Cunninghamella is a genus of the order Mucorales which includes saprophytic species, rarely causing mycoses. The most frequently reported in human mycoses is the thermophilic species Cunninghamella bertholletiae. However, this species does not appear to cause mucormycosis in animals, so there is scarce information about C. bertholletiae isolates from animals.
AimsIn this paper we describe the phenotypic and genotypic characterization, and the phylogenetic analysis, of an isolate of C. bertholletiae involved in a central nervous system mucormycosis in a dolphin.
MethodsThe isolate studied in this publication was characterized using the current morphological and physiological identification system for Cunninghamella species. DNA sequencing and analysis of the D1/D2 regions of the 26S rRNA gene and the ITS-5.8S rRNA gene sequences were also performed.
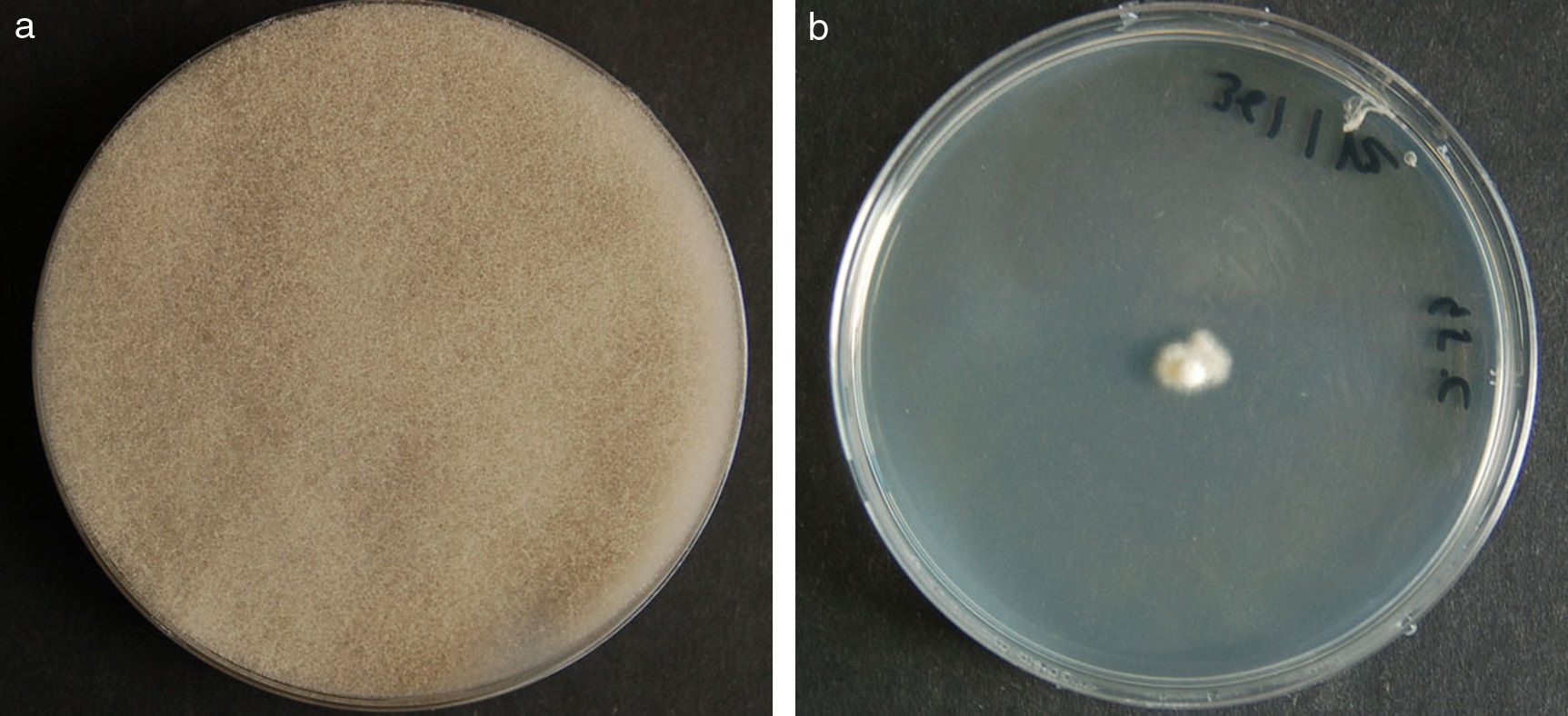
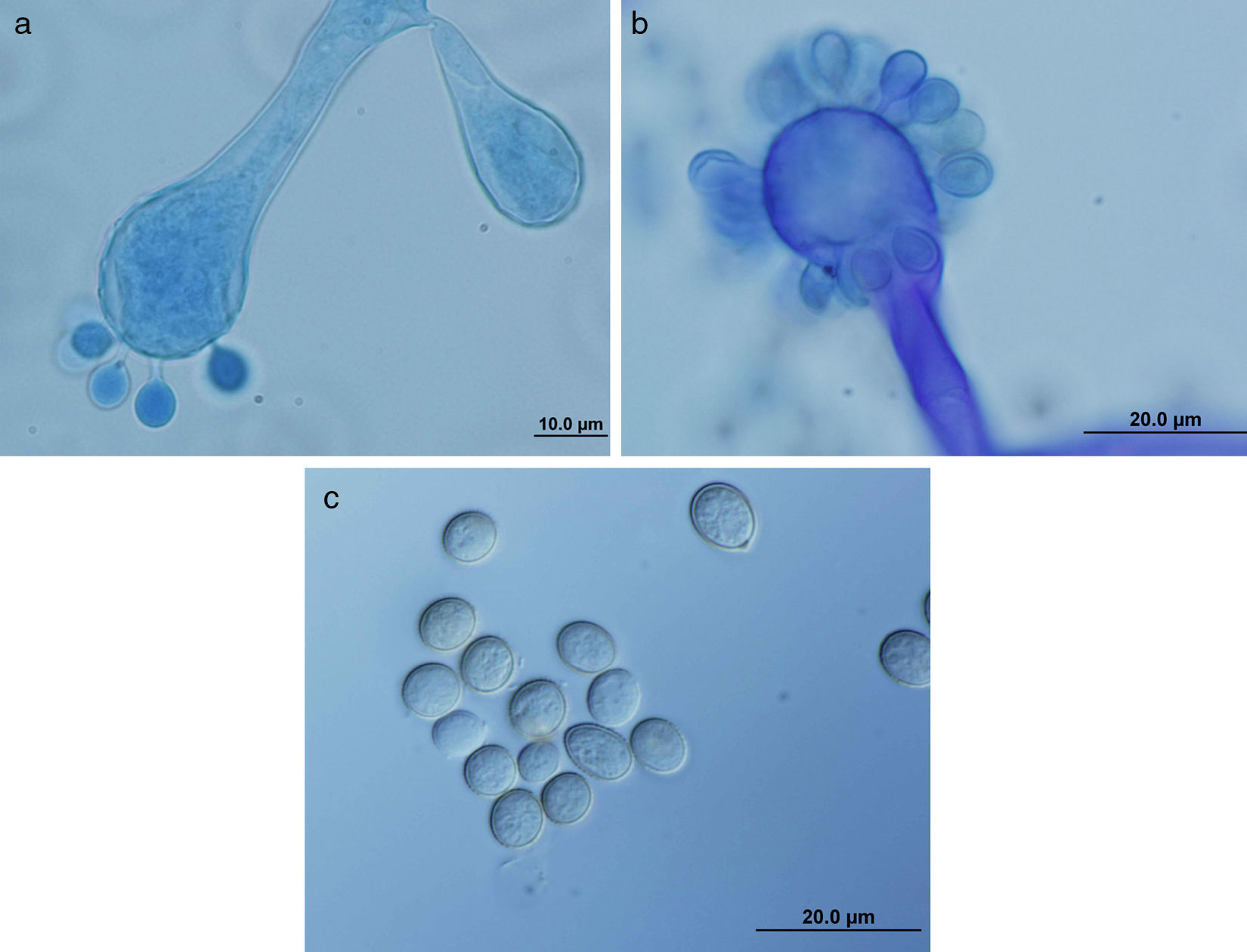
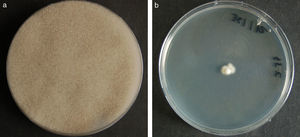
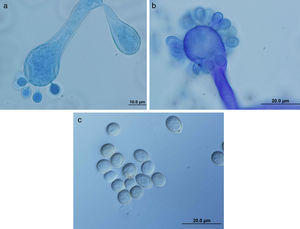
ResultsColonies were fast-growing, white at first, although they became tannish-gray, covering the whole plate after 7 days of incubation at 30 and 40°C. Limited growth was observed after 7 days at 45°C. The