Tackling the dissemination of antibiotic resistance is one of the main global challenges. Manures from animal production are a recognized source of antibiotic resistance genes (ARGs) and mobile genetic elements (MGEs) requiring appropriate treatment methods. One of the main approaches for manure treatment is anaerobic digestion (AD). Meta-analyses have demonstrated that AD can significantly reduce the load of ARGs. However, antibiotics, ARGs and MGEs still remain in the final product (digestate). A sustainable agricultural use of digestates under the One Health framework requires wide assessments of their effects in the soil resistome. The objective of this review was to present the state of the art of digestate effects on ARGs of agricultural soils, focusing exclusively on digestates from animal manures. A systematic review was conducted. The examination of the resulting literature indicated that although temporal decays are observed for a variety of ARGs in single-application and repeated-applications experiments, for certain ARGs the pre-treatment or control levels are not restored. However, the low number of studies and the heterogeneous experimental conditions preclude a clear understanding of the fate of ARGs in soil and their risk for agroecosystems. The inclusion of multiple MGEs and the assessment of the long-term influence of digestates on soil properties and microbial communities could be keystones for a better understanding of the risks associated with digestate-induced changes in the soil resistome.
El abordaje de la diseminación de resistencia a los antibióticos representa uno de los principales desafíos a nivel global. Los estiércoles derivados de la producción animal constituyen una reconocida fuente de genes de resistencia a antibióticos (GRA) y de elementos genéticos móviles (EGM), por lo que requieren tratamientos apropiados. La digestión anaeróbica (DA) es uno de los principales métodos de tratamiento. Los metaanálisis demuestran que la DA puede reducir significativamente la carga de GRA. Sin embargo, en el producto final (digerido), aún permanecen antibióticos, GRA y EGM. El uso agrícola sustentable de digeridos bajo el paradigma «Una Salud» requiere evaluaciones amplias de sus efectos en el resistoma del suelo. El objetivo de esta revisión es presentar el estado del conocimiento acerca del efecto de digeridos sobre GRA en suelos agrícolas, con foco exclusivo en digeridos derivados de estiércoles animales. Se llevó a cabo una revisión sistemática. La revisión de la literatura resultante indicó que, si bien se observan decaimientos temporales de diversos GRA en experimentos de aplicaciones simples y repetidas, ciertos GRA pueden no alcanzar los niveles pretratamiento o control. No obstante, el bajo número de estudios y la heterogeneidad de condiciones experimentales impiden una comprensión clara del destino de los GRA en el suelo y del riesgo para los agroecosistemas. La inclusión de múltiples EGM y la evaluación de la influencia a largo plazo en las propiedades del suelo y en las comunidades microbianas podrían representar piezas claves para un mejor entendimiento de los riesgos asociados a cambios en el resistoma del suelo.
According to the World Health Organization (WHO), the dissemination of antibiotic resistance genes (ARGs) is one of the main challenges to global public health33. The antimicrobials used in food animals represent 73% of all antimicrobials sold globally and the consumption of veterinary antimicrobials at global scale has been estimated to increase 11.5% in 2030 relative to 201729. Simultaneously, manure production has increased 23% in the period 1990–2018, reaching 125 million tonnes N globally5. Considering that many antibiotics used in veterinary medicine have high excretion rates26, one of the main problems associated with this increase is the transfer of antibiotics and ARGs to the soil environment when untreated manures are used as agricultural amendments20. Currently, ARGs are considered emerging contaminants9,21,34.
In this scenario, anaerobic digestion (AD) processes are increasingly used to treat animal wastes while providing several potential benefits to agricultural soils through the application of the byproduct, the anaerobic digestates17. However, the comparison of input and outputs of anaerobic digesters indicate an incomplete removal of antibiotics and even increases in abundance of specific ARGs3,16,22,30 as well as in the diversity of the resistome relative to raw manure3. A recent meta-analysis revealed that although the total level of ARGs can decrease significantly (∼50%), these reductions comprised only 56% of the total number of ARGs that were evaluated in the study. Moreover, when multiple AD-operational parameters were considered, the authors observed significant effects of these modulators on the ability of AD to reduce ARGs levels. Among them, mesophilic temperatures showed a significantly lower reduction of ARG levels than the thermophilic temperatures6. The incomplete removal of antibiotics and ARGs indicate that the agricultural use of digestates requires more attention from the One Health perspective. Different treatment options have been studied to reduce the discharge of ARGs into the soil environment, including compost of the solid fraction of digestates7. However, in several AD facilities, composting of digestate is not a common operation yet.
