Herpes simplex virus (HSV)-based vectors have been used as platforms for the generation of genetic vaccines against different viruses. Some of the advantages of defective HSV-1 vectors include: (a) HSV-1 can infect most cell types, including antigen-presenting cells; (b) HSV-1 vectors have a large transgene capacity (up to 150-kbp); (c) HSV-1 vectors can be designed to cause no toxicity; (d) different studies showed that mice vaccinated with such vectors encoding antigens from different pathogens were partially protected1,3, indicating that HSV-1 based vectors are attractive tools for the development of safe genetic vaccines.
There are two main platforms of HSV-1-derived vectors: defective recombinant viruses (deleted in ICP4, ICP27 and ICP22 genes) and amplicons. Defective HSV-1 viruses can only be propagated using a complementing cell line. Production of amplicons depends on helper functions, provided in trans. In both systems, different genetic elements were intentionally introduced to conduct the heterologous gene expression and to allow the titration of the vector stocks in different cell lines2,4.
In our group, HSV-1 vectors were developed for the expression of several rotavirus genes. Genes from other pathogens were also expressed, thus generating a set of tools to be tested as new vaccine candidates against different diseases.
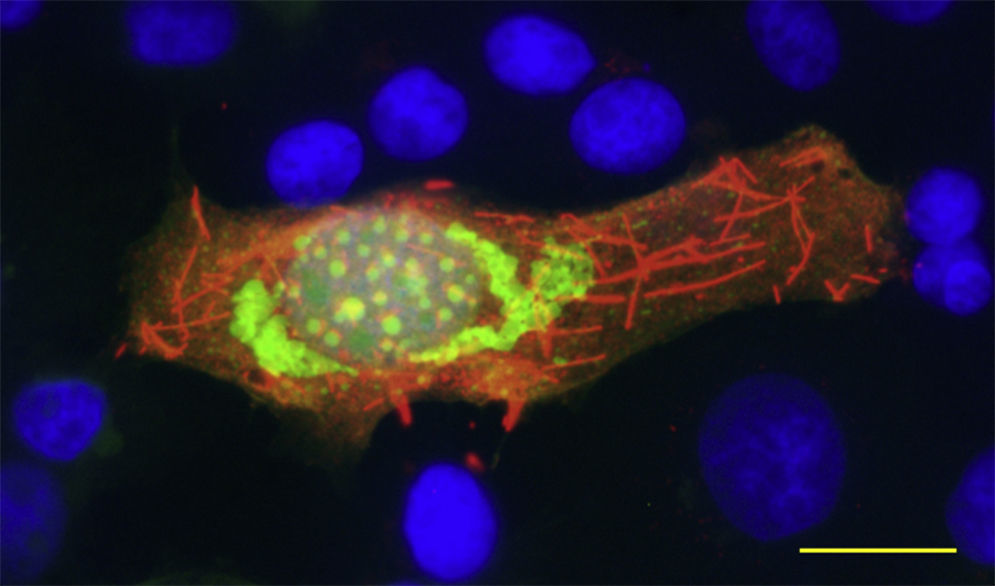
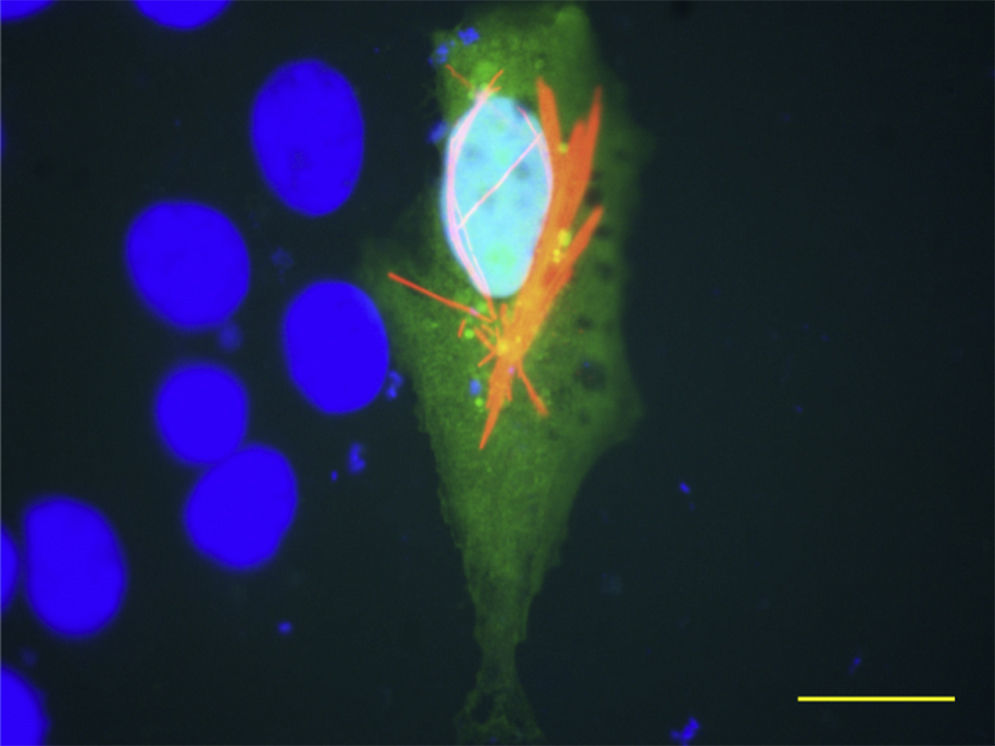
In this section, we show two different HSV-1 vectors that express the VP6 protein from different rotavirus strains. Figure 1 shows a defective recombinant virus expressing simultaneously Enhanced Green Fluorescent Protein (EGFP) and VP6 from murine rotavirus strain EC, in Figure 2 and movie S1, an amplicon vector overexpressing VP6 from a Wa strain of human rotavirus is shown. In both cases, the VP6 proteins were recognized by a monoclonal antibody and were found extensively expressed forming filaments along the cells, showing a typical intra-cellular pattern, in the absence of other rotavirus protein. These results show that the recombinant proteins maintain the specific capacity of self-assembly of the native protein and, therefore, are potential vaccine candidates to be explored. Recently, results obtained by our group and in collaboration with others, have demonstrated the feasibility of these systems to induce protective immune responses against different viral infections1,3.