micromorphology showed characteristic erect sporangiophores. The identification of the isolate was confirmed by DNA sequencing of the D1/D2 regions of the 26S and the ITS-5.8S (ITS) rRNA gene sequencing.
ConclusionsIn the phylogenetic study, the isolate clustered in the same clade as C. bertholletiae neotype strain although some differences were observed in the ITS sequences. In the cetacean cases, the possible sources of infection are unclear. The reasons why this pathogen has been found only in cetaceans and not in other domestic or wild animals are at the moment unknown and need further study.
Cunninghamella es un género perteneciente al orden Mucorales, que incluye especies saprófitas que raramente causan micosis. De este género, Cunninghamella bertholletiae es la especie termófila más frecuentemente citada en micosis humanas. No obstante, no parece que sea una causa habitual de mucormicosis en animales, ya que es escasa la información sobre cepas de esta especie procedentes de estos.
ObjetivosEn esta publicación describimos la tipificación fenotípica, genotípica y el análisis filogenético de una cepa de C. bertholletiae causante de una mucormicosis del sistema nervioso central en un delfín.
MétodosLa cepa fue tipificada mediante los criterios morfológicos y fisiológicos actualmente utilizados para la identificación de estas especies. También se llevó a cabo la secuenciación y el análisis de los fragmentos génicos D1/D2 26S e ITS-5.8S del ARN ribosómico.
ResultadosLas colonias presentaron un crecimiento rápido; eran blanquecinas al principio y se volvieron de color marrón agrisado con el tiempo, y cubrieron totalmente las placas a los 7 días de incubación a las temperaturas de 30 y 40°C. A 45°C, después de 7 días de incubación, el crecimiento fue limitado. Al microscopio se pudieron observar los característicos esporangióforos de esta especie. La identificación de la cepa se confirmó mediante la secuenciación de los fragmentos génicos D1/D2 26S e ITS-5.8S del ARN ribosómico.
ConclusionesEn el estudio filogenético, la cepa se agrupó en el mismo clado que la cepa neotipo de C. bertholletiae, aunque se detectaron algunas diferencias en las secuencias correspondientes a los ITS. En los casos causados por esta especie en cetáceos, se desconocen las posibles fuentes de infección. Tampoco se conoce por el momento por qué este patógeno ha sido aislado solo de cetáceos y no de otros animales domésticos o salvajes.
