M27-A3 and M27-S4 documents. Of the 287 isolates, 4 C. dubliniensis and one C. africana strains (1.39% and 0.35% prevalence, respectively) were identified. This is the first description of C. africana in Argentina and its identification was confirmed by sequencing the ITS2 region and the hwp1 gene. C. dubliniensis and C. africana strains showed very low MIC values for all the tested antifungals. Fluconazole-reduced-susceptibility and azole cross-resistance were observed in 3.55% and 1.41% of the C. albicans isolates, respectively. These results demonstrate that antifungal resistance is still a rare phenomenon in this kind of isolates.
La clasificación taxonómica de Candida africana está en discusión, es considerada una nueva especie dentro del complejo C. albicans o una variedad inusual de C. albicans. La prevalencia de las especies relacionadas a C. albicans (C. dubliniensis y C. africana) como agentes de vulvovaginitis en Argentina se desconoce. Los objetivos de este trabajo fueron determinar la prevalencia de C. dubliniensis y C. africana en muestras vaginales y evaluar la sensibilidad a los antifúngicos de aislamientos vaginales de las especies del complejo C. albicans. Para diferenciar C. dubliniensis y C. africana utilizamos un método molecular asociado a un nuevo método de extracción de ADN. Se utilizó una colección de 287 cepas originalmente identificadas como C. albicans aisladas durante 2013 en un hospital de Argentina. Se evaluó la sensibilidad a fluconazol, clotrimazol, itraconazol, voriconazol, nistatina, anfotericina B y terbinafina utilizando los documentos M27-A3 y M27-S4 del CLSI. De los 287 aislamientos, se identificaron 4 C. dubliniensis y 1 C. africana (1,39 y 0,35% de prevalencia, respectivamente). Esta es la primera descripción de C. africana en Argentina. Su identificación fue confirmada por secuenciación de la región ITS2 y del gen hwp1. Las cepas identificadas como C. dubliniensis y C. africana mostraron valores de CIM muy bajos para todos los antifúngicos probados. En los aislamientos de C. albicans, la sensibilidad reducida al fluconazol y la resistencia cruzada a todos los azoles se observó en el 3,55% y el 1,41%, respectivamente. Estos resultados demuestran que la resistencia a los antifúngicos es todavía un fenómeno raro en este tipo de aislamientos.
More than twenty different species of Candida have been reported as human pathogens14,20. However, Candida albicans is the most common human fungal pathogen and the most studied of all Candida species8,10,15,17,20. C. albicans has a great degree of variability and some of the firstly known as “atypical” C. albicans isolates were later considered as related species. That is the case with the well-known Candida dubliniensis30. Other “atypical” C. albicans considered as separated species are now described as varieties (e.g. C. albicans var. stellatoidea) and included as one of the 173 C. albicans synonyms10. The most recently described C. albicans related species is Candida africana31. The first C. africana strain was isolated from African patients suffering from vulvovaginal candidiasis (VVC) and firstly considered “atypical” C. albicans strains32. Later, it was proposed as a new species based on morphological, biochemical and physiological differences which include its inability to assimilate glucosamine and N-acetylglucosamine and its impossibility to form chlamydospores on corn meal agar24,31. Odds et al. in 2007 included C. africana in C. albicans clade 13 and supported a varietal status (C. albicans var. africana)16. One year later, the same group included several C. africana strains in a group highly distinct from the majority of C. albicans together with Candida stellatoidea type I and other sucrose-negative atypical C. albicans isolates9. These facts support the controversial taxonomic status of these isolates.
The prevalence of these C. albicans related species as agents of VVC is not known in Argentina. Moreover, data on antifungal susceptibility patterns of isolates causing VVC are scarce12.
The aim of this study was to establish the prevalence of C. dubliniensis and C. africana in a collection of yeasts originally identified as C. albicans and isolated from VVC using a molecular-based method coupled with a pooled DNA extraction methodology. Additionally, we evaluated the antifungal susceptibility patterns of these strains.
