The fungus Alternaria is an important allergen in asthma pathogenesis. A number of studies have shown that Alternaria sensitivity is a risk factor for asthma development8,9,12,23,24, for persistence of asthma in children10, for emergency room treatment of asthma20, and for asthma severity2,21. Alternaria exposure in sensitized individuals has been associated with the development of severe exacerbations associated with respiratory arrest22. Clearly, Alternaria allergen exposure is a significant risk for the development, persistence, lability, and severity of asthma in sensitized individuals.
DNA-based immunotherapeutics offer a novel therapy for allergic disease and are entering human trials6,7,28. A number of studies have investigated the use of DNA from bacterial or plasmid sources that contain unmethylated cytosine-guanine dinucleotide (CpG) motifs14,29,30, and have shown that treatment with CpG-based therapy is effective in preventing or reversing the allergen-induced inflammatory airway response in animals3,11,13,15-19,25,27. To date, relatively few studies have used allergens that have been highly associated with human asthma15,19.
In order to explore innovative therapies that may eventually have therapeutic use in humans, we have developed an animal model of Alternaria sensitivity to evaluate DNA-based vaccine therapy. We used a recombinant major Alternaria allergen, rAlt a 24, to conduct these studies because of the strong link between Alternaria sensitivity and asthma persistence and severity. We also employed direct delivery of the DNA-based vaccine to the airway to maximize its effectiveness. Using this model, we hypothesized that a DNA-based vaccine would significantly ameliorate the sequelae of allergen challenge in Brown Norway rats sensitized to the Alternaria allergen, rAlt a 2.
Methods
Animals
The studies were approved by the Animal Care and Use Committee of the University of Wisconsin, Madison. Brown Norway male (RijHsd strain, Harlan Sprague Dawley, USA) rats, initially 10 weeks of age, were used in the study.
Allergen sensitization
Sensitization of animals to rAlt a 2 was performed by intraperitoneal injections of a mixture containing 0.5 ml of aluminum hydroxide suspension (Pierce, USA) and 0.2 ml of saline containing 20-40 mg/ml of rAlt a 2. In addition, the animals also received 0.5 ml of Bordetella pertussis antigen intraperitoneally (Difco, USA).
DNA vaccine preparation
DNA vaccine for rAlt a 2 was prepared by ligating the rAlt a 2 gene obtained via polymerase chain reaction into a plasmid DNA 3.1/V5-HIS TOPO-TA vector (Invitrogen, Carlsbad, CA); the vector was used to transfect Escherichia coli-competent cells (Invitrogen, USA). After growth, selected bacterial colonies were isolated, plasmid DNA was purified, and sequence analysis was conducted to confirm the proper orientation and correct reading frame for the rAlt a 2 gene. Vaccine was administered by insufflating it into the trachea in anesthetized animals through an endotracheal tube at a dose of 200μg of DNA in 0.1 ml of phosphate-buffered saline (PBS). As a control, a group of animals received the plasmid DNA vector with an out-of-frame rAlt a 2 gene.
Study design
The study design is summarized in Table 1. The following groups of animals were studied. A control group (C) did not receive any treatment, but 4 weeks after the study was initiated it did receive an airway challenge with rAlt a 2. The second group was sensitized (S) at week 2; then challenged at week 4 with rAlt a 2. The third group (CpG-S) received the DNA vaccine containing an out-of-frame rAlt a 2 gene at the beginning and at 1 week after initiation of the study. Animals were then sensitized to rAlt a 2 on the second week and challenged on week 4 after study initiation. The fourth group received rAlt a 2-DNA-based vaccine (VS). This group was then sensitized 2 weeks after initiation of the study and challenged at 4 weeks. The fifth group (SV) was sensitized to rAlt a 2, received rAlt a 2-DNA-based vaccine at 2 and 3 weeks, and were challenged on week 4 after initiating the study.
Pulmonary physiology measurements
We chose pulmonary physiology changes as the primary endpoint to determine the effectiveness of our DNA-based vaccine. To ascertain the effect of allergen challenge and the protective effects of DNA-based vaccines on pulmonary physiologic responses, animals were sedated with pentobarbital, and entubated. Rats were placed in a total body plethysmograph, and pressure and flow signals were monitored using a Buxco pulmonary mechanics analyzer (USA). The apparatus was programmed to deliver 3 lung inflations to 30 cmH2O pressure with passive exhalation, then an inflation from an endexpiratory volume to total lung capacity (TLC) over 1.5-2 s, followed by a 1-s breath hold, and then a rapid lung deflation to residual volume using -40 cmH2O pressure applied to the tracheal tube. TLC was defined as the volume at plateau pressure of 30 cmH2O after a 1-s breath hold. The resulting lung volume versus time relationship is analogous to a voluntary forced expiratory maneuver in humans, and the variables, inspiratory capacity (IC), forced vital capacity (FVC), and forced expiratory volume in 0.2 s (FEV0.2; analogous to FEV1 in human spirometry), were determined for a minimum of 2 maneuvers, and an average from all acceptable maneuvers was recorded. Maneuvers were excluded if there was an erratic flow-volume waveform (usually caused by secretions that were then removed) or if the values for the IC or FVC varied from the other maneuvers by greater than 10%. After completing the forced expiratory maneuvers, the plethysmograph was changed from a constant-pressure to constant-volume mode to increase sensitivity to small-volume changes. Following three lung inflations to 30 cmH2O pressure, the airway opening was occluded at end-expiratory lung volume for 8 s while recording the changes in airway pressure and thoracic volume during inspiratory efforts; functional residual capacity (FRC) was computed from these data and the ambient barometric pressure by the Boyle's law method (Pulmonary Maneuvers Software, Buxco Electronics). TLC was computed as TLC = FRC + IC.