Cunninghamella is a genus of the order Mucorales which includes saprophytic species, rarely causing mycoses. The more common pathogens of this order are included in the genera Rhizopus, Mucor and Lichtheimia.5 Currently 12 species are accepted in Cunninghamella,13 of which the most frequently reported in human mycoses is the thermophilic species Cunninghamella bertholletiae. This species remains a rare cause of human mucormycosis and has been described almost exclusively for immunosuppressed hosts.5 Infections by other species such Cunninghamella blakesleeana, Cunninghamella echinulata, and Cunninghamella elegans are highly exceptional.12 In the same way this species is not often reported as an animal pathogen. A comprehensive review of opportunistic mycoses of animals (humans included) was published by Smith9 in the late eighties of the last century. This book reviews important aspects of the pathogenesis of these mycoses and includes a detailed list of probable cases of mucormycosis in more than 40 animal species other than man. Although this fungus is considered in this book as a well documented causal agent of mucormycosis in the human being, none of the cases mentioned in animals was caused by C. bertholletiae. So this species does not appear to cause mucormycosis in animals.
Surprisingly, more recently this fungus has been involved in two cases of mucormycosis in cetaceans. C. bertholletiae has been reported as the cause of fatal pneumonia in a captive-held killer whale (Orcinus orca)1 and as the cause of meningoencephalomyelitis in a freeranging bottlenose dolphin (Tursiops truncatus), which was found stranded and dead on the Spanish Mediterranean coast.6 Until now, to our knowledge, no other cases of C. bertholletiae infection, neither in domestic nor in wild animals, have been published.
