Bovine mastitis poses a significant threat to global dairy production, resulting in substantial losses in milk production. Streptococcus bacteria, particularly Streptococcus uberis, Streptococcus agalactiae, and Streptococcus dysgalactiae, are commonly implicated in this condition. An accurate diagnosis is crucial for implementing effective treatment and minimizing its impact on production. This study examined 115 Streptococcus strains isolated from bovine mastitis cases in Uruguay using PCR for species identification. Additionally, the resistance to tetracycline, erythromycin, and penicillin was assessed in 81 of the bacterial strains under study. Significant disparities between phenotypic and genotypic detection were evident across all three species, with only 31% of strains identified phenotypically aligning with PCR results. Phenotypic prevalence indicated S. dysgalactiae as the most prevalent (44.35%), followed by S. uberis (24.34%) and S. agalactiae (6.09%). However, the genotypic identification revealed S. uberis as the most prevalent, followed by S. dysgalactiae, while S. agalactiae remained the least prevalent. The high sensitivity and speed of PCR suggest its potential routine implementation for diagnosing bovine mastitis caused by Streptococcus in any laboratory. Although, penicillin resistance was practically nonexistent, tetracycline and erythromycin exhibit higher resistance levels across all three species studied. In conclusion, the study underlines the importance of early diagnosis, highlights variations in bacterial prevalence, and proposes PCR as a valuable diagnostic tool for Streptococcus species responsible for bovine mastitis.
La mastitis bovina constituye una amenaza significativa para la producción láctea global, al ocasionar considerables pérdidas en la producción de leche. Streptococcus uberis, Streptococcus agalactiae y Streptococcus dysgalactiae son las especies comúnmente asociadas con esta enfermedad. El diagnóstico preciso es fundamental para implementar un tratamiento eficaz y minimizar su impacto en la producción lechera. Se analizaron 115 cepas de Streptococcus spp. aisladas de casos de mastitis bovina, que se identificaron a nivel de especie mediante PCR convencional. Además, se evaluó la resistencia a tetraciclina, eritromicina y penicilina en 81 de estas cepas. Se observaron disparidades significativas entre la detección fenotípica de rutina y genotípica en las tres especies, con solo un 31% de coincidencia entre ambos procedimientos. La prevalencia fenotípica indicó que S. dysgalactiae fue la especie más común (44,35%), seguida por S. uberis (24,34%) y S. agalactiae (6,09%). Sin embargo, la identificación genotípica ubicó como más prevalente a S. uberis, seguida por S. dysgalactiae, mientras que S. agalactiae permaneció como la menos prevalente. La PCR demostró una alta sensibilidad y rapidez como herramienta diagnóstica. A pesar de la escasa resistencia a la penicilina detectada, se constataron niveles más altos de resistencia a tetraciclina y eritromicina en las tres especies estudiadas. En conclusión, los hallazgos de este estudio evidencian variaciones en la prevalencia bacteriana según el método de identificación empleado y proponen el uso de PCR como una herramienta valiosa para establecer con precisión y precozmente la especie de Streptococcus responsable de la mastitis bovina.
Uruguay is one of the largest milk producers in Latin America, with some 2.2 billion liters per year. Notably, 70% of this production is exported to over 60 countries, positioning Uruguay as the 7th largest exporter of milk and dairy products worldwide17. This prominent economic position emphasizes the importance of preserving the health and productivity of the dairy cattle population in the country.
However, a persistent challenge within the dairy industry is bovine mastitis (BM), one of the most prevalent diseases in dairy cattle. BM manifests itself in clinical and subclinical forms, posing substantial threats to animal welfare and milk quality21,26. Streptococcus agalactiae (S. agalactiae), Streptococcus dysgalactiae (S. dysgalactiae), and Streptococcus uberis (S. uberis) emerge as critical pathogens contributing to the prevalence of mastitis, with their distinct characteristics and implications for dairy farming18. A study published in 2002 determined that in Uruguay, the most commonly isolated bacteria in order of prevalence were Staphylococcus aureus, coagulase-negative Staphylococcus, S. agalactiae, S. dysgalactiae, S. uberis, Corynebacterium bovis, Escherichia coli, Klebsiella spp., and Trueperella pyogenes14. A more recent study from 2022 continues to position S. aureus as the main pathogen causing bovine mastitis10.