Soil is also a vast reservoir of ARGs4, several of which can be horizontally transferred through mobile genetic elements (MGEs). Among them, plasmids are major drivers of horizontal gene transfer (HGT) within bacterial communities and have the greatest influence on the dissemination of ARGs in microbial ecosystems31. Plasmids can transfer themselves or even mobilize smaller plasmids by conjugation4,8. Bacteriophages are additionally involved in HGT of ARGs in soil10 while the role of natural transformation in the acquisition of adaptive genes remains to be clarified8. Field studies of manure-treated soils indicate that MGEs have a key influence on the soil resistome after years of applications12.
Given the complexity of the soil habitat, its microbiomes and the multiple HGT processes that can potentially mobilize ARGs, the fate of ARGs in this environment cannot be predicted from the levels reported in digestates. The sustainable use of digestates requires understanding the ARGs response in soils to improve agricultural practices and post-AD treatments that alleviate the enrichment of ARGs. Bacterial taxa that include relevant human and animal pathogens such as Clostridium, Acinetobacter and Pseudomonas were confirmed as major players in the enrichment of ARGs in manure-treated soils, indicating that the risks of transfer to pathogenic species should not be overlooked11. The aim of this review was to disentangle the effects of anaerobic digestate from animal manures on the soil resistome when used as biofertilizer and discuss relevant points that should be addressed for a sustainable use under the One Health paradigm.