In this paper we describe the phenotypic and genotypic characterization, and the phylogenetic analysis of the isolate of C. bertholletiae involved in the first case of a central nervous system mucormycosis in a bottlenose dolphin.
Materials and methodsMorphological and physiological characterizationThe morphological and physiological characterization of the isolate was carried out based on macroscopic and microscopic characteristics of the colonies on Sabouraud glucose agar (SGA; Oxoid, Basingstoke, UK) and potato dextrose agar (PDA; Becton Dickinson) cultures after incubation at 25°C for 7 days. Other tests, such as the isolate temperature tolerance (30, 40 and 45°C) on SGA and PDA for 7 days, were also performed.4,13
DNA extraction, gene amplification, sequencing and phylogenetic analysisMolecular characterization of the isolate was carried out based on the D1/D2 regions of the 26S (D1/D2) and the ITS-5.8S (ITS) rRNA gene sequencing. DNA was extracted and purified directly from a seven day old culture in SGA according to the FastDNA Spin kit protocol with the FastPrep FP-24 instrument. The procedures for the amplification and sequencing were according to the protocols described previously.2 The resulting sequences were aligned using Clustal X v2.0.127 and regions of ambiguous alignment were removed with Gblocks.3 Maximum likelihood trees were inferred with the server version of RAxML-HPC2 v8,10 as implemented on the Cipres portal, using the GTR model. The robustness of the trees was estimated by a bootstrap analysis with 1000 replicates. For the phylogenetic analyses sequences of representative strains from the rest of the Cunninghamella species deposited in GenBank were also included.