Although S. dysgalactiae was initially considered less virulent than S. agalactiae, it has been observed that S. dysgalactiae can cause severe mammary gland infections. Some authors consider S. dysgalactiae to be an environmental pathogen; however, it manages to persist in the mammary gland, using it as a reservoir. Therefore, it is also classified as a contagious pathogen6,32. Most strains of S. dysgalactiae are non-hemolytic (although there are some alpha-hemolytic strains), CAMP (Christine-Atkinson-Munch-Peterson test) negative, do not degrade esculin and are usually classified into Lancefield group C18. S. agalactiae is closely associated with subclinical mastitis; it is a contagious pathogen capable of colonizing the gastrointestinal tract of dairy cows and can survive indefinitely in the mammary gland forming biofilm7. It is considered beta-hemolytic, but non-hemolytic strains have been observed; it is CAMP-positive and belongs to group B of the Lancefield classification30. Finally, S. uberis is mainly environmental, although cases of transmission have been observed from an infected animal to a healthy one18. This species is primarily alpha-hemolytic, but has also been shown to be non-hemolytic in some cases; CAMP and esculin degradation are variable and also present variability concerning the Lancefield group it belongs to, which can be E, G, P, or U18.
The Streptococcus genus can be identified through growth on blood agar media and a negative catalase test. However, this diagnostic method has notable drawbacks, including a high rate of false negative results (27–50%) and the need for laborious and time-consuming analyses (48–72h)3. Traditional phenotypic methods face limitations, with approximately 30% of mastitis cases remaining unidentified due to the reliance on detecting only viable pathogens3.
Due to the considerable phenotypic variability exhibited by these pathogens, molecular biology techniques such as polymerase chain reaction (PCR), mass spectrometry (MALDI/TOF), and next-generation sequencing (NGS) have started to be employed for their identification. These methods have proven to be faster, more sensitive, and more specific than conventional culturing for diagnosing mastitis-causing pathogens3,6,12,15,33,34. It is suggested that the PCR technique can be combined with culture-based diagnostic techniques to identify bacteria responsible for mastitis5,19,31.
Antimicrobials are often used for both the treatment and prophylaxis of bovine mastitis22. It is estimated that more than 50% of all antimicrobials used worldwide are employed in veterinary medicine. The most commonly used are penicillins, sulfonamides, ampicillin, cloxacillin, and aminoglycosides7,33. Moreover, antimicrobial therapy has raised significant public health concerns; their administration to animals increases the potential for spreading resistant microorganisms both along the food chain and in various ecosystems2. In Uruguay, there is a great lack of information regarding antimicrobial resistance in Streptococcus strains that cause mastitis. The most current work in 2014 by Gianneechini et al.13 reported that S. agalactiae and S. dysgalactiae were mainly resistant to tetracycline and enrofloxacin and that S. uberis showed high sensitivity to all antibiotics tested. Another study revealed that some strains of S. uberis and S. dysgalactiae were primarily resistant to tetracycline and erythromycin. In contrast, S. agalactiae strains were sensitive to these antibiotics24. In addition, sensitivity of all the Streptococcus strains to β-lactam antibiotics was observed. However, an increase in the minimum inhibitory concentration (MIC) was noted, which could reflect a future change in the susceptibility of these microorganisms23.
This study aims to address the phenotypic and genotypic identification of S. agalactiae, S. dysgalactiae, and S. uberis, employing traditional phenotypic methods in routine diagnostic laboratories and advanced molecular biology techniques, such as multiplex PCR. Additionally, the resistance profiles of the identified strains were analyzed.
Materials and methodsPhenotypic identification and DNA extractionA total of 127 strains isolated from milk samples of cows with clinical mastitis referred to two private laboratories located in the departments of Colonia and San José during the years 2019–2023 were analyzed. The isolates were identified phenotypically using biochemical tests following the National Mastitis Council guidelines25. The milk samples belonged to different dairy herds in the southern dairy basin of Uruguay, comprising 23 commercial dairy farms, all equipped with milking machines. As a first step, colony morphology was observed, and Gram staining, the catalase test, and the hemolysis production test by growth on blood agar base with 5% sheep blood (blood agar) were performed. Subsequently, the tests available in the laboratory that were performed to identify the species were the CAMP test, growth in Edwards medium with esculin, and determination of the Lancefield group. These tests identified the strains as S. agalactiae, S. uberis, and S. dysgalactiae.
All strains identified in this study were stored in a −80°C freezer. They were grown in 1.5ml microtubes in trypticase soy broth (TSB) and incubated with agitation for 48h at 37°C. The tubes were then centrifuged for 7min at 5000rpm, and the pellet was resuspended in 20% glycerol broth (20ml glycerol per 80ml TSB).
Pure culture obtained on blood agar after 48h of incubation at 37°C was used for genomic DNA extraction, which was performed using the thermal lysis method with achromopeptidase enzyme at a concentration of 10U/μl (Sigma-Aldrich®, St. Louis, MO)11. Briefly, 95μl of diethyl pyrocarbonate-treated water (DEPC-treated water) was incorporated into a 1.5ml tube, and then 5μl of achromopeptidase solution was added. Two or three colonies were taken from the plate and incorporated into the microtube containing the enzyme by vigorous vortexing. The microtubes were thermoblocked for 10min at 50°C and then at 94°C. They were then centrifuged for 7min at 17000g, and 80μl of the supernatant was stored at −20°C until further use11. The purity and amount of extracted DNA were evaluated in NanoDrop™ One (Thermo Scientific™), using the absorbance ratio 260/280 and DNA concentration measured in ng/μl.