MDBK cells were transduced with T0EC-VP6 recombinant vector. Permeabilized cells were probed with monoclonal antibody (MAb) 2F, directed against the UK strain rotavirus VP6, followed by Alexa-594 anti-mouse antibody (Molecular Probes, USA). Nuclear DNA was stained with 4′,6,-Diamidino-2-Phenylindole, Invitrogen (DAPI). Wa-VP6 and EGFP expression were observed under the microscope and the obtained images were merged into the same picture. Bar, 20μm.
Vero cells were transduced with HSV [Wa-VP6] amplicon vector. Permeabilized cells were probed with MAb 2F, followed by Alexa-594 anti-mouse antibody (Molecular Probes, USA). Nuclear DNA was stained with 4′,6,-Diamidino-2-Phenylindole, Invitrogen (DAPI). Wa-VP6 and EGFP expression were observed under the microscope, and the obtained images were merged into the same picture. The bar indicates 20μm.
The authors declare that the procedures followed were in accordance with the regulations of the relevant clinical research ethics committee and with those of the Code of Ethics of the World Medical Association (Declaration of Helsinki).
Confidentiality of dataThe authors declare that no patient data appear in this article.
Right to privacy and informed consentThe authors declare that no patient data appear in this article.
Conflict of interestThe authors declare that they have no conflicts of interest.





![Vero cells were transduced with HSV [Wa-VP6] amplicon vector. Permeabilized cells were probed with MAb 2F, followed by Alexa-594 anti-mouse antibody (Molecular Probes, USA). Nuclear DNA was stained with 4′,6,-Diamidino-2-Phenylindole, Invitrogen (DAPI). Wa-VP6 and EGFP expression were observed under the microscope, and the obtained images were merged into the same picture. The bar indicates 20μm. Vero cells were transduced with HSV [Wa-VP6] amplicon vector. Permeabilized cells were probed with MAb 2F, followed by Alexa-594 anti-mouse antibody (Molecular Probes, USA). Nuclear DNA was stained with 4′,6,-Diamidino-2-Phenylindole, Invitrogen (DAPI). Wa-VP6 and EGFP expression were observed under the microscope, and the obtained images were merged into the same picture. The bar indicates 20μm.](https://static.elsevier.es/multimedia/03257541/0000004700000001/v4_201504110401/S0325754115000127/v4_201504110401/en/main.assets/thumbnail/gr2.jpeg?xkr=ue/ImdikoIMrsJoerZ+w96p5LBcBpyJTqfwgorxm+Ow=)