ResultsMorphology and physiologyColonies on SGA and PDA were fast-growing, white at first, although they became tannish-gray, covering the whole plate (85mm in diameter) after 7 days of incubation at 25, 30 and 40°C. Limited growth was observed after 7 days at 45°C (20mm in diameter) (Fig. 1). The micromorphology showed characteristic erect sporangiophores showing the typical branching patterns. The sporangiophores had short lateral branches in the apical region which ended in globose to pyriform vesicles covered with globose one-spored sporangioles. Sporangiola born on pedicels, and vesicles were ovoid to subglobose, 6.4–11.2×5.4–8.2μm, hyaline to pale brownish and smooth to very shortly echinulate (Fig. 2). Chlamydospores were not observed. The isolate was identified as C. bertholletiae on the basis of its morphological and thermophilic characteristics.4,13
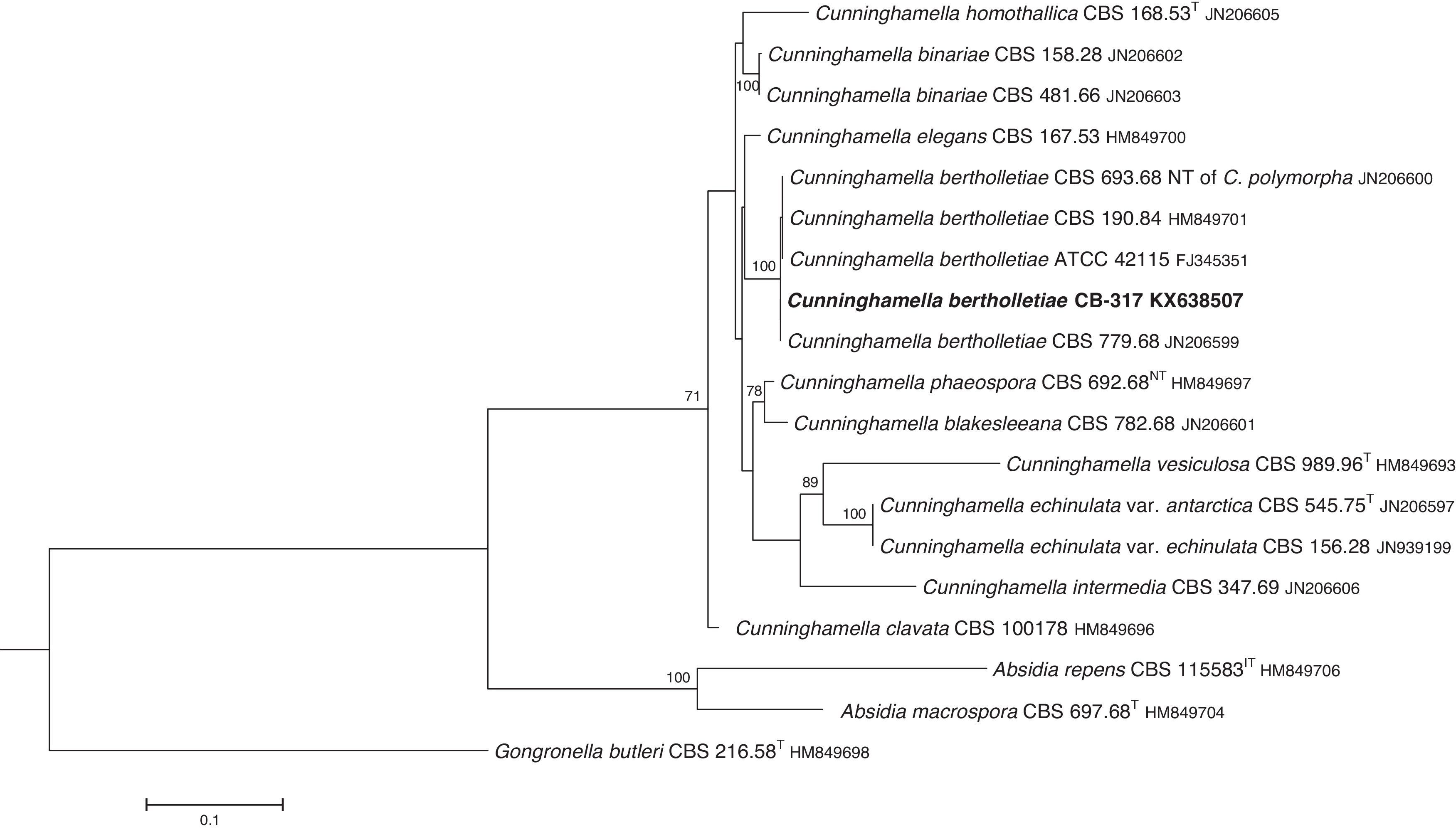
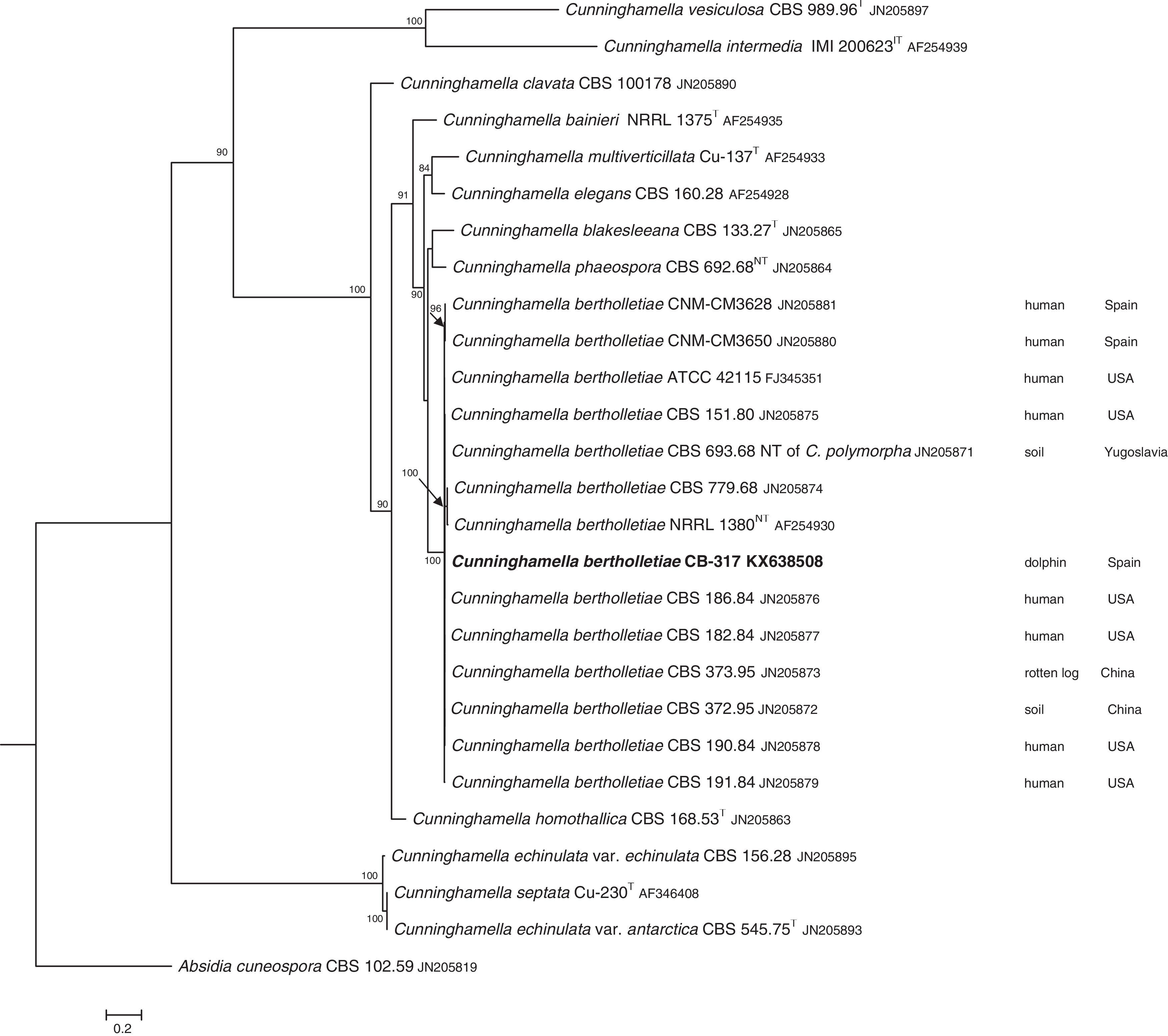
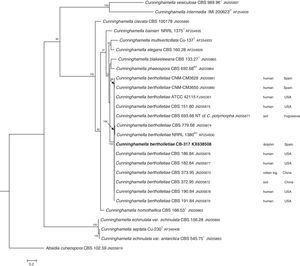
The D1/D2 and ITS sequences included 693 and 688 base pairs, respectively. The nucleotide sequences of the D1/D2 and ITS determined in this study were deposited in the GenBank and their accession numbers are given in Figs. 3 and 4. In the phylogenetic trees of D1/D2 (Fig. 3) and ITS (Fig. 4), our isolate clustered with other C. bertholletiae strains forming a well supported clade (100% bootstrap). Our strain and C. bertholletiae CBS 779.68 showed the same D1/D2 sequence and differed at 1–2bp from the rest of the strains analyzed. Intraspecific variability in ITS sequences was greater. Our strain differed from neotype strain NRRL 1380 of C. bertholletiae at 44 nucleotide positions whereas only differed at 3 nucleotide positions from C. bertholletiae ATCC 42115.