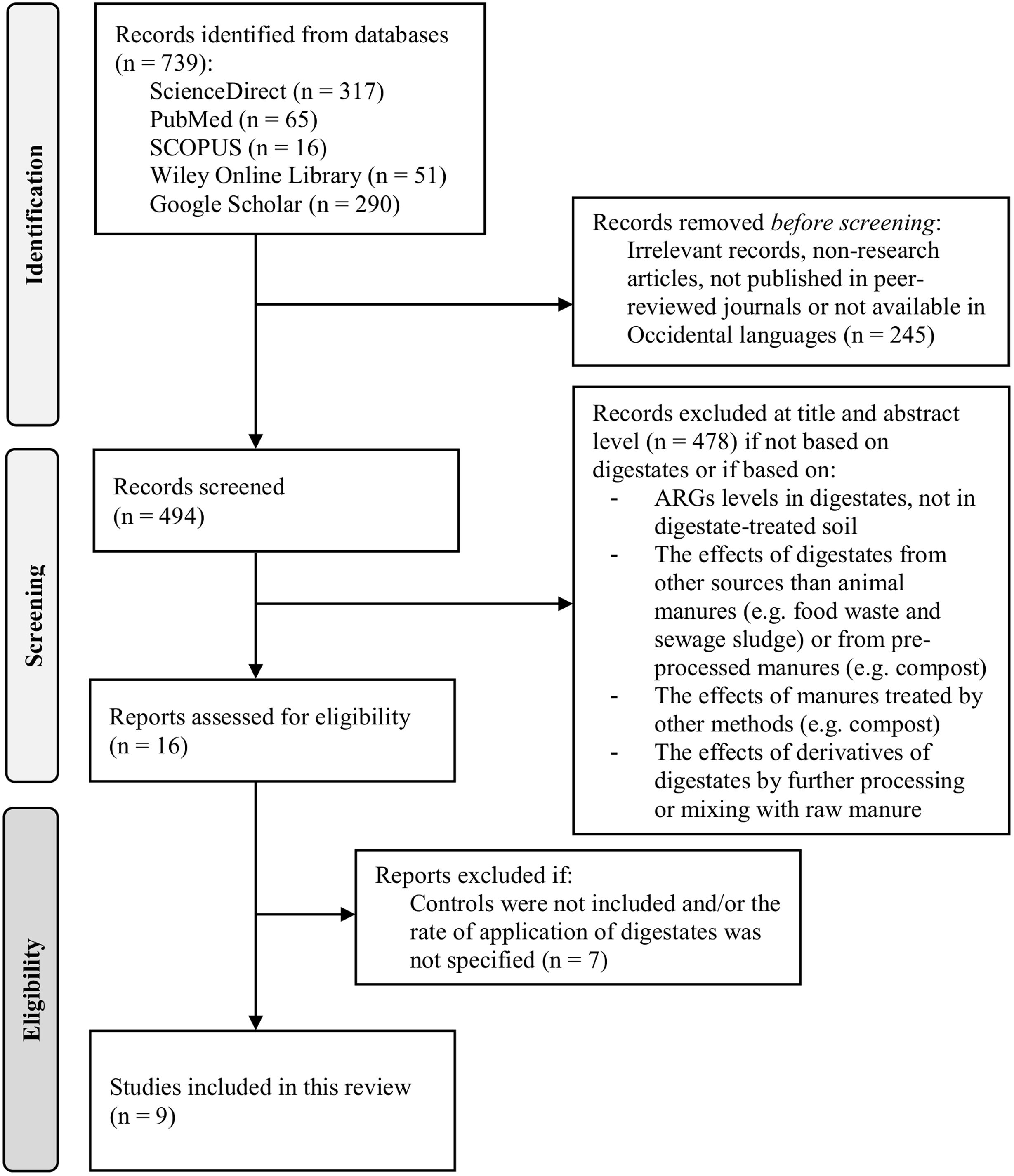
MethodsSearch strategyScientific literature search was performed on July 25, 2023 with no restrictions in date. The main search was conducted on ScienceDirect and was followed by additional searches in SCOPUS, PubMed, Wiley Online Library and Google Scholar. The following keywords and syntax were used in the first four databases: (ARGs OR RGs OR “antibiotic resistance genes”) AND (digestates OR “biogas slurry”) AND soil. In Google Scholar, we use the additional term (“quantitative PCR”) in the syntax to make a more stringent search due to the notably higher number of irrelevant results relative to the other scientific databases. The results of the initial search were then filtered by inspection of title and abstract, excluding articles not related with digestates (e.g., based exclusively on raw manures or processing methods different to AD) as well as digestate-related studies that met any of the following criteria: (1) exclusively assessed the effect of the AD process on ARGs, (2) assessed the effects of products derived from the processing of digestates (e.g., compost, struvite, constructed wetland or digestate mixed with raw manures), and (3) assessed the effect of digestates obtained from sources other than manures (e.g., sewage sludge and food