The molecular reclassification of the order Trichosporonales placed the medically relevant Trichosporon species into three genera of the family Trichosporonaceae: Cutaneotrichosporon, Trichosporon, and Apiotrichum. From the clinical and epidemiological standpoint, it is important to identify any species of the family Trichosporonaceae because they present different antifungal susceptibility profiles. In Mexico, little is known about trichosporonosis etiology because the fungi are identified through phenotypic methods.
AimsTo identify at a molecular level 12 yeast isolates morfologically compatible with Trichosporon, obtained from patients with superficial infections.
MethodsThe yeast isolates were obtained from patients with white piedra, onychomycosis, and hand and foot dermatomycosis, and were identified morphologically and genotypically (sequencing of the IGS1 region and phylogenetic analysis using the Maximum Likelihood Method). The phylogenetic analysis included 40 yeast sequences from the order Trichosporonales and one from Cryptococcus neoformans as outgroup.
ResultsBased on the molecular analysis, we identified three (25%) Trichosporon inkin isolates, two (16.7%) Trichosporon asteroides, two (16.7%) Cutaneotrichosporon mucoides, and one each (8.3%) of Trichosporon aquatile, Trichosporon asahii, Apiotrichum montevideense, Cutaneotrichosporon cutaneum, and Cutaneotrichosporon jirovecii.
ConclusionsThe molecular characterization of the isolates showed a broad diversity of species within the order Trichosporonales, particularly among onychomycosis. It is essential to identify these yeasts at the species level to delve into their epidemiology.
La reclasificación molecular del orden Trichosporonales ubicó las especies de Trichosporon de importancia médica en tres géneros en la familia Trichosporonaceae: Cutaneotrichosporon, Trichosporon y Apiotrichum. Desde la perspectiva clínica y epidemiológica es importante la identificación de las especies dentro de la familia Trichosporonaceae debido a sus diferentes perfiles de sensibilidad a los antifúngicos. En México, la etiología de la trichosporonosis es poco conocida porque los hongos son identificados por métodos fenotípicos.
ObjetivosIdentificar molecularmente 12 aislamientos de levaduras, morfológicamente compatibles con el género Trichosporon, recuperados de pacientes con infección superficial.
MétodosLos 12 aislamientos, procedentes de pacientes con piedra blanca, onicomicosis y dermatomicosis de las manos y los pies, fueron identificados morfológica y genotípicamente (secuenciación de la región IGS1 y análisis filogenético con el método de máxima verosimilitud). En el análisis filogenético se incluyeron 40 secuencias de levaduras del orden Trichosporonales y una de Cryptococcus neoformans como grupo externo.
ResultadosEn base al análisis molecular se identificaron tres (25%) aislamientos de Trichosporon inkin, dos (16,7%) de Trichosporon asteroides, dos (16,7%) de Cutaneotrichosporon mucoides y uno (8,3%) de cada una de las siguientes especies: Trichosporon aquatile, Trichosporon asahii, Apiotrichum montevideense, Cutaneotrichosporon cutaneum y Cutaneotrichosporon jirovecii.
ConclusionesLa caracterización molecular de los aislamientos mostró una diversidad de especies dentro del orden Trichosporonales, especialmente en las onicomicosis. Es importante identificar estas levaduras a nivel de especie para profundizar en su epidemiología.