Materials and methodsYeast strainsA total of 287 strains were included in this study. All isolates were obtained from patients having vaginal infection symptoms attending the José María Cullen Hospital (Santa Fe – Argentina) during 2013. All the strains were identified as C. albicans by germ tube formation in human serum and their ability to form green color colonies on CHROMAgar Candida™ (Biomerieux – Medica-Tec SRL, Buenos Aires, Argentina). All yeast isolates were referred to the “Micología y Diagnóstico Molecular” Laboratory (CONICET Universidad Nacional del Litoral) where they were stored and identified in accordance with morphology and carbohydrate assimilation and fermentation11 and by a molecular procedure as described below. C. albicans ATCC 90028 and C. dubliniensis NCPF 3949 were used as control strains.
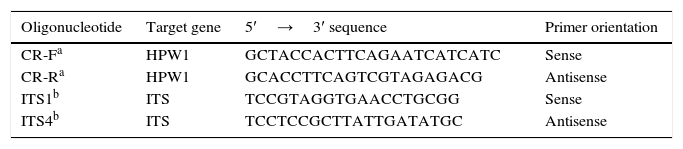
Molecular identification by using hwp1 gene amplicon lengthA PCR method based on hwp1 gene amplification to differentiate C. albicans, C. africana and C. dubliniensis was used. This method identified the three species through the differences in sizes of the obtained PCR fragments: 569bp, 700bp and 941bp, respectively, when the DNA of C. dubliniensis, C. africana and C. albicans DNA was used as template23. Primers are displayed in Table 1.
Individual and pooled DNA extractionThe isolates were individually grown in 1ml of YPD broth (1% yeast extract, 2% Bacto peptone, 2% dextrose) at 35°C for 24h and constant shaking (200rpm). Then, 200μl of 10 individual cultures were mixed to obtain 2ml yeast cells pools from which the pooled DNA was extracted. The remaining 800μl of yeast cultures were kept at −20°C and used later to obtain individual genomic DNAs if necessary. Yeast DNAs (individual and pooled) were extracted using a phenol-based procedure26.
High-throughput screening to rapidly identify C. africana and C. dubliniensis in a strain collectionOriginally, Romeo et al.23 designed their method to identify strains using individual DNAs. In an effort to establish a high throughput screening method to study fungal collections we decided to use the same PCR technique but using pooled DNAs (containing DNAs isolated from ten different strains). The presence of C. africana or C. dubliniensis DNA was suspected when more than one PCR band was observed. To uncover C. dubliniensis and C. africana, we extracted individual DNA from all the pools showing multiple bands (941bp plus 569bp and/or 700bp) and the hwp1 PCR was repeated using individual DNAs as templates. Additionally, we extracted individual DNA for 10 randomly chosen pools showing one 941bp band (100 DNAs in total) and repeated the hwp1 PCR to confirm the specificity of the technique.
Identification confirmationAll the strains identified as C. dubliniensis or C. africana by hwp1 gene amplification were subsequently identified by sequencing the 5.8S RNA gene and adjacent internal transcribed spacer 1 and 2 regions (ITS1 and ITS2) and the hwp1 gene2,35. Primers are displayed in Table 1.