Streptococcus spp. strain identification by multiplex PCRThe Streptococcus spp. strains were identified by multiplex PCR32 (Table 1). A DNA mixture, at equal concentration, of ATCC strains of the three Streptococcus species (ATCC, Washington, DC, USA): S. uberis (ATCC 9927), S. agalactiae (ATCC 13813) and S. dysgalactiae (ATCC 12394) was used as a positive control, and DEPC-treated water was used as a negative control.
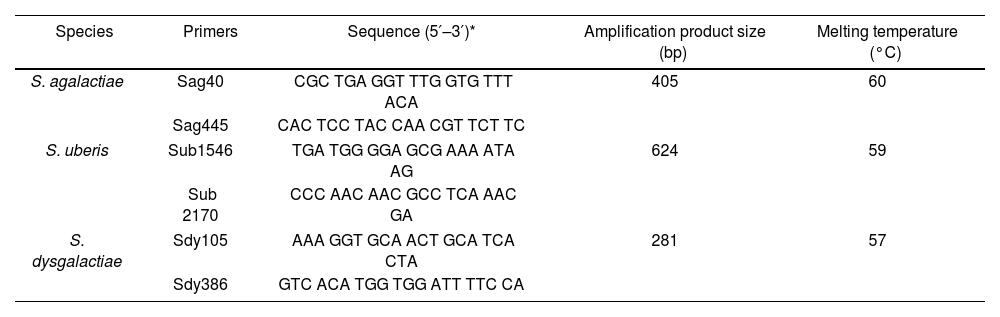
Primers for identifying S. dysgalactiae, S. uberis, and S. agalactiae by multiplex PCR.
| Species | Primers | Sequence (5′–3′)* | Amplification product size (bp) | Melting temperature (°C) |
|---|---|---|---|---|
| S. agalactiae | Sag40 | CGC TGA GGT TTG GTG TTT ACA | 405 | 60 |
| Sag445 | CAC TCC TAC CAA CGT TCT TC | |||
| S. uberis | Sub1546 | TGA TGG GGA GCG AAA ATA AG | 624 | 59 |
| Sub 2170 | CCC AAC AAC GCC TCA AAC GA | |||
| S. dysgalactiae | Sdy105 | AAA GGT GCA ACT GCA TCA CTA | 281 | 57 |
| Sdy386 | GTC ACA TGG TGG ATT TTC CA | |||
The PCR mixture was performed in a final volume of 25μl: 14μl of Mango Mix (Bioline®, Meridian Life Science, Inc), 0.4μM of each of the primers corresponding to the three species, 100–200ng/μl of DNA, and DEPC-treated water until the final volume was completed. Subsequently, the mixture was thermally cycled once at 94°C for 1min, then by 35 cycles, 45s at 94°C, 1min at 58°C and 2min at 72°C. Finally, once the cycles were completed, an extra extension step of 72°C for 1min was added. The PCR-amplified products were observed by electrophoresis on a 2% agarose gel in 1X Buffer TAE at 100V for 1h, followed by staining using GoodView™ (Beijing SBS Genetech Co., Ltd). A 100-bp molecular weight marker (Thermo Scientific™) was used to identify the products of interest.
Antimicrobial susceptibility testingAntimicrobial sensitivity was assessed in 81 strains previously identified through multiplex PCR using the Kirby-Bauer agar disc diffusion method following the guidelines of the Clinical Laboratory Standard Institute (CLSI VET01S ED6:2023)8. Discs containing antibiotics were employed at the following concentrations: penicillin (10U), tetracycline (30μg), and erythromycin (15μg). The isolates were cultured at 37°C on trypticase soy agar plates supplemented with 5% sheep blood. The cultured bacteria were suspended in normal saline, and the suspensions were adjusted to a 0.5 McFarland standard. The suspension was inoculated with a sterile swab onto Mueller-Hinton agar (Oxoid) supplemented with 5% sheep blood, followed by incubation at 37°C for 48h.
Data analysisIndividual samples were assigned unique identifiers using Microsoft Excel spreadsheets. The identification results for each bacterial strain were categorized as positive or negative at both phenotypic and molecular levels. To assess potential statistically significant differences in species determination between phenotypic and molecular genotypic methods, the chi-squared contingency test was employed. In addition, Fisher's exact test was applied to identify any significant differences between the three species when comparing the two methods, and Cohen's test to determine the concordance between the two methodologies.
ResultsStrain viability control and confirmation genusOut of the 127 strains isolated and stored at −80°C, 115 were used in this study. They were cultured on 5% blood agar, and Gram stain and catalase tests were performed to confirm viability and purity.