Physiologic studies were conducted prior to and 72 h after administration of the airway challenge with rAlt a 2. After completing the baseline physiologic studies, a solution of 0.1 mg rAlt a 2 in 0.1 ml sterile PBS was insufflated into the airway by advancing a PE 50 catheter to the distal end of the tracheal tube, and the solution was forcefully expelled from a syringe loaded with 0.9 ml air behind the 0.1-ml allergen solution. After the insufflation, the tracheal tube was removed, and the rat was held vertically for 1 min, then positioned supine with the head slightly elevated until it awakened.
Lung histopathology
The effects of DNA-based vaccine on Alternaria-allergen-induced pulmonary inflammation was evaluated by histopathologic examination of the lung tissues. The right lung of the animal was fixed with 10% buffered formalin and embedded in paraffin. Thin layer sections were stained with Giemsa stain and examined histologically by one investigator (LP) in a blinded manner. A total inflammatory score was obtained that included both interstitial and intra-alveolar inflammatory cells of all cell lines and scored as: 0 = none, 1 = 5-10 cells, 2 = 11-20 cells, 3 = 21-30 cells, and 4 = > 30 cells. A scoring system for consolidation was based on the following: 0 = none, 1 = occasional isolated discrete foci, 2 = increased numbers of discrete foci, 3 = coalescing foci, and 4 = extensive solid areas.
Specific antibody responses
To determine the effects of DNA vaccine on specific antibody responses, enzyme-linked immunoassays were performed. Serum was collected from isoflurane-anesthetized animals from the tail artery for enzyme-linked immunoassays for the detection of specific IgE (Th2 response) and specific IgG2b (Th1 response) using modifications of methods published by Coligan et al5. Measurements of antibody levels by ELISA were conducted by coating immulon plates (Costar®, Corning, USA) with rAlt a 2 at a concentration of 10 µg/ml in PBS; coated plates were blocked with 1% bovine serum albumin (BSA). Rat sera were added and diluted 1:100 in PBS. The plates were incubated for 2 h; after washing, biotinylated rat IgG subclass-specific antibodies (Biosource International, USA) were added. After 1.5 h of incubation, plates were washed, and avidin-alkaline phosphatase detection solution at a dilution of 1-5000 (Sigma, St. Louis, USA) was added to each well. One hour later, the plates were washed and the substrate, p-nitro-phenyl-phosphate (SigmaFastTM, Sigma), was added, for an additional 1 h of incubation. The plates were then read on an ELISA plate reader (Biotech, Winooski, USA) at an absorbance of 405 nm. Samples were run in duplicate.
Cytokine production by peribronchial lymph node cell suspensions (PBLN)
To ascertain the effects of DNA vaccine on Th1 and Th2 cytokine production, enzyme-linked immunoassays for interferon-g (IFNg) and interleukin-13 (IL-13) were performed on PBLN cell suspension supernatants. Following the post-challenge physiologic studies, animals were euthanized and PBLN were removed surgically under aseptic conditions to obtain cell suspensions for cytokine analysis. PBLN-derived single-cell suspensions were prepared in PBS and the cells were cultured at 5× 105 cells/well in RPMI 1640 with fetal bovine serum, glutamine, HEPES, 2-? mercaptoethanol, penicillin, and streptomycin for 5 days at 37 °C in 96-well round-bottom plates in the presence or absence of rAlt a 2 (10μg/ml). Supernatant fluids were collected for measurements of IFN-γ and IL-13 levels by commercially available enzyme-linked immunoassays (BD Pharmigen, USA, and Biosource International, respectively). Samples were run in duplicate.
Data analyses
Specific antibody titers and cytokine levels were log transformed to conform to parametric assumptions. The general linear model was employed for one-way ANOVA analysis and planned pair-wise comparisons were done with the Fisher least square differences method (SYSTATv10® Software, USA)1.