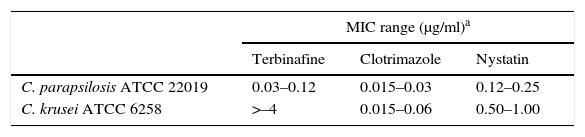
Antifungal susceptibility testingAntifungal drugs tested were fluconazole (FLC), itraconazole (ITC), voriconazole (VRC), clotrimazole (CLT), terbinafine (TRB), anfotericin B (AMB) and nystatin (NYS) (all purchased from Sigma–Aldrich Química – Buenos Aires, Argentina). Drug selection was performed based on the treatment options (topical, vaginal ovules and systemic presentations) available at José María Cullen Hospital (Santa Fe, Argentina). Although AMB is not used to treat vaginal infections, it was added to the list of tested drugs to assess if it was possible to use it as an in vitro surrogate marker for NYS resistance since AMB susceptibility testing is standardized by CLSI and both are polyenes5,6. Inoculums of all the isolates were obtained according to CLSI document M27-A3. Result interpretations were performed according to CLSI documents M27-A3 and M27-S4. FLC, ITC and AMB MIC microtitration plates were produced following CLSI M27-A3 guidelines5,6. In addition, TRB, CLT and NYS were diluted in dimethyl sulfoxide and the final concentrations ranged from 8 to 0.015μg/ml. Since there are no MIC limit ranges for microdilution tests for these antifungal agents, the quality control strains Candida parapsilosis ATCC 22019 and Candida krusei ATCC 6258 were subjected to twenty MIC repetitions on different days (results are displayed in Table 2). Afterwards, this MIC data was added to earlier MIC results obtained by our group8 and were used as a control of the produced plates.
Terbinafine, clotrimazole and nystatin MIC ranges for control strains obtained following CLSI M27-A3 and M27-S4 document recommendations
| MIC range (μg/ml)a | |||
|---|---|---|---|
| Terbinafine | Clotrimazole | Nystatin | |