Trichosporonosis is an opportunistic infection caused by some yeasts of the genus Trichosporon.5 These yeasts, which are occasionally part of the microbiota of skin, mucous membranes and nails of mammals, can cause superficial trichosporonosis (white piedra, onychomycosis, and foot dermatomycosis), genitocrural piedra and invasive disease (peritonitis, endocarditis, among others), as well as allergic pneumonia.5,17 Invasive trichosporonosis primarily affects immunosuppressed individuals, while superficial infections and allergic pneumonia are mainly diagnosed in immunocompetent hosts.4,5 The species most commonly associated with hematogenous and disseminated infections is Trichosporon asahii, whereas Trichosporon asteroides and Trichosporon cutaneum are predominant in superficial infections, Trichosporon inkin in white piedra of the hair and Trichosporon ovoides in the genital area. However, any species can cause an invasive infection with a bad prognosis, which is particularly unfavorable for immunosuppressed patients.10,14,19
Until recently, Trichosporon domesticum, Trichosporon montevideense, T. cutaneum, Trichosporon dermatis, Trichosporon jirovecii, Trichosporon mucoides, T. asahii, T. asteroides, Trichosporon coremiiforme, Trichosporon faecale, T. inkin, Trichosporon japonicum, Trichosporon lactis, T. ovoides, Trichosporon loubieri, and Trichosporon dohaense were known as the medically relevant species of the genus Trichosporon (class Tremellomycetes, order Trichosporonales, and family Trichosporonaceae).5 However, tremellomycetes were recently reclassified based on phylogenetic analyses using sequences from different genes: the small subunit (SSU or 18S), the D1/D2 domains of the large subunit (LSU or 26S) rDNA, and the rDNA internal transcribed spacer (ITS1 and ITS2, including 5.8S) regions. Two subunits of the RNA polymerase II (RPB1 and RPB2), the translation elongation factor 1-α (TEF1) and the mitochondrial cytochrome b gene (CYTB) were also considered.15,23 This reclassification caused changes in the Trichosporon genus since the medically relevant species were relocated in three genera: Cutaneotrichosporon (Cutaneotrichosporon cutaneum, Cutaneotrichosporon jirovecii, Cutaneotrichosporon dermatis, Cutaneotrichosporon mucoides); Trichosporon (T. asahii, T. asteroides, T. coremiiforme, T. faecale, T. inkin, T. japonicum, T. lactis, T. ovoides, and T. dohaense), and Apiotrichum (Apiotrichum domesticum, Apiotrichum montevideense, and Apiotrichum loubieri).15 Therefore, the identification of any fungus in the order Trichosporonales is necessary from both clinical and epidemiological perspectives to determine the specific pathological agent and to implement a proper antifungal therapy since different susceptibility profiles have been reported depending on the species.10,21
In clinical laboratories, yeasts’ identification is often based on morphological (observation of arthroconidia, hyphae, blastospores, and trichoid bodies) and biochemical criteria using the API 20C AUX and Vitek 2 Compact YST (bioMérieux, Marcy l’Etoile, France) systems. Nonetheless, several authors have demonstrated that these methods are inadequate to identify the species within the order Trichosporonales since these methods only allow to identify three species (T.asahii, T.inkin, and C.mucoides).10,14 For this reason, the sequencing of the intergenic spacer 1 (IGS1) of the D1/D2 domain and the ITS regions is now being used for the molecular identification of Trichosporon species, being the IGS1 region the most relevant for this purpose.10,22 These molecular methods enhance the knowledge about the species distribution and their genotypes, providing a better understanding of the trichosporonosis epidemiology.10 In several countries, including Mexico, both the incidence and the specific etiology of this disease are poorly recorded, mainly because the pathogens are identified only phenotypically. For this reason, this study was designed to re-identify through sequence analysis of the IGS1 region, 12 isolates recovered from patients with superficial infections in Mexico City that were identified previously as Trichosporon spp. by their phenotypic characteristics.
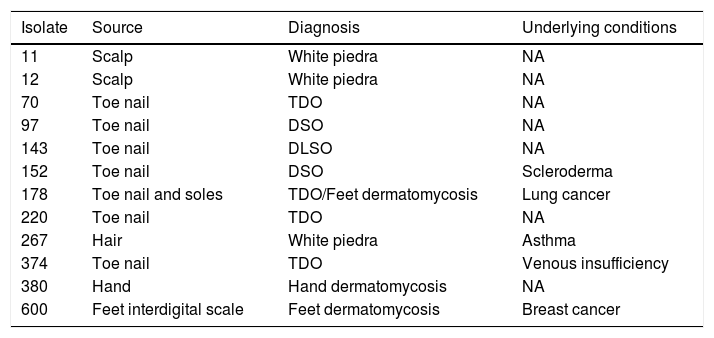
Material and methodsClinical isolatesThis study included 12 clinical yeasts obtained between January and December 2018 in a third-level hospital. The isolate source, diagnosis, and underlying conditions of the patients are included in Table 1. In order to improve the isolation of any fungus from nails the technique described by Fernandes-Meireles et al. was carried out.8 Samples of scales and hair were collected according to Del Boz et al.6
Isolates according to the type of infection and underlying condition of every patient.