waste). Among the filtered articles, two exclusion criteria were considered after full text analysis: (1) articles with no specification of fertilizer rate in each fertilization event (as total load of ARGs depends on the quantity of digestate applied); (2) articles without a control (unfertilized control) or without comparison to other fertilizers or amendments (inorganic fertilizers or raw manure). The flow diagram (Fig. 1) resumes the above-mentioned steps.
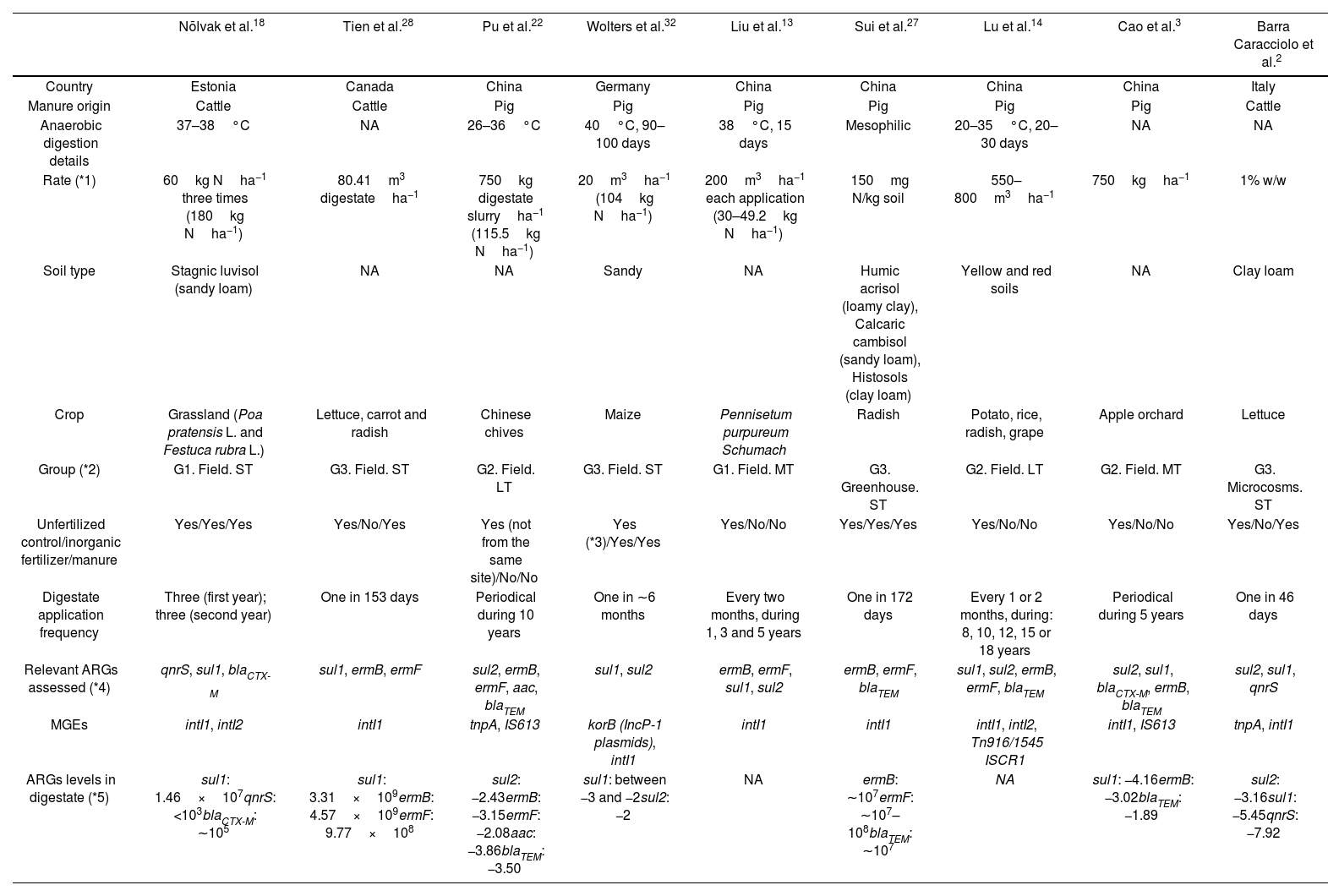
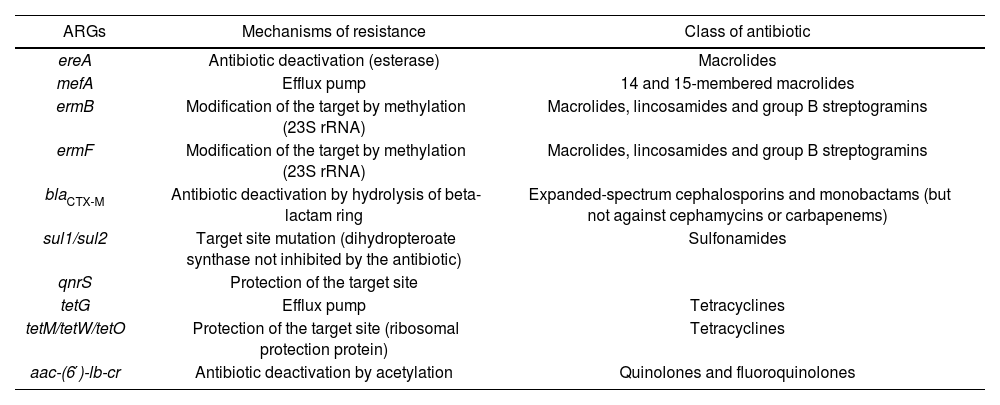
ResultsThe initial search yielded 739 records: Google Scholar (290), ScienceDirect (317), SCOPUS (16), PubMed (65) and Wiley Online (51). Only 16 peer-reviewed journal articles passed the first filtering step (Fig. 1). Results indicated that the vast majority of the research related with ARGs and AD has been focused on the fate of ARGs during the digestion process, e.g. the comparison of samples from the input and output of the reactors. A comparatively lower number of studies have focused on the fate of ARGs in the soil environment. A total of 9 articles satisfied the eligibility conditions and were included (Fig. 1). The main categories for the description of the studies are listed in Table 1. A general overview indicated that most of them (7 of 9) were conducted at field level and with digestates from pig-manure (6 of 9). Table 2 indicates the type of antibiotic resistance encoded by the ARGs discussed in the next section.