| C. parapsilosis ATCC 22019 | 0.03–0.12 | 0.015–0.03 | 0.12–0.25 |
| C. krusei ATCC 6258 | >–4 | 0.015–0.06 | 0.50–1.00 |
24h-reading. MIC ranges include 20 repetitions performed in this work plus 20 repetitions performed in a previous work conducted by our group8.
The MIC data presented here are expressed as geometric means (GMs) of three experiments performed on different days. The off-scale MICs were converted to the next concentration up or down in order to be included in the analysis. MIC values were approximated to a normal distribution by transforming them into log2 values. GMs were used to statistically compare MIC values and MIC log2 values were used to establish susceptibility differences between strains. To establish significant levels of MIC differences, a one-way ANOVA test with Bonferroni's correction for multiple comparisons was used. A P value ≤0.05 was considered significant. This study was approved by the School of Biochemistry (Universidad Nacional del Litoral) and JM Cullen ethics committees.
Nucleotide sequence accession numbersThe C. africana strain hwp1 gene sequence was deposited in GenBank under accession number KR704898.
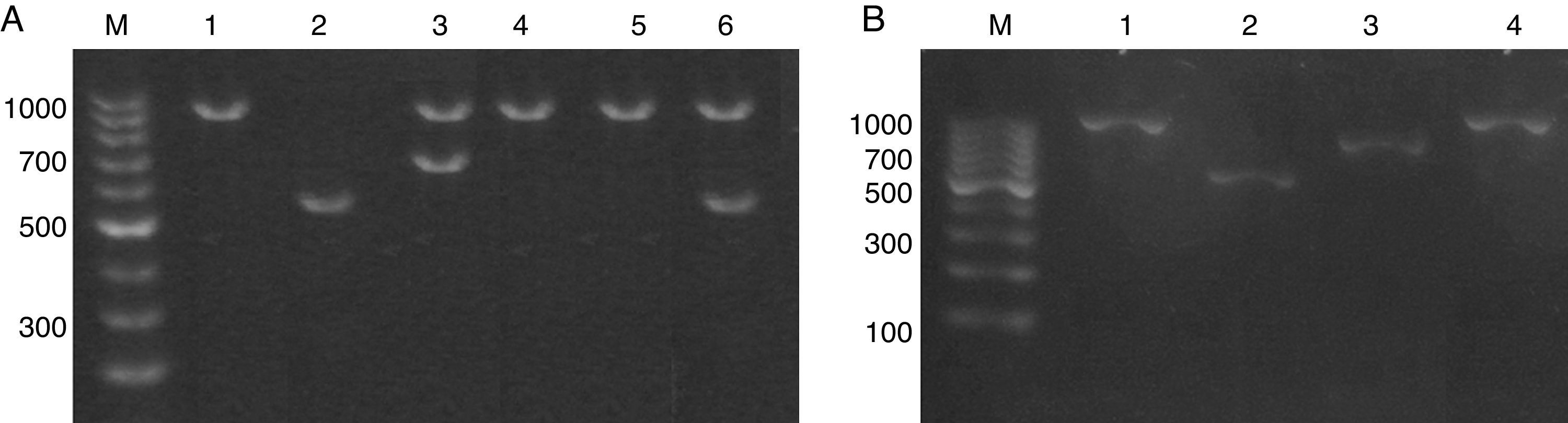
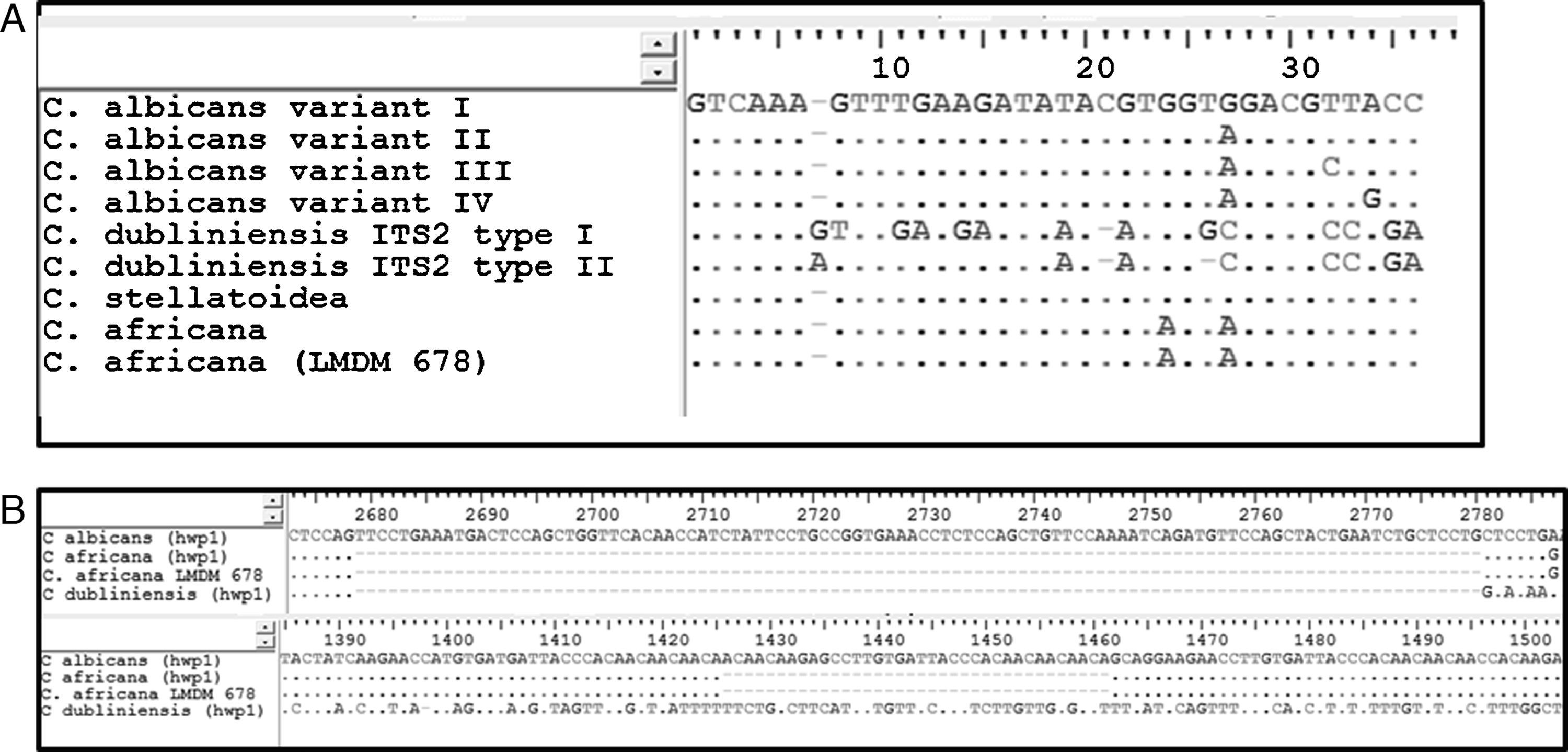
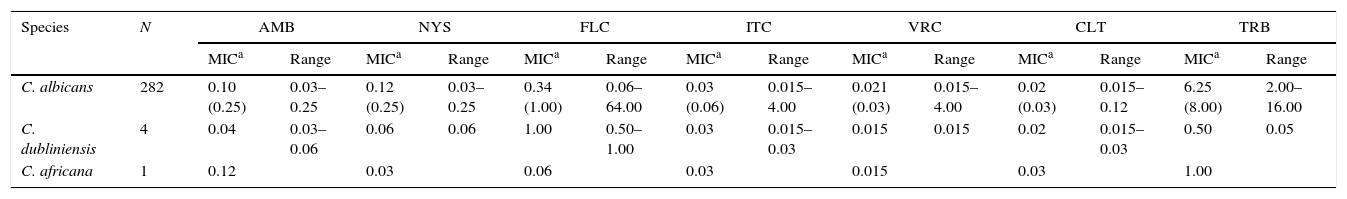
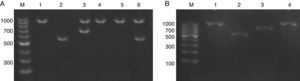
ResultsIdentification and prevalence of C. dubliniensis and C. africanaWith the intention of evaluating the prevalence and potential clinical significance of C. dubliniensis and C. africana in vaginal samples from Santa Fe city, we molecularly identify the 287 isolates received (previously identified by phenotypic methods as C. albicans) by using the amplification of the hwp1 gene using a single primer pair23. We employed a higher throughput methodology