| Isolate | Source | Diagnosis | Underlying conditions |
|---|---|---|---|
| 11 | Scalp | White piedra | NA |
| 12 | Scalp | White piedra | NA |
| 70 | Toe nail | TDO | NA |
| 97 | Toe nail | DSO | NA |
| 143 | Toe nail | DLSO | NA |
| 152 | Toe nail | DSO | Scleroderma |
| 178 | Toe nail and soles | TDO/Feet dermatomycosis | Lung cancer |
| 220 | Toe nail | TDO | NA |
| 267 | Hair | White piedra | Asthma |
| 374 | Toe nail | TDO | Venous insufficiency |
| 380 | Hand | Hand dermatomycosis | NA |
| 600 | Feet interdigital scale | Feet dermatomycosis | Breast cancer |
NA: not available; TDO: total dystrophy onychomycosis; DSO: distal subungual onychomycosis; DLSO: distal and lateral subungual onychomycosis.
The isolates were cultured in Sabouraud agar (BD Bioxon™, MX) tubes at 28°C, and their macroscopic and microscopic morphologies were observed.18
Molecular identificationGenomic DNA extraction: The yeasts were cultured in 15ml of YEPD broth (yeast extract 10%, peptone 10%, dextrose 20%), with shaking (150rpm), until the OD600 reached the value 10. The cells were recovered by centrifugation at 7500rpm for 10min. Afterwards, DNA was extracted according to Bolano et al.3 Then, the DNA was eluted with 50μl of sterile MiliQ water. The quality and concentration of the obtained DNA were determined using spectrophotometry at 260nm (Spectrophotometer DS-11, DeNovix, USA).
PCR: The IGS1 region was amplified by PCR using the primers 26SF (5′-ATCCTTTGCAGACGACTTGA-3′), and 5SR (5′-AGCTTGACTTCGCAGATCGG-3′).21 The PCR was performed in a thermocycler (model T100; Bio-Rad Laboratories Inc., USA) with a first denaturation at 94°C for 3min, followed by 30 cycles of 94°C for 30s, 57°C for 30s, and at 72°C for 1min, and a final extension at 72°C for 10min. The amplification products were separated by electrophoresis in 1,5% agarose gels in TBE 0,5X buffer (45mM Tris-Base, 45mM boric acid, 1mM EDTA and pH 8.3) at 90V for 60min, and visualised by GelRed 30 X stain (Biotium, USA). The 100bp DNA ladder (Jena Bioscience™, Jena, GE) was used as a molecular size marker. The Molecular Imager® Gel Doc™ XR (Bio-Rad Laboratories) was used to capture the gel images. The expected size of the amplicons was in the range of 195–719bp, depending on the species.22
Sequencing: The amplicons were purified with the Agarose Gel Extraction Kit (Jena Bioscience™) and sequenced with the 26SF and 5SR primers at the Genomic Services Unit in LANGEBIO (CINVESTAV, MX). The obtained sequences were deposited in the GenBank database (accession no: MT531540–MT531551).
Phylogenetic analysis: The amplicon sequences were edited with the program BioEdit ver. 7.2.512 and compared with registered sequences in GenBank through the BLAST program (http://www.ncbi.nim.nih.gov/).2 The alignment of the sequences was performed with the program Muscle implemented in MEGA 6.0.24 With the maximum likelihood statistical method a phylogenetic tree was constructed, and the groups’ node significance was determined with 1000 repetitions (bootstrap). The analysis included 40 yeast sequences (available in the GenBank) that belong to yeasts within the order Trichonosporales. A Cryptococcus neoformans (KX721893) sequence was also incorporated as outgroup.