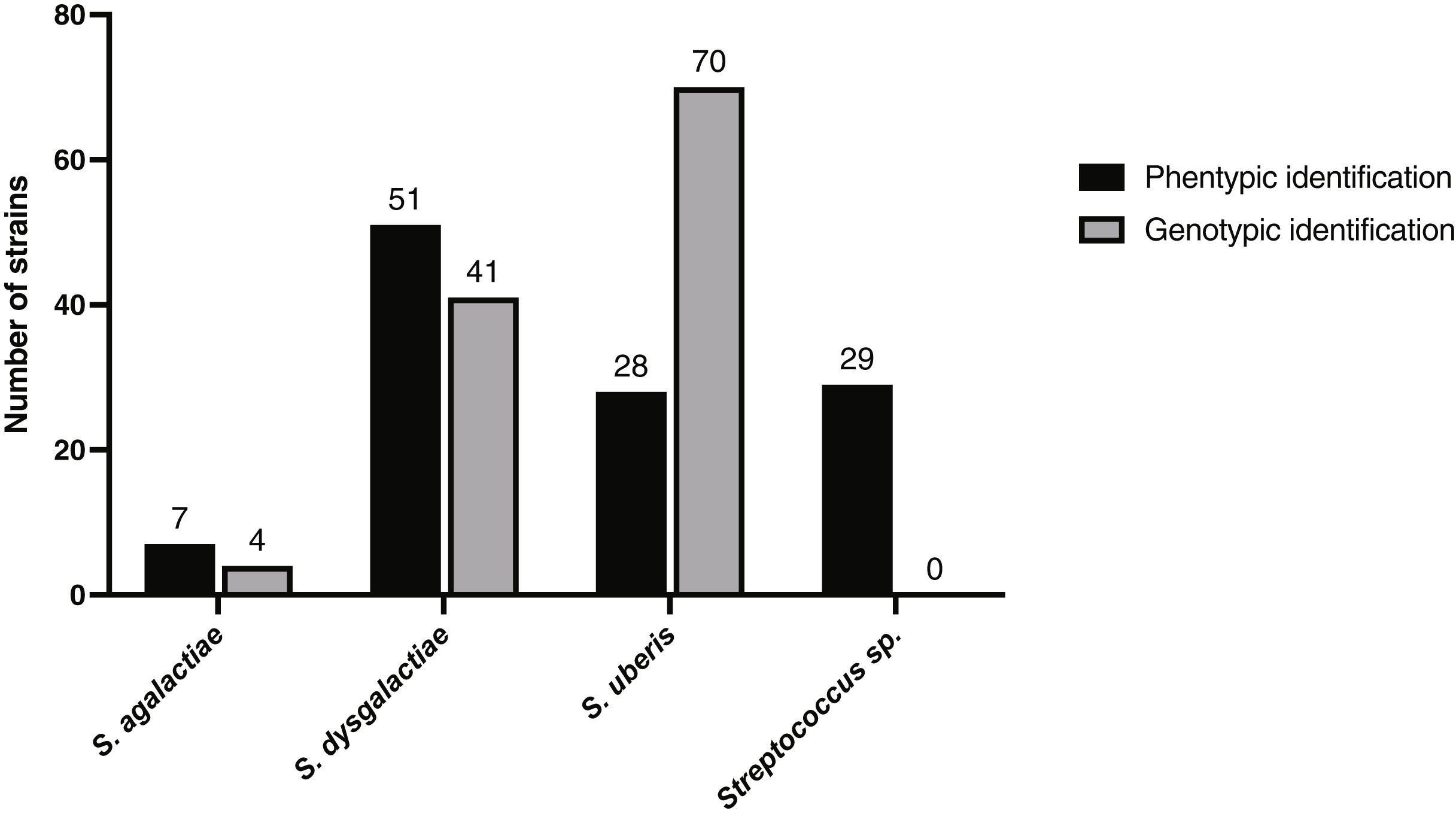
Genotypic identification of strains and comparison with phenotypic identificationSignificant differences were found between the results regarding the phenotypic and genotypic identification of the 115 strains (p-value=0.0018). The phenotypic identification suggested that S. dysgalactiae was the predominant pathogen, and the genotypic identification revealed that S. uberis was the most common. Moreover, it was observed that both S. dysgalactiae and S. uberis identification by molecular methods differ significantly from the phenotypic methods (p-value=0.0059 for S. dysgalactiae and p-value=0.00007 for S. uberis). Multiplex PCR showed that 60.87% (70/115) of strains corresponded to S. uberis, 3.48% (4/115) to S. agalactiae, and 35.65% (41/115) to S. dysgalactiae (Fig. 1). Based on the phenotypic identification, of the total 115 strains, 24.34% (28/115) were identified as S. uberis, 6.09% (7/115) as S. agalactiae, 44.35% (51/115) as S. dysgalactiae, and 25.22% (29/115) could not be categorized by species through phenotypic identification, reported as Streptococcus spp. (Fig. 1).