using DNA pools obtained from ten different isolates as templates. When pooled DNAs were used, the presence of non-C. albicans was suspected when two or more bands appeared in the electrophoresis. A 941bp fragment along with a 569bp fragment demonstrated the presence of at least one C. albicans and one C. dubliniensis strain in the DNA pool while the presence of a 700bp fragment proved the existence of at least one C. africana strain in the pool (Fig. 1). The presence of two PCR bands was observed in five out of the 29 DNA pools. Four showed a 941–569bp-double band and the fifth showed a 941–700bp-band pair representing the presence of at least four C. dubliniensis and one C. africana strains. These five DNA pools were subsequently studied to confirm the results. Individual DNAs were used as PCR template plate to amplify the hwp1 gene. Four C. dubliniensis and one C. africana were identified. Altogether, we obtained the same results using pooled and individual DNA extraction: four C. dubliniensis and one C. africana strains out of 287 studied isolates (1.39% and 0.35% prevalence, respectively). The identification of these strains was confirmed by ITS and hwp1 sequencing. The ITS obtained sequences were compared with the published ITS signature sequences from C. albicans (variants I to IV) (Gene bank aa. numbers: KP675686.1, HQ876051.1, KP675529.1 and KP675666.1, respectively), C. dubliniensis (variants I and II) (aa. numbers: AB035589.1 and KP131696.1, respectively), C. albicans var. stellatoidea (aa. numbers: AJ853768.1) and C. africana (aa. numbers: AY342214.1)2 (Fig. 2A). The hwp1 gene sequences of our non-albicans strains showed the characteristic nt. gaps (372bp and 241bp difference for C. dubliniensis and C. africana, respectively) (Fig. 2B).