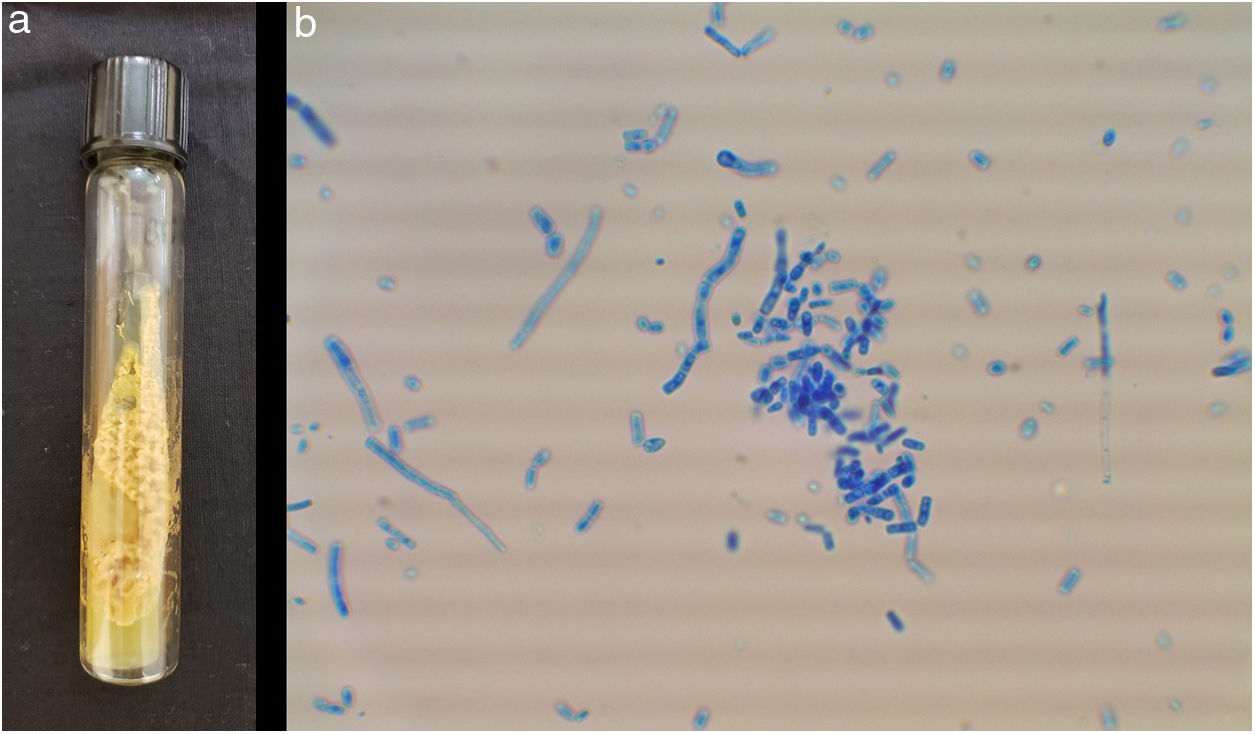
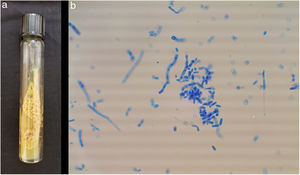
ResultsThe studied isolates showed the typical morphology of the yeasts within the order Trichosporonales. The colonies were whitish, waxy, and cerebriform (Fig. 1a). The amplification of the IGS1 region showed fragments within the range 328–716bp.
