Apophysomyces is a monotypic genus belonging to the order Mucorales. The species Apophysomyces elegans has been reported to cause severe infections in immunocompromised and immunocompetent people. In a previous study of Álvarez et al.3 [J Clin Microbiol 2009;47:1650–6], we demonstrated a high variability among the 5.8S rRNA gene sequences of clinical strains of A. elegans.
Material and methodsWe performed a polyphasic study based on the analysis of the sequences of the histone 3 gene, the internal transcribed spacer region of the rDNA gene, and domains D1 and D2 of the 28S rRNA gene, as well as by evaluation of some relevant morphological and physiological characteristics of a set of clinical and environmental strains of A. elegans.
Results and conclusionsWe have demonstrated that A. elegans is a complex of species. We propose as new species Apophysomyces ossiformis, characterised by bone-shaped sporangiospores, Apophysomyces trapeziformis, with trapezoid-shaped sporangiospores, and Apophysomyces variabilis, with variable-shaped sporangiospores. These species failed to assimilate esculin, whereas A. elegans was able to assimilate that glycoside. Amphotericin B and posaconazole are the most active in vitro drugs against Apophysomyces.
Apophysomyces es un género monoespécifico perteneciente al orden Mucorales. La especie Apophysomyces elegans, ha sido reportada como causante de infecciones severas en pacientes inmunocomprometidos e inmunocompetentes. En un estudio previo (Álvarez et al., J Clin Microbiol. 2009;47:1650–6 ), demostramos la elevada variabilidad dentro de las secuencias del gen 5.8S del ARNr en un grupo de cepas clínicas de A. elegans.
Material y métodosHemos realizado un estudio polifásico basado en el análisis de las secuencias del gen de la histona 3, la región de los espaciadores internos del ADNr y los dominios D1 y D2 del gen 28S del ARNr, así como la evaluación de caracteres morfológicos y fisiológicos relevantes de un grupo de cepas clínicas y ambientales de A. elegans.
Resultados y conclusionesHemos demostrado que A. elegans es un complejo de especies. Proponemos como nuevas especies para la ciencia Apophysomyces ossiformis, caracterizada por sus esporangiosporas con forma de hueso; Apophysomyces trapeziformis, con esporangiosporas trapezoidales; y Apophysomyces variabilis, con esporangiosporas de formas variables. Las nuevas especies no asimilan la esculina, en tanto que A. elegans fue capaz de asimilar dicho glicósido. La anfotericina B y el posaconazol fueron los antifúngicos más activos frente a Apophysomyces.
The genus Apophysomyces, belonging to the subphylum Mucoromycotina (oldest phylum Zygomycota),11 was erected by Misra et al.18 in 1979 to accommodate the only species of the genus, Apophysomyces elegans, which was isolated from soil samples in northern India. This fungus was characterized by pyriform sporangia, conspicuous funnel- and/or bell-shaped apophyses, and subhyaline, thin-, and smooth-walled sporangiospores that are mostly oblong with rounded ends. It is a thermotolerant fungus that grows rapidly between 26 and 42°C.6,18A. elegans is not only isolated from soil, decaying vegetation, and as an environmental contaminant,17,18,24 but it is also able to cause severe human infections.8 Unlike other members of Mucorales, this fungus primarily infects immunocompetent hosts.17 The infection typically follows traumatic implantation of the agent, but may also result from inhalation of spores into the sinus.5,6,12,15,17,21 This disease is more common in tropical and subtropical climates. Cases have been reported in Australia,6,20 India,5,13,14,26 the United States,3,15 Sri Lanka,4 Thailand,24 and in Central and South America.21,22
The genetic population structure of A. elegans remains largely unknown and may be due in part to the lack of preservation of strains for study. In a recent survey on the spectrum of species of Mucorales from clinical sources in the United States, we demonstrated a high intraspecific 5.8S rRNA gene sequence diversity in A. elegans.3 Additionally, in a typing study of A. elegans using microsatellites markers, it was demonstrated that, in a set of clinical strains, mainly from India, different banding patterns exist.5 These data suggest that more than one phylogenetic species may be present within the morphospecies A. elegans.