Results
Effects of DNA-based vaccine on pulmonary physiologic changes
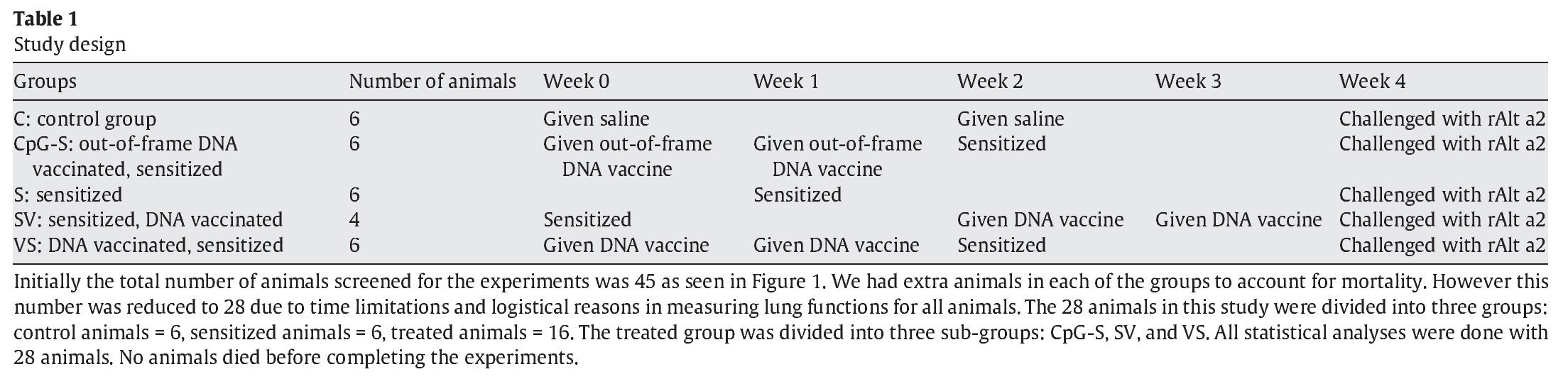
The effects of the DNA-based vaccine on pulmonary responses are shown in Figure 1. These data summarize the FEV0.2, FVC, and TLC at 72 h after the rAlt a 2 challenge in the study animals as a percent of the pre-challenge measurement. All three variables were significantly lower (p < 0.001) in the sensitized, untreated group (S) compared to the non-sensitized controls (C). Animals that received DNA vaccine after sensitization (SV) appeared to be protected from the pulmonary effect of an rAlt a 2 challenge with their FEV0.2, FVC, and TLC each being significantly higher (p≤ 0.03) compared to the sensitized untreated group (S), while not statistically different (p > 0.4) compared to the control (C) animals. Animals treated with the out-of-frame DNA-based vaccine (CpG-S) had pulmonary physiologic changes that were similar to those of the sensitized, untreated (S) group.
Figure 1. FEV (A), FVC (B), and TLC (C) 72 h after administration of rAlt a 2, expressed as % of pre-challenge baseline in the study groups. Each symbol represents one rat. The line at 100% represents no change from baseline values. C: controls; CpG-S: vaccinated with out-of-frame DNA vaccine, sensitized; S: sensitized; SV: sensitized, vaccinated; VS: vaccinated, sensitized.
Because FEV0.2 and FVC may be altered by changes in TLC, we also assessed the FEV0.2/FVC ratio (to detect reduced airway caliber) and the FVC/TLC ratio (to detect premature airway closure). There were no statistically significant differences among study groups with regard to these measures of airway dysfunction, indicating that the alterations of pulmonary physiology were primarily due to loss of air space (atelectasis and inflammatory infiltrates) as opposed to reduced airway caliber.
Effect of DNA-based vaccine on lung histopathology
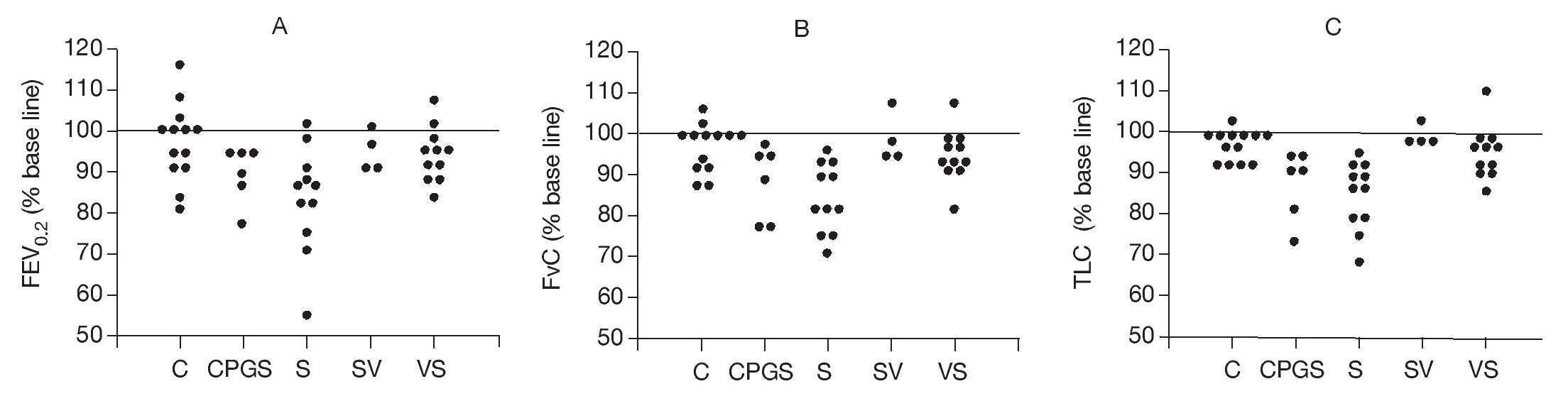
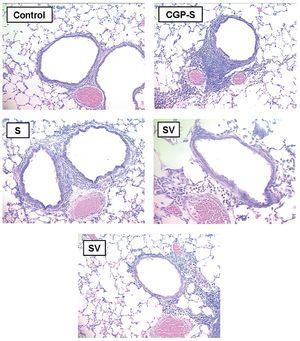
Histopathologic examination of the lung tissue from animals receiving the DNA-based vaccine (Fig. 2) demonstrated reductions in inflammatory infiltrates compared to the untreated sensitized controls. Examples of control, sensitized, and treated animals are shown. Control animals (C) showed minimal peribronchial cellular infiltrates and airway wall thickening. In contrast, marked peribronchial inflammatory cells and submucosal airway wall thickening were observed in sensitized, untreated animals (S) as well as loss of airspace due to consolidation and atelectasis. Increases in inflammatory cells were also observed in the CpG-S group. In contrast, the group that received DNA vaccine (SV) showed attenuation of the inflammatory response and less airspace changes compared to the sensitized, untreated (S) group.