Excluding the 29 samples with phenotypically unidentifiable species, only 30.23% (26/86) had coincident results. There was minimal concordance in the strains identified through phenotypic and genotypic methods.
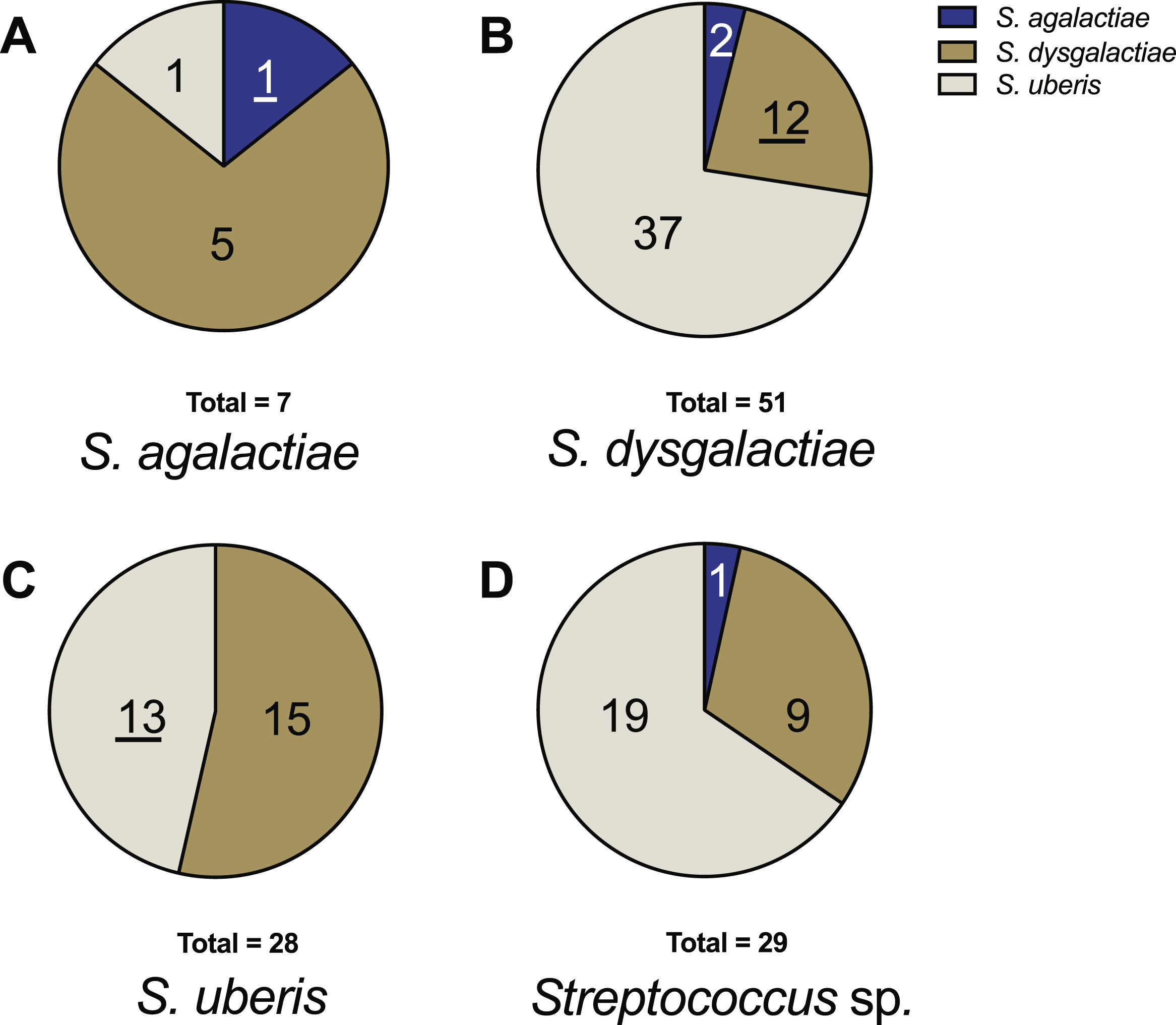
Concerning strains identified phenotypically as S. agalactiae, only 14.3% (1/7) coincided with both genotypic and phenotypic identification results. For non-coincident identifications, 71.4% (5/7) were identified as S. dysgalactiae and 14.3% (1/7) as S. uberis by multiplex PCR (Fig. 2). For S. dysgalactiae, only 23.5% (12/51) of the phenotypic results coincided with the genotypic results, while 72.5% (37/51) corresponded to S. uberis and 3.9% (2/51) to S. agalactiae (Fig. 2). Regarding S. uberis strains, 46.4% (13/28) coincided in phenotypic and genotypic identification. Conversely, 53.6% (15/28) of strains initially identified as S. uberis phenotypically were identified as S. dysgalactiae through molecular methods. None of these strains were identified as S. agalactiae during molecular identification (Fig. 2). For strains identified phenotypically as Streptococcus spp. (29 samples), the genotypic analysis categorized them as S. agalactiae 3% (1/29), S. dysgalactiae 31% (9/29), and S. uberis 66% (19/29) (Fig. 2).