To determine possible cryptic species in A. elegans, we performed a polyphasic study on a diverse panel of strains, based on a multilocus sequence analysis of three loci (the histone 3 gene (H3), internal transcribed spacer region of the rDNA (ITS), and domains D1 and D2 of the 28S rRNA gene) and the evaluation of different morphological and physiological characters.
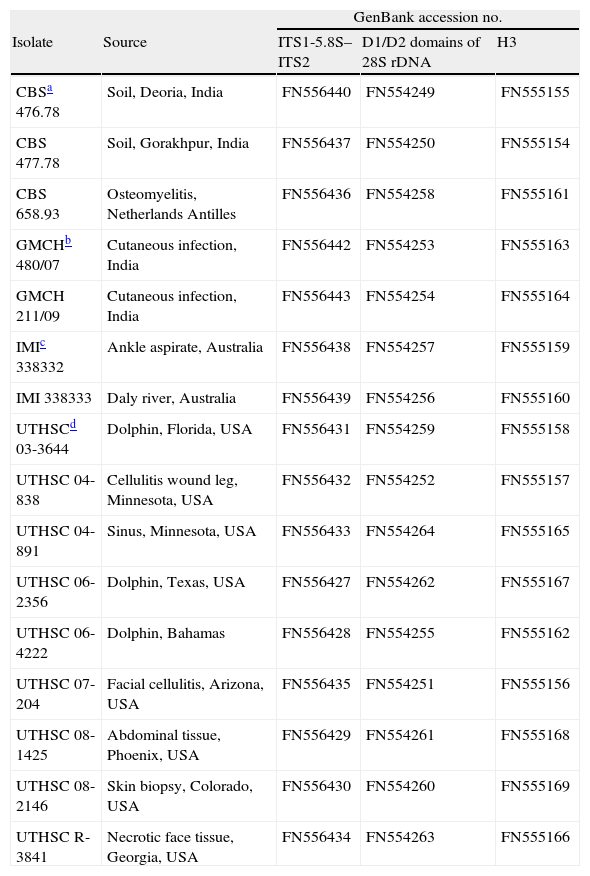
Materials and methodsFungal strainsA total of 16 strains from different origins were included in the study (Table 1). The fungi were cultured on potato dextrose agar (PDA, Pronadisa, Madrid, Spain) and incubated at 35±1°C for 2–5 days.
Origin of Apophysomyces strains included in the study
| GenBank accession no. | ||||
| Isolate | Source | ITS1-5.8S–ITS2 | D1/D2 domains of 28S rDNA | H3 |
| CBSa 476.78 | Soil, Deoria, India | FN556440 | FN554249 | FN555155 |
| CBS 477.78 | Soil, Gorakhpur, India | FN556437 | FN554250 | FN555154 |
| CBS 658.93 | Osteomyelitis, Netherlands Antilles | FN556436 | FN554258 | FN555161 |
| GMCHb 480/07 | Cutaneous infection, India | FN556442 | FN554253 | FN555163 |
| GMCH 211/09 | Cutaneous infection, India | FN556443 | FN554254 | FN555164 |
| IMIc 338332 | Ankle aspirate, Australia | FN556438 | FN554257 | FN555159 |
| IMI 338333 | Daly river, Australia | FN556439 | FN554256 | FN555160 |
| UTHSCd 03-3644 | Dolphin, Florida, USA | FN556431 | FN554259 | FN555158 |
| UTHSC 04-838 | Cellulitis wound leg, Minnesota, USA | FN556432 | FN554252 | FN555157 |
| UTHSC 04-891 | Sinus, Minnesota, USA | FN556433 | FN554264 | FN555165 |
| UTHSC 06-2356 | Dolphin, Texas, USA | FN556427 | FN554262 | FN555167 |
| UTHSC 06-4222 | Dolphin, Bahamas | FN556428 | FN554255 | FN555162 |
| UTHSC 07-204 | Facial cellulitis, Arizona, USA | FN556435 | FN554251 | FN555156 |
| UTHSC 08-1425 | Abdominal tissue, Phoenix, USA | FN556429 | FN554261 | FN555168 |
| UTHSC 08-2146 | Skin biopsy, Colorado, USA | FN556430 | FN554260 | FN555169 |
| UTHSC R-3841 | Necrotic face tissue, Georgia, USA | FN556434 | FN554263 | FN555166 |
DNA was extracted and purified directly from fungal colonies following a slightly modified Fast DNA kit protocol (Bio101, Vista, CA, USA), consisting of a homogenization step repeated three times with a FastPrep FP120 instrument (Thermo Savant, Holbrook, NY, USA). DNA was quantified by the GeneQuant pro (Amersham Pharmacia Biotech, Cambridge, England). The internal transcribed spacer (ITS) region of the nuclear rDNA was amplified with the primer pair ITS5 and ITS4, the D1–D2 domains of the 28S rRNA gene were amplified with the primer pair NL1–NL4, and histone 3 (H3) gene was amplified with the primer pair H3-1a–H3-1b.10