Figure 2. Representative histopathologic features of rat lung 72 h following allergen challenge. Tissues are stained with Giemsa and photographed at 40x magnification. C: control animals; S: sensitized animals; CpG-S: treated animals; VS: DNA-vaccine-sensitized animals; SV: sensitized/DNA-vaccine-treated animals.
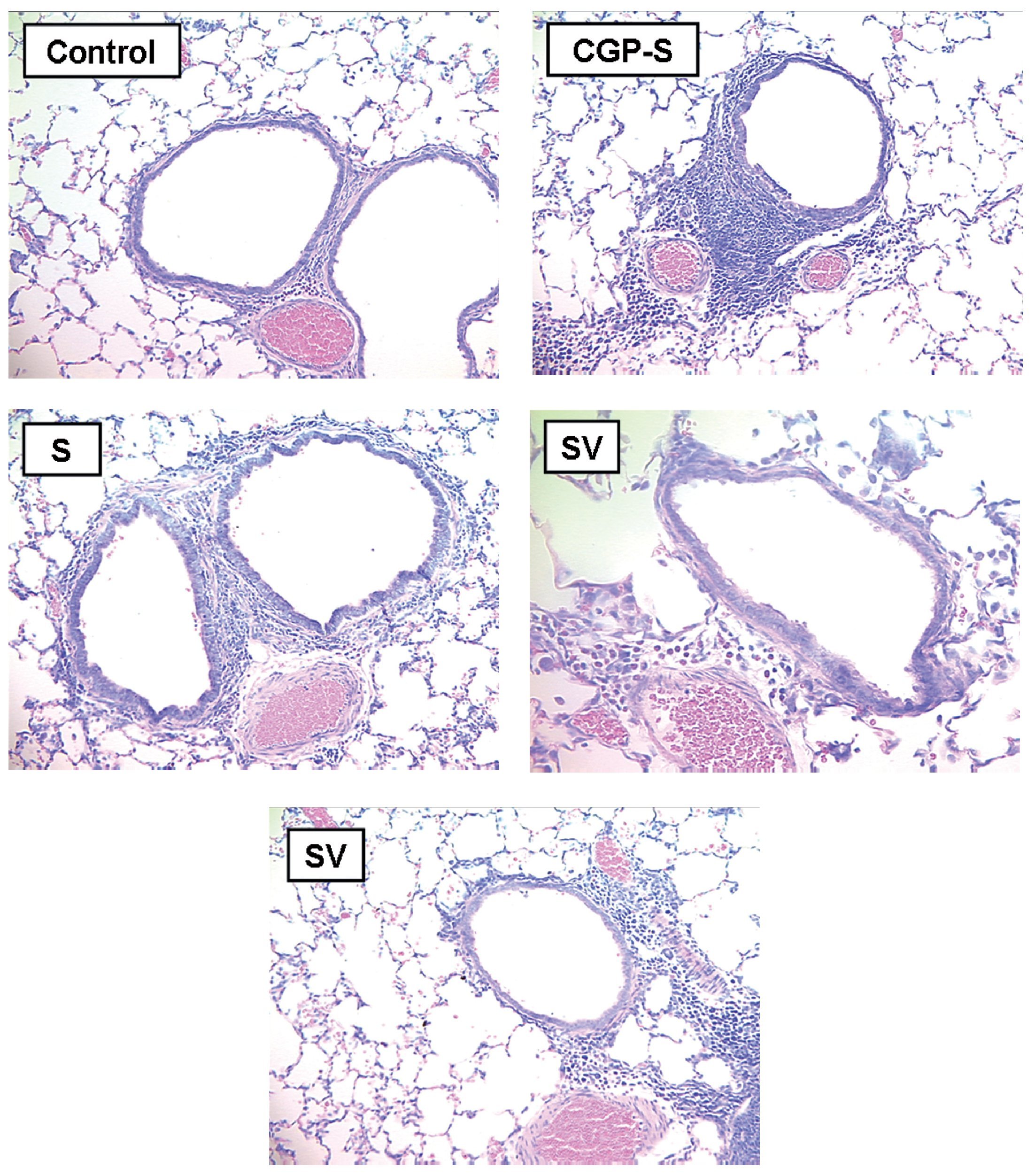
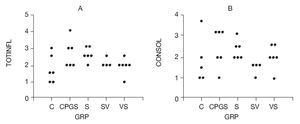
Total inflammatory scores were lesser in DNA-vaccine-treated animals (SV and VS) compared to sensitized animals (S) and those receiving the out-of-frame DNA vaccine (CpG-S; Fig. 3). Consolidation in the alveolar spaces was also reduced in DNA-vaccine-treated animals (SV and VS) compared to sensitized animals (S) and those receiving the out-of-frame DNA vaccine (CpG-S).
Figure 3. A) Total inflammation score graded 0-4. B) Consolidation score graded 0-4. C: control animals; CpG-S: animals vaccinated with out-of-frame DNA vaccine and sensitized; S: sensitized animals; VS: animals vaccinated and sensitized; SV: animals sensitized and vaccinated.