Distribution of genotypic identification for each species in relation to their phenotypic identification. (A) Strains phenotypically identified as S. agalactiae. (B) Strains phenotypically identified as S. dysgalactiae. (C) Strains phenotypically identified as S. uberis. (D) Strains phenotypically identified as Streptococcus sp. The underlined numbers represent the strains that matched in both identification methods.
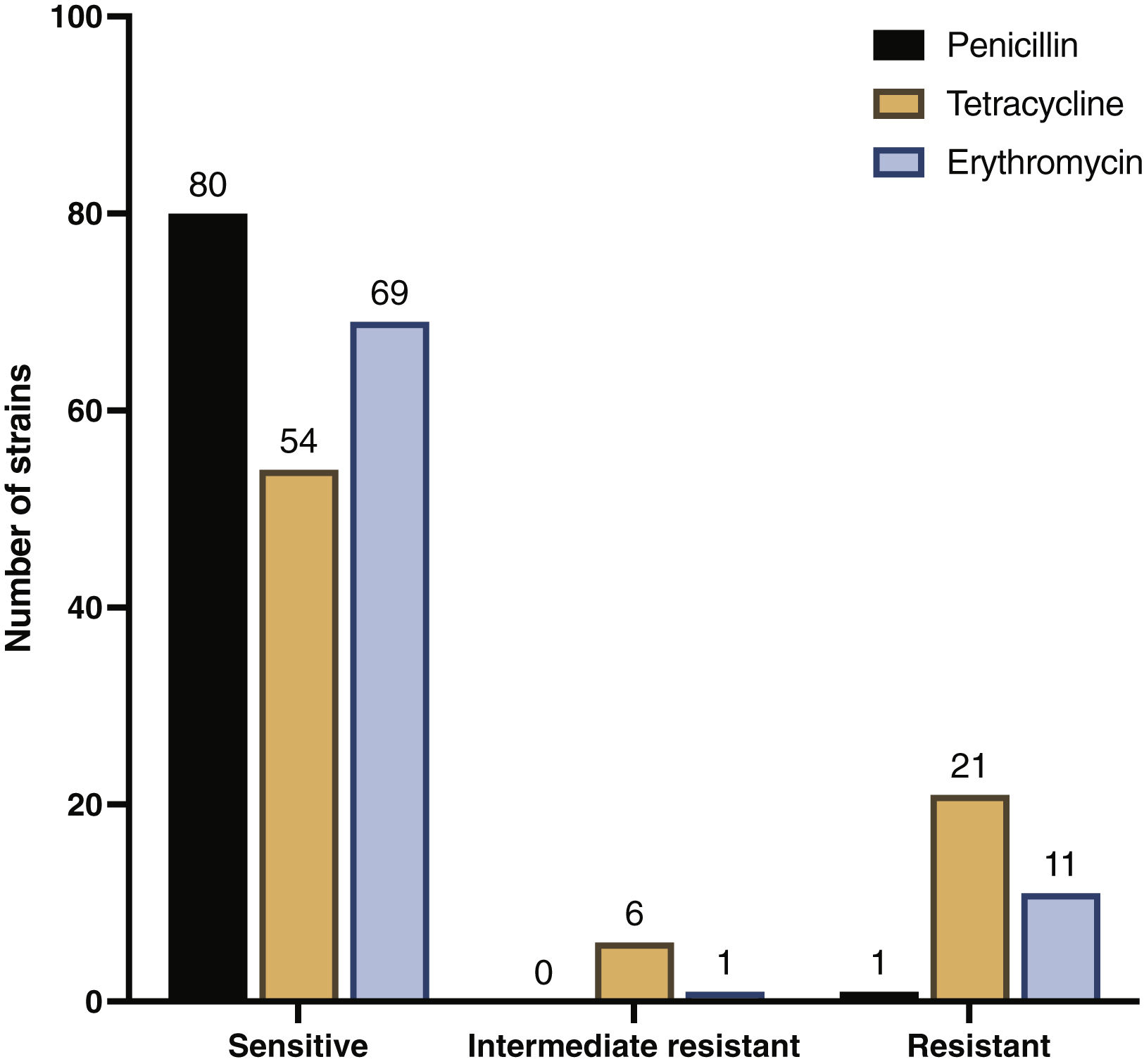
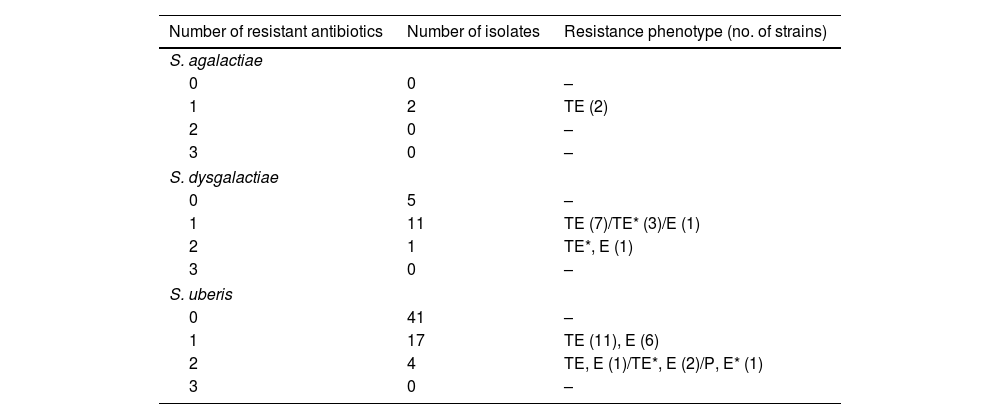
In an overview of the 81 samples tested, 98.8% (80/81) were sensitive to penicillin, and only 1.2% (1/81) were resistant. Tetracycline sensitivity was observed in 66.7% (54/81) of strains, while 25.9% (21/81) displayed resistance, and 7.4% (6/81) exhibited intermediate resistance. For erythromycin, 85.2% (69/81) of strains were sensitive, 13.6% (11/81) were resistant, and 1.2% (1/81) displayed intermediate resistance (Fig. 3).
Table 2 shows the phenotypic resistance profiles of the strains by species. All evaluated S. agalactiae strains were susceptible to penicillin and erythromycin but resistant to tetracycline. Most S. dysgalactiae strains were similarly susceptible to penicillin and erythromycin, with some showing resistance or intermediate resistance to tetracycline. Almost all S. uberis strains were sensitive to penicillin, with a small fraction resistant to it, and some exhibited resistance to tetracycline and erythromycin (Table 2 and Supplementary Figure 1). Finally, it is noteworthy that none of the strains analyzed from any of the three Streptococcus species exhibited resistance to all three antimicrobials simultaneously, indicating the absence of multi-drug resistance (MDR) strains.
Resistance profiles of S. agalactiae, S. dysgalactiae and S. uberis strains tested.
| Number of resistant antibiotics | Number of isolates | Resistance phenotype (no. of strains) |
|---|---|---|
| S. agalactiae | ||
| 0 | 0 | – |
| 1 | 2 | TE (2) |
| 2 | 0 | – |
| 3 | 0 | – |
| S. dysgalactiae | ||
| 0 | 5 | – |
| 1 | 11 | TE (7)/TE* (3)/E (1) |
| 2 | 1 | TE*, E (1) |
| 3 | 0 | – |
| S. uberis | ||
| 0 | 41 | – |
| 1 | 17 | TE (11), E (6) |
| 2 | 4 | TE, E (1)/TE*, E (2)/P, E* (1) |