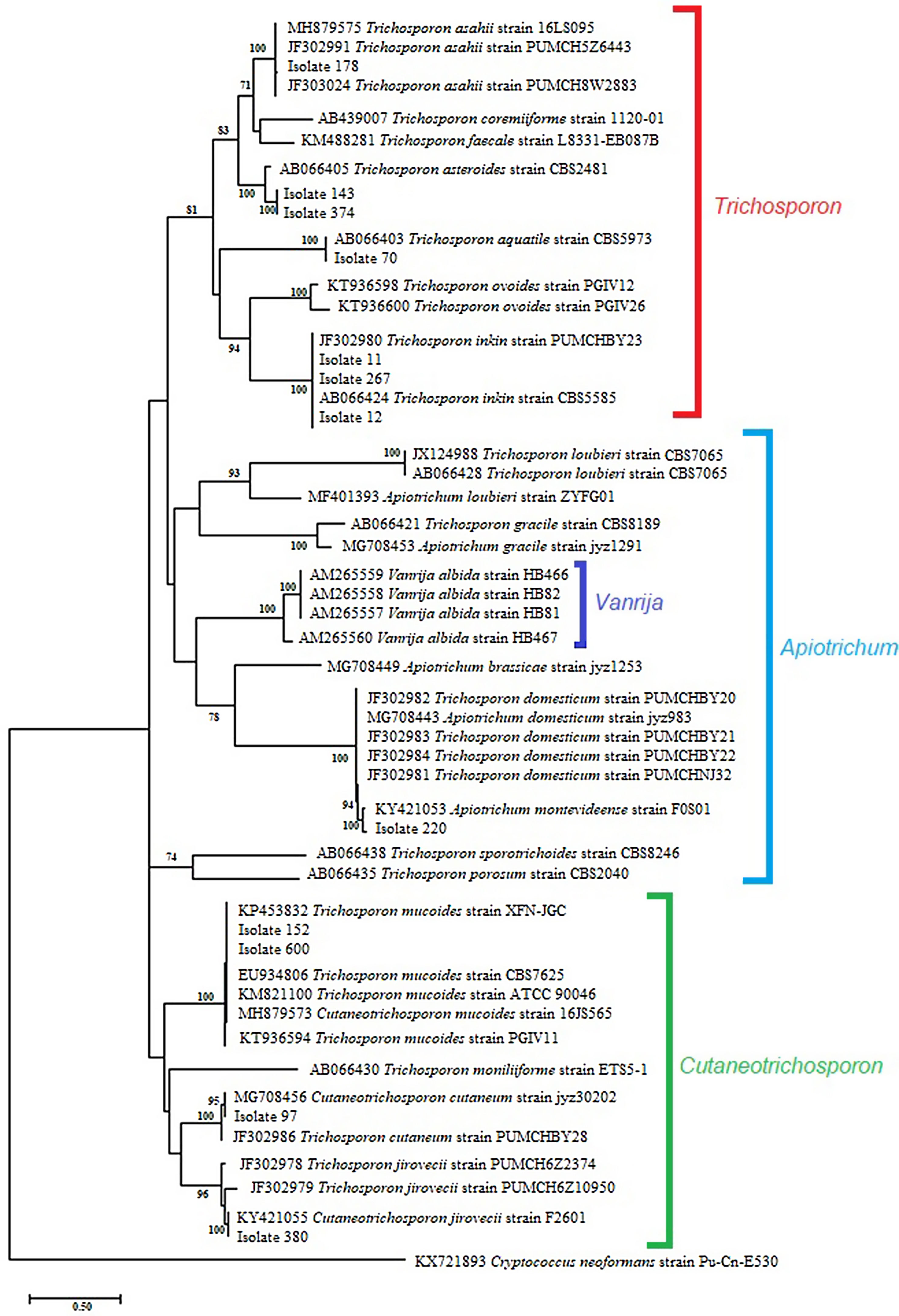
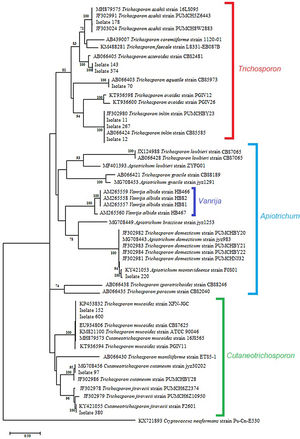
The sequence analysis of the 26S-5S region of the rRNA intergenic spacer showed a high percentage identity with the Trichosporon, Cutaneotrichosporon and Apiotrichum genera. (Table 2). In the phylogenetic tree constructed with the IGS1 sequences the studied isolates and the reference sequences were grouped according to the genus: Trichosporon, Cutaneotrichosporon, and Apiotrichum. The genus Vanrija was placed with Apiotrichum.