Effect of DNA-based vaccine on humoral immunologic responses
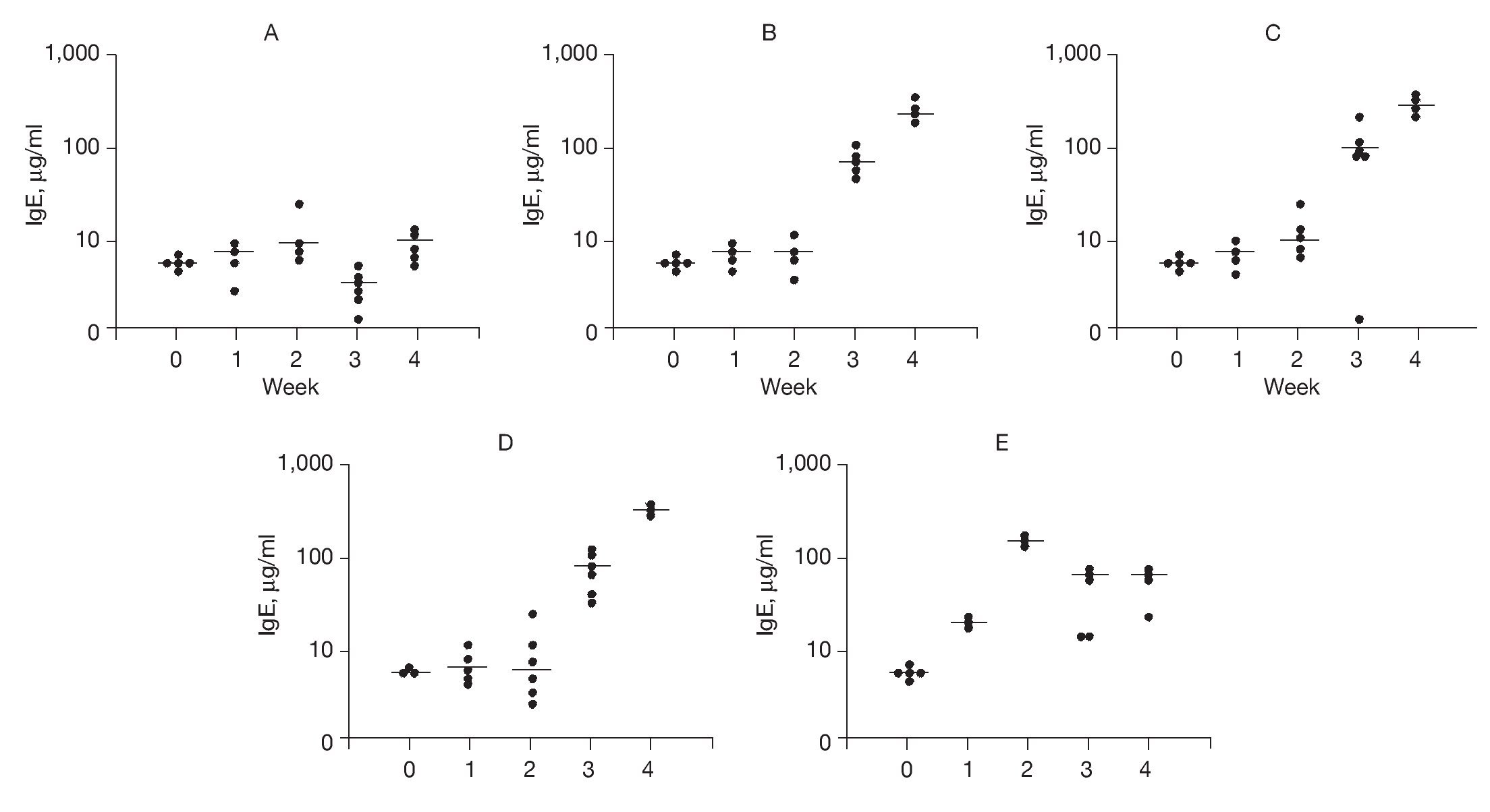
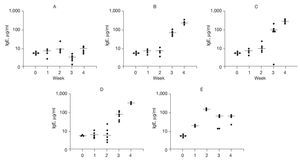
To examine possible mechanisms to account for the effects of DNA-based vaccines we examined specific IgE and IgG2b antibody titers. The results of DNA-based vaccine treatment on specific IgE levels to rAlt a 2 are shown in Figure 4. Control animals typically had a background level of < 10 µg of IgE per milliliter, which did not change over the course of the study (Fig. 4A). Sensitized animals showed a progressive increase in specific IgE levels at weeks 3 and 4 compared to week 2 (p < 0.002; Fig. 4B). Animals receiving out-of-frame DNA vaccine (CpG-S) that were sensitized at week 2 showed an increase in specific IgE serum concentration at 3 and 4 weeks (p < 0.001) compared to week 2 (Fig. 4C). Animals that were vaccinated and then sensitized (VS) also showed progressive increases in specific IgE levels at weeks 3 and 4 compared to week 2 (p < 0.001; Fig. 4D). Animals that were sensitized and then received the DNA vaccine (SV) showed an initial increase in specific IgE with a peak of 171μg/ml within 1 week of sensitization (Fig. 4E). On administration of the DNA-based vaccine, there was a significant reduction in the median specific IgE levels, ranging from 44 to 55μg/ml at the end of the 4 weeks of study. This represents a 70% reduction from the peak level (p < 0.001), and indicates that the DNA-based vaccine effectively reduced specific IgE antibody titers in previously sensitized animals.