The C. bertholletiae isolate studied was unambiguously identified by its morphology and genotypic methods. Morphological and physiological characters matched those described for this species.4,13C. bertholletiae is quite different morphologically from Cunninghamella intermedia and Cunninghamella multiverticillata which have a similar temperature tolerance. They are able to grow at 41–43°C (maximum growth temperature).13 Despite the fact that the latter two are thermophilic species, C. intermedia and C. multiverticillata are not involved in human or animal infections. However C. bertholletiae grows at 45°C4 and is the most frequently reported Cunninghamella species in human mycoses.
The identification of the isolate was confirmed by DNA sequencing of the D1/D2 and ITS. In the phylogenetic study, the isolate clustered in the same clade as C. bertholletiae NRRL 1380 neotype strain although some differences were observed in the ITS. It has been reported that ITS regions are highly variable within the genus Cunninghamella, showing an intraspecific variability that ranges from 2.9% to 14.4%, sometimes even greater than interspecific distances between closely related species.12 The intraspecific variability of ITS sequences described for C. bertholletiae is 1%,11 although the neotype strain NRRL 1380 was not included in the study. The strain in our study had an identity of 99% when compared to different strains of C. bertholletiae isolated from different human patients, as well as strain ATCC 42115. This strain showed 100% identity with a clinical strain isolated from a pulmonary case of mucormycosis in a killer whale.1
Mucormycosis in animals is frequently recorded in association with erosion and/or ulceration of the alimentary tract, placentitis and acute pneumonia, among other sites of infection. In ruminants, increased gastric acidity, malnutrition and the feeding of rough, dry food may lead to the development of gastrointestinal mucormycotic lesions.9 However, in animals, as in some human cases, diagnosis of mucormycosis is often still made at necropsy with histopathological evidence of tissue invasion by characteristic non-septate broad hyphae, but no microbiological analyses are performed to identify the etiological agent. C. bertholletiae is recognized as a cosmopolitan soil organism which has been also found in a wide variety of nuts, seeds, and plants. The small size of sporangiospores allows them to remain airborne for prolonged periods, which can increase the exposure risk. The predominant mode of acquisition of C. bertholletiae infections is presumed to be via the respiratory tract.5
In the cetacean cases, the possible sources of infection are unclear. However, the entry via aerosol into the laryngeal tonsillar tissue and hematogenous dissemination to the central nervous system was suspected in the dolphin case, based on the severe involvement of arterioles at the basal arteriolar network of the brain.6 On the other hand, in the killer whale case a large number of stones were found in the stomach compartments; they might act as a facilitating factor for the onset of the mucormycosis in this immunocompromised animal.1
The reasons why this pathogen has been isolated only in cetaceans and not in other domestic or wild animals are at the moment unknown and need further study. However, C. bertholletiae has been recently recovered from gastric fluid and blowhole bottlenose dolphin samples.8 Surprisingly, it was the only mucoralean fungus isolated from these animals.
Conflict of interestThe authors declare that they have no conflicts of interest
The authors thank Carolina Gómez from the Veterinary Mycology Group of the Universitat Autònoma de Barcelona (UAB) for her valuable technical assistance. Financial support came from Servei Veterinari de Bacteriologia i Micologia and Servei de Diagnòstic de Patologia Veterinària of the UAB.