Main properties of studies that passed the eligibility criteria.
| Nõlvak et al.18 | Tien et al.28 | Pu et al.22 | Wolters et al.32 | Liu et al.13 | Sui et al.27 | Lu et al.14 | Cao et al.3 | Barra Caracciolo et al.2 | |
|---|---|---|---|---|---|---|---|---|---|
| Country | Estonia | Canada | China | Germany | China | China | China | China | Italy |
| Manure origin | Cattle | Cattle | Pig | Pig | Pig | Pig | Pig | Pig | Cattle |
| Anaerobic digestion details | 37–38°C | NA | 26–36°C | 40°C, 90–100 days | 38°C, 15 days | Mesophilic | 20–35°C, 20–30 days | NA | NA |
| Rate (*1) | 60kg Nha−1 three times (180kg Nha−1) | 80.41m3 digestateha−1 | 750kg digestate slurryha−1 (115.5kg Nha−1) | 20m3ha−1 (104kg Nha−1) | 200m3ha−1 each application (30–49.2kg Nha−1) | 150mg N/kg soil | 550–800m3ha−1 | 750kgha−1 | 1% w/w |
| Soil type | Stagnic luvisol (sandy loam) | NA | NA | Sandy | NA | Humic acrisol (loamy clay), Calcaric cambisol (sandy loam), Histosols (clay loam) | Yellow and red soils | NA | Clay loam |
| Crop | Grassland (Poa pratensis L. and Festuca rubra L.) | Lettuce, carrot and radish | Chinese chives | Maize | Pennisetum purpureum Schumach | Radish | Potato, rice, radish, grape | Apple orchard | Lettuce |
| Group (*2) | G1. Field. ST | G3. Field. ST | G2. Field. LT | G3. Field. ST | G1. Field. MT | G3. Greenhouse. ST | G2. Field. LT | G2. Field. MT | G3. Microcosms. ST |
| Unfertilized control/inorganic fertilizer/manure | Yes/Yes/Yes | Yes/No/Yes | Yes (not from the same site)/No/No | Yes (*3)/Yes/Yes | Yes/No/No | Yes/Yes/Yes | Yes/No/No | Yes/No/No | Yes/No/Yes |
| Digestate application frequency | Three (first year); three (second year) | One in 153 days | Periodical during 10 years | One in ∼6 months | Every two months, during 1, 3 and 5 years | One in 172 days | Every 1 or 2 months, during: 8, 10, 12, 15 or 18 years | Periodical during 5 years | One in 46 days |
| Relevant ARGs assessed (*4) | qnrS, sul1, blaCTX-M | sul1, ermB, ermF | sul2, ermB, ermF, aac, blaTEM | sul1, sul2 | ermB, ermF, sul1, sul2 | ermB, ermF, blaTEM | sul1, sul2, ermB, ermF, blaTEM | sul2, sul1, blaCTX-M, ermB, blaTEM | sul2, sul1, qnrS |
| MGEs | intI1, intI2 | intI1 | tnpA, IS613 | korB (IncP-1 plasmids), intI1 | intI1 | intI1 | intI1, intI2, Tn916/1545 ISCR1 | intI1, IS613 | tnpA, intI1 |
| ARGs levels in digestate (*5) | sul1: 1.46×107qnrS: <103blaCTX-M: ∼105 | sul1: 3.31×109ermB: 4.57×109ermF: 9.77×108 | sul2: −2.43ermB: −3.15ermF: −2.08aac: −3.86blaTEM: −3.50 | sul1: between −3 and −2sul2: −2 | NA | ermB: ∼107ermF: ∼107–108blaTEM: ∼107 | NA | sul1: −4.16ermB: −3.02blaTEM: −1.89 | sul2: −3.16sul1: −5.45qnrS: −7.92 |
NA: not available.
*1 Rates in kg Nha−1 were not available for all studies.
*2 Group definition is indicated in the main text. (ST=short-term): less than 1 year. (MT=medium-term): between 1 and 5 years. (LT=long-term): more than 5 years.
*3 The unfertilized control corresponds to time 0 (before application).
*4 Among the ARGs suggested for monitoring (Luby et al.)15. Description of resistance mechanisms are indicated in Table 2.
*5 ARG levels are indicated as absolute abundance (copiesg−1 dry weight). For Pu et al. (2018), Wolters et al. (2018), Cao et al. (2022) and Barra Caracciolo et al. (2022) the log10 of the relative abundance values (copies/16S rRNA gene) are shown.