Figure 4. Serum-specific IgE levels (μg/ml) to rAlt a 2. Each dot represents one animal. Mean value indicated by (-) A) Control group showed no significant changes. B) Sensitized group showed significant elevation of specific IgE levels (compared to baseline) at weeks 3 and 4 (p < 0.001). C) CPG-S group showed significant elevation of specific IgE level at 3rd and 4th weeks (p < 0.001). D) VS group showed significant elevation in specific IgE levels at 3rd and 4th weeks (p < 0.001). E) SV group showed elevated IgE at week 2 compared to baseline (p < 0.001) but a significant decline at weeks 3 and 4 (p < 0.001) compared to week 2.
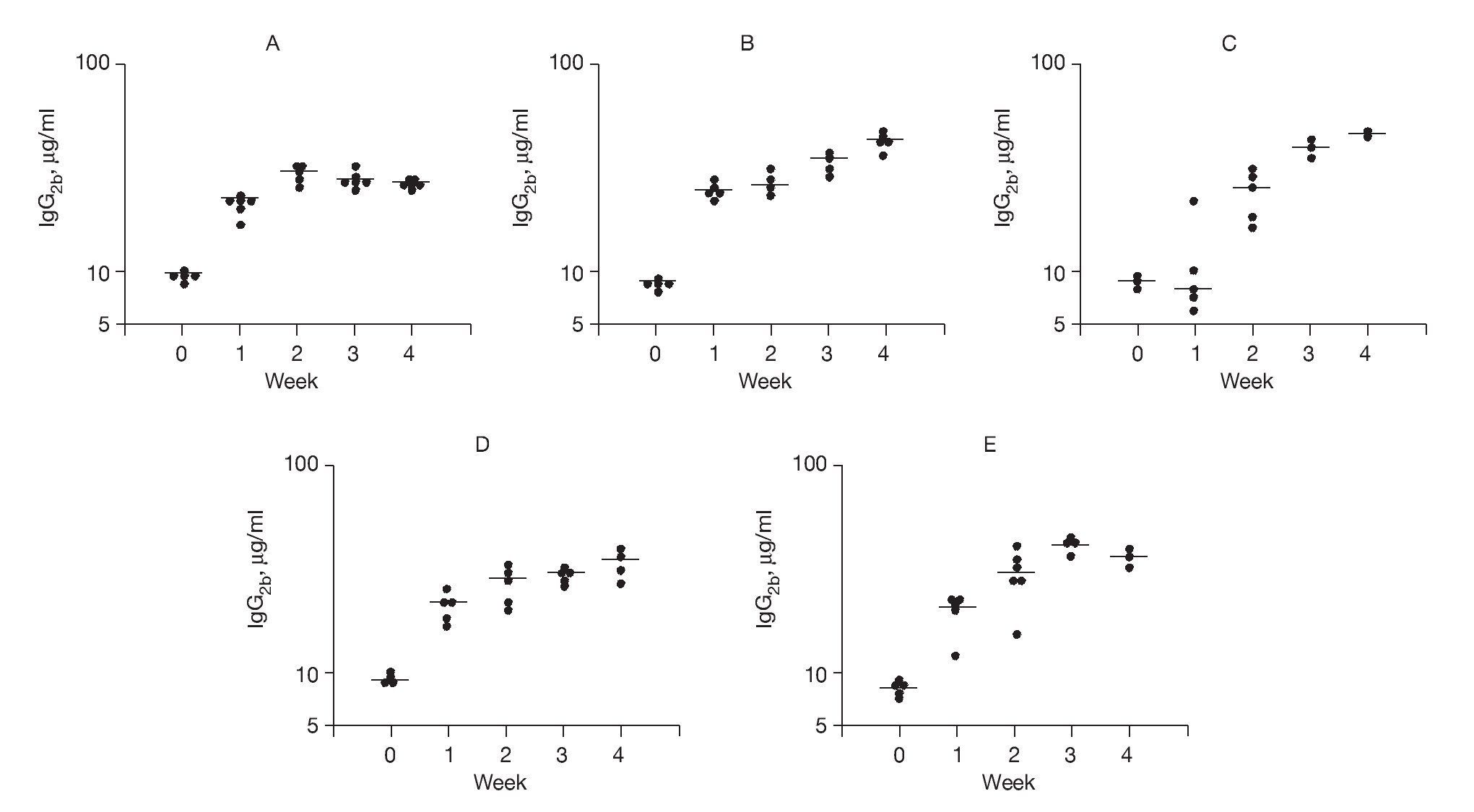
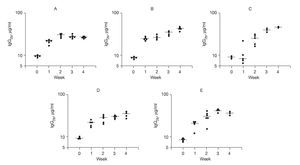
In order to study Th1-driven responses, specific IgG2b concentrations to rAlt a 2 were measured (Fig. 5). The SV group had a progressive rise in specific IgG2b levels (p < 0.001) compared to baseline at all time points following vaccine administration, which were significantly higher (p < 0.03) at week 4 compared to the other study groups. These latter findings suggest an immunotherapeutic effect of the DNA vaccine on previously sensitized animals.
Figure 5. Serum-specific IgG levels (μg/ml) to rAlt a 2. Each dot represents one animal mean value indicated by (-) A) Control group showed a 2-fold increase from baseline at week 1 but no further change. B) Sensitized group showed a progressive elevation over the 4 weeks of the study (p < 0.001) compared to baseline. C) CpG-S group showed significant elevations at week 2-4 (p < 0.001) compared to baseline. D) VS group showed elevations at week 1-4 (p < 0.001) compared to baseline. E) SV group showed elevation at week 4, which was significantly higher (p < 0.03) compared to those of other treatment groups.