The PCR mix (25μl) included 10mM Tris–HCl (pH 8.3), 50mM KCl, and 2.5mM MgCl2 (10× Perkin-Elmer buffer II plus MgCl2 solution Roche Molecular Systems, Branchburg, NJ, USA), 100μM of each dNTP (Promega, Madison, WI, USA), 1μM of each primer, and 1.5U of AmpliTaq DNA polymerase (Roche). The amplification program for the three DNA fragments included an initial denaturation at 94°C for 5min, followed by 35 cycles of denaturation at 95°C for 30s, annealing for 1min at 54°C, and extension for 1min at 72°C. The products were purified with an Illustra GFX™ PCR DNA and Gel Band Purification Kit (General Electric Healthcare, Buckinghamshire, UK) and stored at −20°C until they were used in sequencing. PCR products were sequenced using the same primers employed for amplification and following the Taq DyeDeoxy Terminator Cycle Sequencing Kit protocol (Applied Biosystems, Gouda, Netherlands). Reactions were run on a 310 DNA sequencer (Applied Biosystems). Consensus sequences were obtained using the Autoassembler program (Perkin-Elmer-Applied Biosystems) and Seqman software (Lasergene, Madison, WI).
Phylogenetic analysesThe sequences were aligned using Clustal X (version 1.8) computer program, followed by manual adjustments with a text editor. Most-parsimonious tree (MPT) analyses were performed using PAUP* version 4.0b10. One hundred heuristic searches were conducted with random sequence addition and tree bisection–reconnection branch-swapping algorithms, collapsing zero-length branches, and saving all minimal-length trees (MulTrees). Saksenaea vasiformis (FMR 10131) was chosen as the outgroup. Support for internal branches was assessed using a heuristic parsimony search of 1000 bootstrapped data sets. The combined data set of the ITS, D1–D2, and H3 was tested for incongruence with the partition homogeneity test (PHT) as implemented in PAUP*. The Kishino–Hasegawa test was performed to determine whether trees differed significantly. Gaps were treated as missing data.
Morphological studiesThe strains were subcultured on PDA, Czapek agar (CZA; Difco, Becton Dickinson, France), malt extract agar (MEA; 10g of malt extract, 20g of agar, and 1000ml of distilled water), and starch agar (SA; 5g of soluble starch, 15g of agar, and 1000ml of distilled water), and incubated at 37 and 42°C. The microscopic features were determined on the sixth day in wet mounts on water and on lactic acid, which were examined under a light microscope. The strains were identified using schemes based on morphological characters.8,18
Physiological studiesGrowth rates at 4, 15, 24, 30, 35, 37, 42, and 50°C were determined on PDA, MEA, CZA, and SA for each of the strains included in the study. The Petri dishes were inoculated in the center, incubated in darkness, and the colony diameters (in millimeters) were measured daily.