Antibiotic resistance mechanisms of the ARGs indicated in Table 1 and discussed in this review.
| ARGs | Mechanisms of resistance | Class of antibiotic |
|---|---|---|
| ereA | Antibiotic deactivation (esterase) | Macrolides |
| mefA | Efflux pump | 14 and 15-membered macrolides |
| ermB | Modification of the target by methylation (23S rRNA) | Macrolides, lincosamides and group B streptogramins |
| ermF | Modification of the target by methylation (23S rRNA) | Macrolides, lincosamides and group B streptogramins |
| blaCTX-M | Antibiotic deactivation by hydrolysis of beta-lactam ring | Expanded-spectrum cephalosporins and monobactams (but not against cephamycins or carbapenems) |
| sul1/sul2 | Target site mutation (dihydropteroate synthase not inhibited by the antibiotic) | Sulfonamides |
| qnrS | Protection of the target site | |
| tetG | Efflux pump | Tetracyclines |
| tetM/tetW/tetO | Protection of the target site (ribosomal protection protein) | Tetracyclines |
| aac-(6′)-lb-cr | Antibiotic deactivation by acetylation | Quinolones and fluoroquinolones |
For the 9 selected articles, we detected clear heterogeneity in the comparisons included in the experimental designs. Four of them included a single comparison of the digestate treatment (to a control soil without fertilizer)3,13,14,22 while other three articles included more comparisons, i.e., to the inorganic fertilizer and to the manure treatment in addition to the non-fertilized control (or the pre-treatment condition)18,27,32. In the remaining studies, digestate-treated soils were compared to manure along with a non-fertilized control2,28 (Table 1).
Several studies reported shifts in digestate-treated soils relative to non-fertilized controls of the same site. During the second year of a two year-field experiment with repeated applications, a significantly higher abundance [copies/g dry weight soil (dws)] of blaCTX-M genes was observed (1.53×105) relative to the control soil (<5×104). Increased levels were also observed when compared to applications of inorganic fertilizer and undigested manure at the same rate. Instead, the ARG sul1 and the integrase gene intI1 showed significantly higher values compared to the control and the inorganic fertilizer but not to the soil treated with undigested manure18. A higher abundance of sul1 was also observed relative to the control at most sampling dates by Tien et al.28 with values higher than the limit of detection (LOD) for the digestate and manure-treated soil and values below LOD for the control soil (except for day 93) (LOD: between 103 and 104 copies/g wet weight soil). In a sampling period of 172 days, Sui et al.27 reported a higher abundance until day 82 of both tetG and ermF in one of the digestate-treated soils (∼105 to 107 and 107 to 108 copies/g dws, respectively) relative to the control soil (∼103 to 104 copies and 105 to 106 copies/g dws, respectively). Among the studies with longer duration, Liu et al.13 observed that, after 5 years of repeated applications, the abundance (log10 copies g−1 dws) of most genes remained significantly higher in the digestate-treated soil (ereA: 4.151, ermF: 2.957, mefA: 2.726, sul1: 6.225, sul2: 6.010, tetG: 6.841 and tetO: 4.103) than in the control soil (ereA: 2.573, ermF: 1.878, mefA: 1.795, sul1: 3.68, sul2: 4.157, tetG: 4.574 and tetO: 2.547). Similarly, when including a pristine soil as control, Cao et al.3 also observed a higher relative abundance (copies per 16S rRNA gene) of several genes in the soil with cumulative applications (5 years) of either the biogas slurry or the biogas residue, including the sul2 and blaCTX-M genes. A significantly higher relative abundance of sul1 and sul2 (digestate-treated soils: 5×10−3 to 6.6×10−2; control soils: 7×10−5 to 4.73×10−3) was also observed after 8–18 years of repeated applications in different soil types14. Overall, these studies provide evidence that several genes could be enriched above baseline levels of the unfertilized treatments in the following weeks or months after the applications (e.g., blaCTX-M genes, tetG and ermF) or at the end of long periods of repeated applications (e.g., sul1, sul2, tetG, tetO and blaCTX-M genes). However, among the studies that included comparisons to raw manure, two of them reported a lower abundance of several ARGs in digestate-treated soils 6 days after the application27,32.