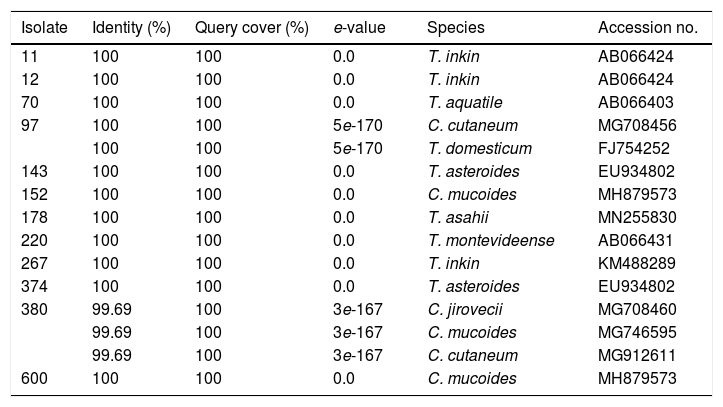
Identification of the studied isolates based on the BLAST analysis.
| Isolate | Identity (%) | Query cover (%) | e-value | Species | Accession no. |
|---|---|---|---|---|---|
| 11 | 100 | 100 | 0.0 | T. inkin | AB066424 |
| 12 | 100 | 100 | 0.0 | T. inkin | AB066424 |
| 70 | 100 | 100 | 0.0 | T. aquatile | AB066403 |
| 97 | 100 | 100 | 5e-170 | C. cutaneum | MG708456 |
| 100 | 100 | 5e-170 | T. domesticum | FJ754252 | |
| 143 | 100 | 100 | 0.0 | T. asteroides | EU934802 |
| 152 | 100 | 100 | 0.0 | C. mucoides | MH879573 |
| 178 | 100 | 100 | 0.0 | T. asahii | MN255830 |
| 220 | 100 | 100 | 0.0 | T. montevideense | AB066431 |
| 267 | 100 | 100 | 0.0 | T. inkin | KM488289 |
| 374 | 100 | 100 | 0.0 | T. asteroides | EU934802 |
| 380 | 99.69 | 100 | 3e-167 | C. jirovecii | MG708460 |
| 99.69 | 100 | 3e-167 | C. mucoides | MG746595 | |
| 99.69 | 100 | 3e-167 | C. cutaneum | MG912611 | |
| 600 | 100 | 100 | 0.0 | C. mucoides | MH879573 |
Within the genus Trichosporon, sequences from isolates 11, 12 and 267 were grouped with reference sequences of T. inkin (AB066424 and JF302980); isolate 70 was grouped with the reference sequence of T. aquatile (AB066403); isolates 143 and 374 were grouped together, and with the reference sequence of T. asteroides (AB066405); and the isolate 178 was grouped with the reference sequences of T. asahii (MH879575, JF302991, and JF303024). All these groupings were supported by a 100% bootstrap value (Fig. 2). In the Apiotrichum genus, isolate 220 was grouped with the reference sequence of A. montevideense (KY421053) with a 100% bootstrap. Finally, for the Cutaneotrichosporon genus, isolates 152 and 600 were grouped with the reference sequences of C. mucoides (MH879573) and T. mucoides (KP453832, EU934806, KM821100 and KT936594) with 100% bootstrap; isolate 97 was grouped with the reference sequence of C. cutaneum (MG708456) (bootstrap 95%), and isolate 380 was grouped with the reference sequence of C. jirovecii (KY421055) (bootstrap 100%).