Carbon source assimilation profiles were determined with the commercial kit API 50CH (bioMérieux, Marcy, l’Etoile, France), following protocols described previously but with minor modifications.23 To obtain sufficient sporulation, all strains were cultured for 6 days on CZA at 42°C. A final concentration of 5×105CFU/ml was prepared in 20ml of yeast nitrogen base (7.7g/l; Difco), containing 0.5g/l L-chloramphenicol (Sigma-Aldrich) and 0.1% Bacto agar (Difco), and each well of the strips was inoculated with 300μl of medium. The viability of the conidia was verified by plating 100μl of serial dilutions of each inoculum onto PDA and incubating at 42°C for 6 days. The inoculated API 50 CH strips were incubated for 48–72h at 37°C in darkness. After incubation, the strips were read visually and growth or lack of growth was noted. Weak growth was considered as a positive result. For nitrogen source assimilations, we used the same inoculum described above, but the yeast nitrogen base broth was replaced by carbon nitrogen base broth (Difco), and testing was performed in sterile, disposable, multiwell microplates. The medium with the nitrogen sources was dispensed into the wells in 150μl volumes with a multichannel pipette and each well was inoculated with 50μl of the conidial suspension. The microplates were incubated at 37°C in darkness for 48 and 72h. We also determined growth of the strains on NaCl (2%, 5%, 7%, 10%), MgCl2 2%, and cycloheximide 0.1%.9,27 All tests were performed in duplicate.
The production of urease was determined after incubation on Christensen's urea agar slants at 37°C for 8 days.16
Mating testsSixteen Apophysomyces strains were grown on CZA plates at 37°C in the dark, and then paired in all combinations, including self-crosses, on CZA. Each strain was streaked onto one half of a CZA plate opposite to the streak of another strain, allowing for a central zone of contact as the strains grew. Plates were incubated at 37°C and examined macroscopically each week for up to 6 months for the presence of zygospores. All tests were performed in duplicate.
Antifungal susceptibility testingThe in vitro activity of seven antifungal agents against the 16 strains of Apophysomyces was evaluated according to Clinical and Laboratory Standards Institute guidelines (M38-A2).19 The drugs tested were amphotericin B (USP, Rockville, MD, USA), anidulafungin (Pfizer Inc., New York, NY, USA), caspofungin (Merck & Co., Inc., Rahway, NJ, USA), itraconazole (Jansen Pharmaceutica, Beerse, Belgium), posaconazole (Schering-Plough Ltd., Hertfordshire UK), ravuconazole (Bristol-Myers Squibb Company, New Brunswick, NJ, USA), and voriconazole (Pfizer Inc., New York, NY, USA).