In addition, several studies that incorporated multiple sampling times reported the temporal dynamic of ARGs in digestate-treated samples. For single-application studies, Tien et al.28 reported that genes such as aadA, ermB, ermF and sul1 decreased from the day of application to 153 days post application. Wolters et al.32 also observed a significant decrease for a sulfonamide resistance gene (sul2) from the beginning (6 days after fertilization) and up to the harvest date (several months after application) relative to the pre-treatment sampling, while no significant shifts were observed for other ARGs (sul1, tetW, tetM and tetQ). Conversely, Barra Caracciolo et al.2 reported a significant increase in the relative abundance of several ARGs during the 46 day-period of the study, including sul1, sul2 and aac-(6′)-lb-cr. For repeated applications, Liu et al.13 reported an enrichment of several genes (ereA, ermF, mefA, sul1, sul2, tetG and tetO) at the end of the first year of applications. While a decrease was observed in the following years until the end of the 5-year trial, the abundance did not return to the control levels. In a shorter study, a sudden increase of sul1 was observed after the application events followed by a decrease in abundance in the following weeks, with levels still higher than the unfertilized control at the end of the experimental period18. Although these studies provide a valuable contribution to the understanding of the temporal dynamic, more studies are needed to quantitatively assess the decay of ARGs after applications, in accordance with decay models. To our knowledge, so far, only one study has reported half-lives (first order decay model). However, the fitting was not observed for all genes, and it was also dependent on the soil type27.
Classification of the studies: application frequency and sampling timesA considerable heterogeneity among studies was also detected with regard to the frequency of the fertilization events and the sampling moments. The frequency of perturbations (e.g., fertilization) is recognized as a relevant factor for the stability of microbial communities25. Among the nine studies included in this review, three distinct groups can be clearly differentiated: (1) those with repeated applications and several sampling times during the application period13,18 (G1, n=2); (2) those with repeated applications but a single sampling time after the last application3,14,22 (G2, n=3); (3) those with a single application and several sampling times2,27,28,32 (G3, n=4) (Table 1 and Fig. 1).
The G1 group allows to monitor the dynamic of ARGs in the period under direct influence of the digestates, as well as the effect of cumulative applications. However, these studies are often restricted to assays of several months or a few years due to the labor-intensive sampling required. Nõlvak et al.18 conducted the most intensive sampling of all the studies screened in this review. Throughout the cultivation period, they sampled 11 times over 151 days, before and after applications as well as a few months after the last application (applications separated by 6–7 weeks). The authors demonstrated that the levels (copies/g dws) of sul1 and integrase genes (intI1 and intI2) in a grassland soil are stimulated after each application by direct input from digestate. Due to the limited persistence in the environment, a subsequent decline was reported. However, a return to a baseline was not observed, pointing to a decay that is not completely reversible. Additionally, they found that one year of applications (total rate=180kg N/ha) was enough to establish a background level of blaCTX-M genes significantly higher than the levels found in the control, in the inorganic fertilizer and in the cattle slurry treatment. Liu et al.13 also included multiple sampling times and fertilizer-application events but spanned over longer time periods (up to five years). The results of this study indicated that when digestates were applied every two months, a significant increase in the levels of several ARGs was identified after the first year and a further gradual decrease was observed annually, until the end of the trial (fifth year). Despite this decreasing trend, a return to control levels was not observed. This observation agrees with previous results of repeated applications during a shorter period18, suggesting that continuous applications during several years could increase the abundance of specific ARGs over the background levels. Whether or not a fertilization legacy on the fate of ARGs can be established under repeated applications should be further investigated.
The G2 group provides an end-point measurement of the state of the microbial community and ARGs after the pressure exerted by repeated applications of digestate, possibly reflecting stable shifts in ARGs levels. Lu et al.14 found a significant increase in the absolute abundance of aminoglycosides, sulfonamides, tetracycline and multidrug resistance genes after 8–18 years of applications. Similarly, Pu et al.22 reported a 21-fold higher relative abundance of total ARGs in the soil treated with biogas slurry along 10 years, including different classes of antibiotics (vancomycin, macrolides, aminoglycosides, beta-lactams, tetracyclines and sulfas). Gradual changes in soil physicochemical properties or cumulative increased levels of antibiotics and heavy metals due to repeated applications may influence the soil resistome. Multivariate analysis techniques, mainly redundancy analysis (RDA) and structural equation modeling (SEM), have been used to explore the relationship between environmental variables and ARGs patterns. Liu et al.13 found a significant influence of pH, total N, total P and soil organic carbon on the distribution of ARGs, although a direct selective effect was not demonstrated. With regard to the accumulation of antibiotics, their persistence in soil depends on the adsorption and degradation of different antibiotic classes in each soil type, with tetracyclines exhibiting the highest adsorption and the longest half-life19. Among the filtered articles of this review, inconsistent results have been observed in long-term studies. While no significant differences of antibiotic levels were reported among soils with 1, 3 or 5 years of repeated applications13, an accumulation of tetracyclines and an indirect contribution to increased ARGs levels was reported by Lu et al.14. However, even low environmental levels of antibiotics might not be overlooked, given that subinhibitory levels may enrich pre-existent mutants or promote de novo selection1. Undoubtedly, more field studies are needed to assess the extent to which antibiotics levels in digestates influence ARGs levels. Digestates may also contain heavy metals, which could accumulate under long-term applications18, with potential effects on ARG selection through co-selection23,24.