DiscussionTrichosporonosis is commonly diagnosed based on the phenotypic characteristics of the isolate, something that, oftentimes, does not lead to a correct species identification.9,25 For this reason, and similarly to what has happened with other genera of fungi, molecular methods are recommended for the correct identification of the yeast species previously classified within the genus Trichosporon, currently separated into three genera: Trichosporon, Apiotrichum, and Cutaneotrichosporon. The preferred molecular method is the analysis of the IGS1 region.15,21,22 However, these methods are not used regularly in clinical laboratories, leading to a lack of data about genera and species distribution. This study characterized 12 yeast isolates from patients diagnosed with white piedra, onychomycosis, and hand or foot dermatomycosis. Based on the morphological analysis, all 12 isolates were identical, showing a morphology compatible with the genus Trichosporon. However, only seven isolates (11, 12, 70, 143, 178, 267, and 374) belonged to the genus Trichosporon according to the analysis of the IGS1 region sequences; one isolate (220) belonged to the genus Apiotrichum, and two (152 and 600) to Cutaneotrichosporon. Two isolates showed ambiguous results, as isolate 97 showed 100% similarity with species from two different genera: C. cutaneum and T. domesticum (A. domesticum). Isolate 380 showed a high percentage of similarity with three species of the genus Cutaneotrichosporon: C. jirovecii, C. mucoides, and C. cutaneum. This ambiguity was solved by observing the phylogenetic tree, where isolate 97 groups with C. cutaneum and isolate 380 with C. jirovecii. Based on all the obtained results, we identified three T. inkin (11, 12 and 267) isolates, two T. asteroides (143, 374), two C. mucoides (152 and 600) and one each of T. asahii (178), A. montevideense (220), C. cutaneum (97), T. aquatile (70) and C. jirovecii (380). The three isolates identified as T. inkin were recovered from patients with white piedra, which coincides with literature reports in which T. inkin and T. ovoides are found the most frequent etiological agents of this disease.10,14 On the other hand, the isolates identified as T. asteroids (143, 374), T. aquatile (70), C. cutaneum (97), T. asahii (178), T. montevideense (220) and C. mucoides (152) were obtained from patients with onychomycosis. It is worth mentioning that although these fungi may colonize nails already infected by dermatophytes, our isolates were the primary pathogens. This fact has also been described in other studies where T. asahii, C. mucoides,T. inkin, T. ovoides, T. asteroides, T. faecale, and C. cutaneum were isolated as etiological agents in nail infections.1,13,16,20 It is worth noting that, to our knowledge, no information in the literature was found regarding T. aquatile as an onychomycosis pathogen, so our case could be the first report. Finally, the isolates identified as T. asahii (178), C. mucoides (600), and C. jirovecii (380) were obtained from patients with dermatomycosis on feet or hands. Other authors also found the aforementioned yeasts as the causative agents of dermatomycosis in patients with suspected tinea pedis and tineamanuum.7,11 It is important to highlight that isolates 178 and 600 (T. asahii and C. mucoides, respectively) were obtained from chronically ill patients (breast and lung cancer, and scleroderma) undergoing a treatment with immunosuppressive agents. Those agents are risk factors for developing superficial or invasive infections caused by trichosporonales.17
Despite the small sample size, the diversity of species belonging to the order Trichosporonales found as etiological agents in superficial infections gives us a glimpse about the importance of identifying any yeast at the species level, especially due to potential antifungal resistances. It has been reported in vitro resistance to amphotericin B in T. asahii and resistance to triazoles in non-T. asahiiTrichosporon species.21 It is also well known that superficial infections may spread to the subcutaneous tissue and even deeper, causing severe systemic disease in immunosuppressed and immunocompetent patients.4
ConclusionsThe molecular characterization of the studied isolates revealed a wide variety of trichosporonales as etiological agents of superficial infections, particularly in onychomycosis. Therefore, identifying these yeasts at the species level is essential to broaden the knowledge of their epidemiology.
Conflict of interestThe authors declare that they have no conflict of interest.