(A) Electrophoresis of the hwp1 gene PCR using DNA pools resolved on 1% agarose gel. Lane M, 100bp size marker. Lane 1, C. albicans control strain (ATCC 90028). Lane 2, C. dubliniensis control strain (NCPF 3949). Lanes 3–6, DNA pools showing hwp1 bands. Lane 3: 941bp and 700bp (C. albicans and C. africana). Lanes 4 and 5: 941bp (C. albicans alone). Lane 6: 941 and 569bp (C. albicans and C. dubliniensis). (B) Electrophoresis on 1% agarose gel of the hwp1 PCR using individual DNAs (from the DNA pool displayed in lane 3 – (A)). Lane M, 100bp size marker. Lane 1, C. albicans control strain (ATCC 90028). Lane 2, C. dubliniensis control strain (NCPF 3949). Lane 3, C. africana (Strain LMDM 678). Lane 4, C. albicans (Strain LMDM 679).
(A) Sequence alignments of the 35-bp portion of the ITS2 region allowing to differentiate the four C. albicans variants, the two C. dubliniensis types, C. albicans var. stellatoidea and C. africana2. C. africana strain LMDM 678 showed no differences with the C. africana type strain. Dots indicate conserved nucleotides; gaps are represented as hyphens and nucleotide differences are displayed as letters. (B) Sequence alignments of regions of the hwp1 gene where gaps were encountered. The upper line displays C. albicans hwp1 (GenBank accession number U64206.1); second line: C. africana hwp1 (EU477610.1); third line: C. africana LMDM 678 (KR704898) and fourth line: C. dubliniensis (AJ632273.1).
Phenotypically, our C. africana strain differed from the C. albicans control strains in its inability to form chlamydospores on corn meal agar, to assimilate trehalose and to grow at 42°C. However, it developed green colonies on CHROMagar and formed germ tubes under regular conditions (2h at 37°C). Our C. africana showed a lower rate of germ tube formation when compared to C. albicans and C. dubliniensis control strains.
Antifungal susceptibility testingThe in vitro activities of the tested antifungal drugs are summarized in Table 3. All non-C. albicans strains included in this study (4 C. dubliniensis and the C. africana) showed very low MIC values for all the tested antifungal agents. Similarly, antifungal drugs showed good in vitro activity against the majority of the remaining 282 isolates (C. albicans). The most potent azole was CLT showing the lowest MIC values (GM=0.02μg/ml, MIC90=0.03μg/ml) and only thirteen strains showed CLT MIC value ≥0.25μg/ml. FLC was the tested drug with the widest range of MIC values (0.06–64.00μg/ml). Reduced susceptibility to FLC was observed in ten C. albicans strains out of the 282 studied (3.55%). Of these ten strains, three were FLC susceptible-dose dependent while seven were resistant to FLC (three strains were exclusively resistant to FLC and four strains were azole-cross-resistant). Concerning the oral triazoles, ITC and VRC showed very good in vitro activity (GM=0.03μg/ml and a MIC90=0.06μg/ml for ITC and GM=0.021μg/ml and a MIC90=0.03μg/ml). There were only four (1.41%) C. albicans strains showing high ITC and VRC MIC values (≥4.00μg/ml). These strains showed FLC cross-resistance (FLC MIC ≥16μg/ml) and high CLT MIC values (≥0.25μg/ml).
In vitro susceptibilities of the strains included in the study. MIC values are expressed in μg/ml
| Species | N | AMB | NYS | FLC | ITC | VRC | CLT | TRB | |||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| MICa | Range | MICa | Range | MICa | Range | MICa | Range | MICa | Range | MICa | Range | MICa | Range | ||
| C. albicans | 282 | 0.10 (0.25) | 0.03–0.25 | 0.12 (0.25) | 0.03–0.25 | 0.34 (1.00) | 0.06–64.00 | 0.03 (0.06) | 0.015–4.00 | 0.021 (0.03) | 0.015–4.00 | 0.02 (0.03) | 0.015–0.12 | 6.25 (8.00) | 2.00–16.00 |
| C. dubliniensis | 4 | 0.04 | 0.03–0.06 | 0.06 | 0.06 | 1.00 | 0.50–1.00 | 0.03 | 0.015–0.03 | 0.015 | 0.015 | 0.02 | 0.015–0.03 | 0.50 | 0.05 |
| C. africana | 1 | 0.12 | 0.03 | 0.06 | 0.03 | 0.015 | 0.03 | 1.00 | |||||||
AMB: amphotericin B. NYS: nystatin. FLC: fluconazole. ITC: itraconazole. VRC: voriconazole. CLT: clotrimazole. TRB: terbinafine.