Effect of DNA-based vaccine on cytokine production by PBLN cell suspensions
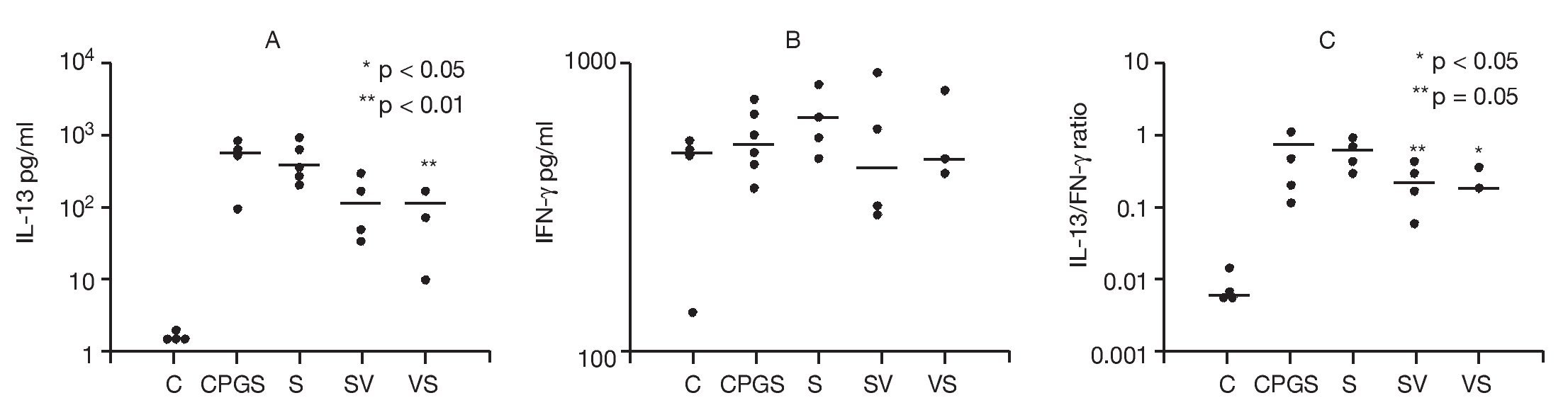
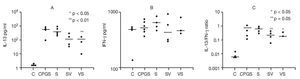
The effects of DNA-based vaccine in skewing cytokine Th1 and Th2 responses were examined. Cytokine concentrations in the supernates from single-cell suspensions of PBLN were compared among study groups in order to assess the effects of DNA-based vaccine treatment on Th1 (IFN-γ) and Th2 (IL-13) responses (Fig. 6). To assess the effects of the in vivo rAlt a 2 challenge on the activation of PBLN cells, we measured spontaneous in vitro cytokine secretion from the explanted cells. Spontaneous IL-13 secretion from PBLN cells was significantly higher in the sensitized groups compared to the control group (p < 0.001). DNA-based vaccine-treated animals (SV, p < 0.05; VS, p < 0.01) had significantly lower IL-13 levels compared to those of the non-treated sensitized (S) group and the group that received the out-of-frame DNA vaccine (CpG-S; Fig. 6A). Spontaneous IFN-γ secretion by PBLN cells was not statistically different among groups (Fig. 6B). IL-13/IFN-γ ratios from spontaneously secreting PBLN cells are shown in Figure 6C. The sensitized groups showed a significantly higher (p < 0.001) ratio compared to that of the control group. The ratio was significantly lower in the group that was sensitized and treated with DNA-based vaccine (SV, p < 0.05) and approached significance (p = 0.05) in the VS group compared to the untreated sensitized group (S) and the group receiving out-of-frame DNA vaccine.
Figure 6. Cytokine analysis of peribronchial lymph node cell suspensions. Each dot represents one animal. Mean values indicated by (-) A) Spontaneous IL-13 secretion. Groups SV and VS showed significantly lower levels (p < 0.05 and p < 0.01, respectively) compared to the CpG-S and S groups. B) Spontaneous IFN-γ secretion was not different between groups. C) IL-13/IFN-γ ratios showed a significant difference in the SV group (p < 0.05) compared to the other groups.
Incubation of PBLN cells with rAlt a 2 did not yield results that were qualitatively different from the spontaneous secretion results, presumably because the PBLN cells had already been activated in vivo by the allergen challenge. Taken together, these results suggest that treatment with the "in-frame" DNA vaccine significantly attenuated IL-13 secretion, skewing the cytokine balance in a Th1 direction.
Discussion
In these studies, Alternaria-induced airway inflammation was used to study the potential therapeutic benefits of a DNA-based vaccine. Selection of Alternaria as the sensitizing agent is highly relevant to human asthma. Sensitivity to this allergen is associated with an increased risk for the development of asthma8,9,12,23,24, with more severe21 and life-threatening episodes of asthma2,22, and a risk factor for the persistence of asthma in children10. Other studies using DNA-based vaccines have employed allergens such as house dust mite15,19, an additional important allergen in asthma, and birch11,13, grass, or ragweed7,28 pollen allergens, whichare more relevant to allergic rhinitis.
The route of DNA vaccine administration we chose has been reported in only one prior study25. Previous studies have typically employed either an intramuscular or intraperitoneal route for vaccine administration3,11,13,15-19,27. DNA vaccine was directly insufflated into the airway, facilitating potentially higher levels at the sites of airway inflammation. The effectiveness of this route of administration was clearly demonstrated in the studies we conducted and are comparable to the study by Shirota et al25. In comparison with previous studies3,11,15-17,27, the direct delivery of DNA vaccine to the airway was as efficacious as intramuscular or intraperitoneal therapy.