| 3 | 0 | – |
TE: tetracycline; E: erythromycin; P: penicillin.
Utilizing Pearson's χ2 statistical test to assess the data derived from phenotypic and genotypic identification, significant differences in the results of conventional tests and the multiplex PCR setup were identified, with a p-value 0.023, which was indicative of statistical significance (p<0.05). Further examination through Fisher's exact test revealed that the most substantial disparities were observed in the identification of S. uberis and S. dysgalactiae, with p-values of 0.0007 and 0.0059, respectively. The Kappa value obtained for S. dysgalactiae and S. uberis through Cohen's test indicated no agreement between the identification methods (−0.22 and 0.13 respectively), while the differences in the agreement of both techniques were insignificant for S. agalactiae (p-value 0.14).
DiscussionTo date, conventional biochemical tests are the Gold Standard method for identifying bacteria causing bovine mastitis. These tests rely on morphological observations of colonies and the performance of the catalase test, Gram staining, and different routine biochemical tests such as the CAMP test, esculin hydrolysis, determination of the Lancefield group, and observation of hemolysis on 4% blood agar. Their effectiveness in accurately identifying Streptococcus spp. has been validated against other methods such as API® 20 Strep and RFLP-16S rRNA27.
In another study, the identification of S. uberis, S. dysgalactiae, and S. agalactiae was compared using phenotypic methods versus multiplex PCR. By combining the outcomes of the 9 phenotypic tests, the efficiencies reported for each pathogen were 92%, 90%, and 100%, respectively. Therefore, they recommend a combined approach involving tests such as catalase and Gram alongside multiplex PCR for accurate identification29,30. It should be noted that treatment, management, and disease characteristics vary epidemiologically based on the specific pathogen present. This holds practical significance as cure rates differ, for instance, with penethamate hydriodide treatment specifically, achieving 87.7% for S. uberis and 64.7% for S. dysgalactiae22.