Both tested polyenes showed a narrow range of MIC values (0.03–0.25μg/ml for both AMB and NYS) and showed no differences in in vitro activity (p=0.43). None of the 287 strains showed high MIC values for NYSor for AMB. Furthermore, TRB was the least active drug against the isolates tested (GM=6.25μg/ml and MIC90=8.00μg/ml). However, the five non-albicans isolates including four C. dubliniensis and one C. africana showed slightly lower TRB MIC values, ranging from 1.00 to 2.00μg/ml (p=0.42).
DiscussionIn the present study, we used the amplification of the hwp1 gene to evaluate the presence of C. africana and C. dubliniensis among vaginal isolates identified as C. albicans in accordance with their phenotypic characteristics. We demonstrated that the amplification of the hwp1 gene using pooled DNAs was as efficient as using individual DNAs to differentiate C. dubliniensis and C. africana from C. albicans. Thus, this methodology can be used as a high throughput screening method to study fungal collections.
Earlier reports from Europe and Africa demonstrated that C. africana represented more than 5% of the C. albicans species complex isolated from female genital specimens and that it was more prevalent than C. dubliniensis in this type of samples2,13,25,31. Conversely, in our patients, C. africana was less prevalent than C. dubliniensis (0.35% and 1.39%, respectively).
To the best of our knowledge, the described C. africana strain is the first autochthonous Argentinian isolate reported so far. The strain was identified as C. africana by two different sequencing-based methods (ITS and hwp1 gene sequencing). Phenotypically, our C. africana strain showed characteristics that coincide with previous reports2,25,33. We observed a lower filamentation rate in serum for the C. africana isolate. This fact was reported also by Borman et al2. These authors linked this lower hyphal formation capacity to the reduced virulence of C. africana and its narrow anatomical niche.
When antifungal susceptibility was analyzed, our C. africana and C. dubliniensis strains showed low MIC values to all the antifungal tested. Moreover, reduced susceptibility was rare in all the Candida strains studied, reinforcing the idea that C. albicans and its related species isolated from VVC patients are usually susceptible to antifungal agents7,8,12,21,22,29. In the case of azoles, CLT and ITC showed the highest potency and FLC MIC GMs values were similar to those obtained in previous reports for yeast causing VVC1,3,22. However, 3.55% (10/282) of the C. albicans strains showed FLC reduced susceptibility (three susceptible-dose dependent), three FLC-resistant and four of these strains were azole cross-resistant. This FLC resistance frequency in C. albicans isolated from VVC was higher than that mentioned in other published reports, where there was no FLC resistance12,21,29.
These susceptibility patterns and the wide range of FLC MIC values observed in our strain collection allow us to infer that in Santa Fe city several molecular mechanism of azole resistance may coexist and antifungal resistance could become a problem in the future4,18,19,27,28,34.
Here we report the first Argentinian C. africana isolate and a high-throughput screening method to uncover the presence of this newly described species variety in a collection of yeasts isolated from VVC. We also demonstrate that C. albicans related species have low prevalence in our region. Moreover, antifungal resistance seems not to be a serious concern in Santa Fe city region (Argentina). However, VVC treatment should be supported by laboratory results.
Ethical disclosuresProtection of people and animals subjectsThe authors declare that no experiments were performed on humans or animals for this study.
Confidentiality of dataThe authors declare that this article does not appear patient data.
Right to privacy and informed consentThe authors declare that this article does not appear patient data.
Conflict of interestThe authors declare that they have no conflicts of interest.
This work was financially supported in part by grants CAI+D prog. RH and PEIS 2011 both from Universidad Nacional del Litoral to G.G.E. and S.G., respectively. C.D. has a predoctoral fellowship from Consejo Nacional de Investigaciones Científicas y Técnicas (CONICET).