The G3 group comprises studies with a single fertilization and multiple sampling times along several weeks to months. These studies provide valuable insights into the time required to return to baseline levels or into the levels reached at specific moments (at harvest or at the time of an upcoming application). Wolters et al.32 observed that the ARGs sul2 and tetW increased their relative abundance in bulk soil compared to the chemical fertilizer in the first week after a single digestate application while no significant differences were observed at harvest.
Mobile genetic elementsA small number of studies have assessed MGEs in soil after treatment with manure-derived digestates to delimitate the actual risk of transfer of ARGs from soil to the food-chain. This is particularly striking considering that changes in the soil resistome can be triggered by either digestate-borne ARGs or an increased transfer of MGEs carrying ARGs13. Integrons are gene-recruiting elements that are not self-mobile and thus require complementary assessments of transposons and plasmids, especially broad-host range plasmids which can spread ARGs among several distant taxa in soil communities. While most studies referenced in this review included class 1 integrons, only one study assessed broad-host range plasmids (of the incompatibility group P-1), reporting non-significant differences relative to manure or the inorganic fertilizer32. Four studies evaluated transposons, which can raise their levels in the soil environment through direct input or through stimulation of indigenous populations. Lu et al.14 analyzed five sites with a different history and observed that 10–12 years of applications can increase the absolute abundance of MGEs by 0.91–1.12 log units in two sites and the relative abundance by 5.71–12.1 log units. The transposable element ISCR1, an insertion sequence that is part of complex class 1 integrons and intimately associates with several ARGs was not detected in any of the control or digestate-treated soil samples, while the transposase gene of Tn916/1545 was detected in soils from different sites only under digestate treatment. Moreover, the authors found a significant positive direct effect of Tn916/1545 on ARGs-associated bacteria which, in turn, were reported as significant drivers of the ARGs profiles. Two other studies analyzed an insertion sequence (IS613)3,22. Pu et al.22 did not find IS613 in an agricultural soil after 10 years of periodical applications of biogas slurry. Conversely, network analyses conducted by Cao et al.3 revealed multiple connections of this IS with several ARGs and a greater relative abundance after 5 years of applications than the pristine soil. This kind of analysis is focused on co-occurrence patterns through multiple samples and allows the simultaneous visualization of the correlations between ARGs and MGEs, suggesting potential ways of mobilization that should be confirmed by whole-genome sequencing of the isolated MGEs.
ConclusionThe results of this review revealed a small number of studies restricted to China and Europe and a remarkable heterogeneity in terms of controls and treatments used to compare manure-derived digestates in each study (unfertilized controls, raw manure or inorganic fertilizer) as well as in application rates and frequency of applications. This heterogeneity underlines the need for more studies sharing similar treatment comparisons and digestate application designs in order to provide conclusive evidence of their effects in the soil resistome. Similarly, the role of soil type and the fertilization legacy in the response of the soil resistome to digestates should be addressed. Due to the complexity of digestate and soil in terms of physiochemical and biological properties, the attention should be directed towards shifts of these properties during digestate-soil interaction. Among them, the physicochemical properties of long-term fertilized soils can significantly influence the structure of microbial communities, which in turn modulates the resistome. The introduction of digestate-borne MGEs and the modification of indigenous MGEs also deserve further research, especially those MGEs with demonstrated correlations with ARGs of clinical relevance for human and animal health. Meanwhile, the reported decrease of several ARGs and antibiotics after AD processes highlights the clear benefits of digestates over raw manure under the current global crisis of antimicrobial resistance.
Conflicts of interestThe authors declare that they have no conflicts of interest.
The authors acknowledge the support received from Consejo Nacional de Investigaciones Científicas y Técnicas of Argentina (CONICET) and Argentinean National Agency for Scientific and Technological Promotion (ANPCyT) through projects Préstamo BID PICT-2021-I-INVI-00467, PICT 2020-002289 as well as the support of Universidad Nacional del Sur through project PGI 24/A250.