This analysis successfully classified each of the 115 studied strains into one of the three Streptococcus species, with no instances of non-amplification recorded. The genotypic identification indicated that S. uberis was the predominant species, followed by S. dysgalactiae and S. agalactiae. These findings align with what is commonly observed in most regions worldwide18,26. However, prevalence studies in Uruguay have reported S. dysgalactaie as the most prevalent among Streptococcus species13. It is crucial to note that, in the present study, samples were exclusively collected from cows with clinical mastitis, and this factor may have influenced the final result, particularly regarding the reported low prevalence of S. agalactiae. Clinical mastitis is primarily caused by ‘environmental’ pathogens, such as S. dysgalactiae and S. uberis, which could explain the observed distribution.
The results obtained from the genotypic identification in this study did not align with those from previously conducted phenotypic tests. According to the latter, S. dysgalactiae was identified as the most frequent species among the studied strains, followed by S. uberis and, finally, S. agalactiae. This concurs with Gianneechini et al.13, who conducted the study using conventional culture methods. This suggests a potential misidentification of some pathogens, and these variations could be attributed to several factors, primarily the inherent characteristics of each test. While PCR relies on amplifying a conserved and species-specific genomic fragment, phenotypic methods involve laboratory tests that, in some cases, may introduce uncertainties in result interpretation and could lead to incorrect clinical treatment.
Moreover, this type of test requires extensive time for the necessary reactions to occur, and the availability of trained personnel is essential for its performance due to the complexity of its manipulation. In the identification of S. uberis by conventional methods, it was found that when determining the Lancefield group, the results can be variable and may suggest that the samples belong to groups E, G, P, or U. Furthermore, S. uberis can also exhibit variable responses to the CAMP test18. On the other hand, for S. dysgalactiae, considered non-hemolytic, there have been reported cases in which it presents with alpha-hemolysis, similar to S. uberis18. Finally, although S. agalactiae is classified as beta-hemolytic, cases have been found where hemolysis does not occur, similar to S. dysgalactiae18. Considering the discrepancies discussed above, conducting further studies to delve deeper into the subject and understand its underlying causes would be advisable. This is particularly important given the practical significance of having an easily applicable, sensitive, and specific technique that complements the currently employed methods.
Our findings compare molecular methodologies with the phenotypic methodologies typically employed in our country. It is worth noting that adding more biochemical tests could enhance the alignment between these two methodologies. Since there are no specific methodological requirements for local authorities to approve routine mastitis diagnostic laboratories, the implementation of a proficiency evaluation system would be advisable to ensure the reliability of diagnoses20,29. However, it is important to remember that molecular methods remain faster and more cost-effective.
With regard to penicillin resistance, most of the Streptococcus strains identified were sensitive to this antimicrobial (only one was resistant), which coincides with what was reported by Gianneechini in 2014 in Uruguay and other European countries such as Germany, France, Switzerland, and Finland4,16,28 but contrary to what has been reported in Asia and Argentina, where penicillin resistance rates exceed 10%1,9,25. Resistance to tetracycline and erythromycin was present in the isolated strains, coinciding with the results obtained by Minst in 2012 in Germany and Uruguay in 201413,23. Although tetracycline is not commonly used to treat mastitis in our country, as it can be used to treat other diseases. Therefore, the presence of resistance in Streptococcus strains represents a potential source of resistance genes to this antimicrobial, which could be transferred to other bacteria. In particular, the epidemiological study of these resistant strains can be useful for understanding how a pathogen spreads in a production system and for evaluating the efficiency of disease mitigation measures, contributing to the design of effective control measures.
For S. uberis, resistant strains (15%) and intermediate resistance strains (2%) to erythromycin were reported, in contrast to those previously reported in Uruguay (0%)13. For the S. agalactiae and S. dysgalactiae strains analyzed, no differences in resistance to this antimicrobial were found with those previously reported.
Finally, the absence of Streptococcus strains with a high degree of multiple resistance (MDR) in the present study should be mentioned, which contrasts with data reported in Germany in 2012, where a 13% incidence of MDR strains was found23. Based on all these results, penicillin continues to be an effective treatment against these pathogens in relation to tetracycline and erythromycin, which showed less effective responses, mainly for S. uberis.
Enhancing mastitis diagnosis at the species level for the genus Streptococcus could be achieved by combining phenotypic techniques with multiplex PCR. These results emphasize the significance of accurate diagnostic methods and reveal potential variations in prevalence when relying on phenotypic and genotypic identifications.
FundingThis work has been financially supported by the Comisión Sectorial de Investigación Científica (CSIC) through the funding of CSIC I+D, PAIE and CIDEC projects.
Conflict of interestThe authors declare that they have no conflicts of interest.