DNA-based vaccine treatment of Alternaria-induced airway dysfunction was effective in influencing a number of outcome measures. The pulmonary physiology data showed that significant declines in TLC occurred in sensitized animals 72 h following antigen challenge (Fig. 1C). Animals receiving an out-of-frame DNA-based vaccine (CpG-S) were likewise unprotected from the antigen challenge whereas control animals (C) and those that had been sensitized and given DNA-based vaccine (SV) or given the vaccine prior to sensitization (VS) were protected without significant change in TLC. Little changes in FEV.02 or vital capacity, after these quantities were adjusted for changes in TLC, were observed (Figs. 1A and B), which may reflect the fact that the antigen challenge was administered by insufflation into the airway directly as opposed to aerosol delivery, resulting in relatively greater alveolar distribution. The observed physiologic changes are most consistent with parenchymal versus airway pathology and no doubt reflect the location and intensity of the histopathologic alterations that we observed (Fig. 2). Indeed, marked inflammation was noted in the airway and parenchymal tissues of the untreated animals that were sensitized and then challenged with rAlt a 2 (Fig. 2). In contrast, animals receiving DNA-based vaccine (VS, SV) had less intense inflammatory changes in the airways and parenchyma (Fig. 2). The inflammatory changes in the airway walls were not sufficient to cause airway obstruction. We did not test airway hyper responsiveness to avoid creating artifacts on lung histology, but it is possible that antigen-induced hyper responsiveness was present in sensitized, untreated rats.
We did observe that animals that received the DNA vaccines had residual inflammation and consolidation, which raises the question that DNA vaccines may be associated with inflammatory changes. In the data presented, animals were given a "control" DNA vaccine that contained out-of-frame rAlt a 2 DNA as well as in-frame rAlt a 2 DNA. All animals were challenged with large doses of allergen intratracheally. The cause of inflammatory changes could be the result of several factors, including contamination of the vaccine with endotoxin (although all efforts are made to reduce this possibility), inability of the vaccine to be completely protective in the face of large antigen challenge, and potentially an adverse effect of the vaccine itself. Further, the large doses of allergen delivered intratracheally led to inflammatory changes in the parenchyma, which may be avoided in future studies by the administration of intranasal or aerosolized allergen.
To study possible mechanisms involved in the alterations in the pulmonary physiologic responses to allergen challenge seen in the DNA-vaccine-treated animals, we examined specific Th1 and Th2 immunologic parameters.
Compared to controls, sensitized animals showed a progressive increase in rAlt a 2 specific IgE antibody titers over the course of the study (Fig. 4B). Use of an out-of-frame DNA-based vaccine (CpG-S) had no effect on specific IgE titers after sensitization (Fig. 4C). Animals treated with DNA vaccine and then sensitized (VS) showed significant increases in specific IgE levels (Fig. 4D). In contrast, animals that had been sensitized and then received the DNA vaccine (SV) showed a marked drop in specific IgE levels after treatment (Fig. 4E). IgG2b antibody formation in rats is considered to be a Th1-mediated response26. Animals that were initially sensitized and then vaccinated (SV) showed a significant (p < 0.03) increase in the specific IgG2b titers at week 4 compared to the other study groups (Fig. 5E), findings that suggest DNA vaccine elicited a skewing of antibody responses toward a Th1-type response in previously sensitized animals.
Experiments utilizing PBLN cell suspension further evaluated whether a skewing in the cytokine balance toward a Th1 response occurred following vaccination. Spontaneous IL-13 secretion was significantly elevated in all the sensitized groups compared to the control group, which indicates that the animals were sensitized (Fig. 6A). Both vaccine-treated groups (SV, VS) had significantly lower IL-13 levels compared to those of non-treated sensitized (S) group and the group that received out-of-frame vaccine (CpG-S; Fig. 6A). However, spontaneous IFN-γ secretion by PBLN cells was not significantly different among groups (Fig. 6B). The IL-13/IFN-γ ratio was significantly lower in the groups that received DNA-based vaccine (SV, VS) compared to untreated sensitized group (S) and the group receiving out-of-frame DNA vaccine (CpG-S; Fig. 6C).
Collectively, these data indicate that the airway delivery of the DNA vaccine resulted in local skewing of cytokine production in a Th1 direction due to reduced IL-13 secretion and support the observations of the humoral immunologic responses (decreased specific IgE levels and increased specific IgG2b levels in DNA-vaccine-treated animals).
In summary, using an aeroallergen highly relevant to the pathophysiology of human asthma, we have shown in an animal model that DNA-based vaccine administration directly into the airways can significantly alter the pulmonary response to a large dose allergen challenge. Beneficial effects were observed when evaluated by a number of outcome measures, including pulmonary physiologic, histologic, and immunologic responses. The fact that these significant effects occurred as a result of the method of vaccine administration utilized in the face of a large amount of allergen directly insufflated into the airway indicates that similar approaches in humans may provide a new way to target organ-specific biologic responses, which may result in enhanced immunotherapeutics for patients with asthma.
Acknowledgement
Support: US Department of Veterans Affairs.
* Corresponding author.
E-mail address: hxs@medicine.wisc.edu (H. Sánchez).
ARTICLE INFO
Article history:
Received September 20, 2007
Accepted December 1, 2008