Nucleotide sequence accession numbersAll the sequences obtained in this study were deposited in GenBank database and assigned the accession numbers listed in Table 1.
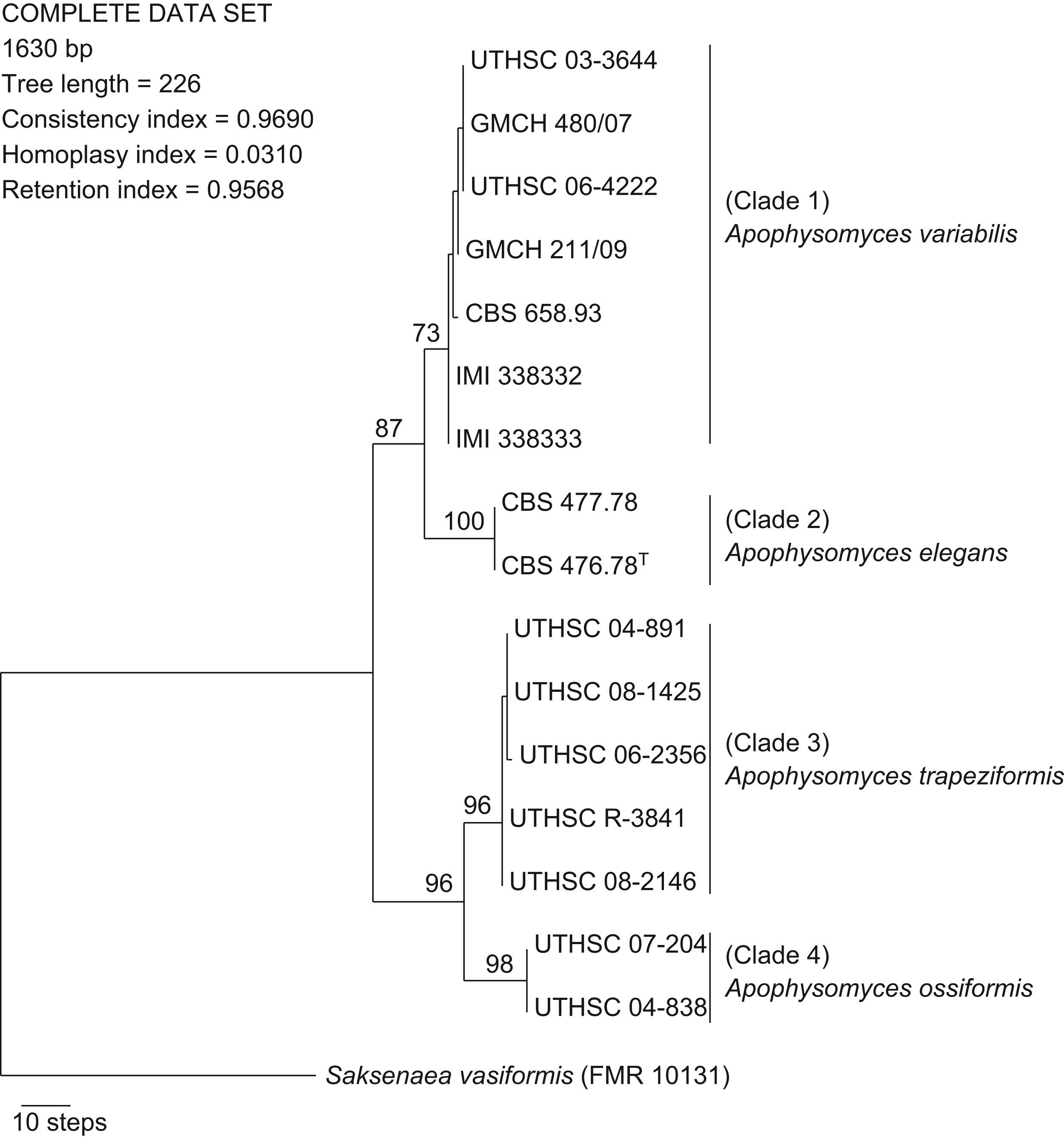
ResultsPhylogenyWith the primers used, we were able to amplify and sequence 684–820, 582–683, and 345–382bp of the ITS, D1–D2, and H3 loci, respectively. Of the 1630 nucleotides sequenced, 48 characters were parsimony informative in the different strains, the lowest number was eight for the H3 gene, and the highest was 27 for the ITS region. Sequences of the three region genes were analyzed phylogenetically as separate (data not shown) and combined datasets. The partition homogeneity test demonstrated that the three loci sequence data sets were congruent (P=0.05) and could therefore be combined. A total of 36 MPT was produced from a heuristic search, using the combined dataset from the three loci (Fig. 1). The trees had a consistency index of 0.969, a retention index of 0.956, and a homoplasy index of 0.031. Clustering was similar to that observed in the particular trees of the different genes analyzed. Most nodes in the combined analysis showed increased clade support as measured by bootstrapping (six nodes with ≥70%). Analyses of the combined partitions support the recognition of four well supported clades (Fig. 1), each of which could be considered separate phylogenetic species. Clade 1 (bootstrap support (bs) 73%) was composed of two strains from India, two from Australia, and one strain each from the United States, Netherlands Antilles, and Bahamas. Within clade 2 (bs 100%) were included the type strain of A. elegans and one strain from Indian soil. In clade 3 five strains (bs 96%) were located, four of them of clinical origin from the United States, and one from a dolphin. Finally, clade 4 (bs 98%) consisted of two clinical strains, both from the United States.
PhysiologyCarbon assimilation profiles of the different strains on API 50CH strips are shown in Table 2. Assimilation patterns of all the strains were positive for 20 tests. Twenty-seven carbon sources were not assimilated by any strain. The profiles of assimilation of two carbon sources, esculin and D-lyxose, were species- and strain-dependent, respectively. Esculin was weakly assimilated only by the strains nested in clade 2. The assimilation of D-lyxose was highly variable among the strains of the different clades. By contrast, the variability in the assimilations of nitrogen sources, and tolerance to NaCl, MgCl2, and cycloheximide was nule among the species (Table 3). All the strains were positive for 11 nitrogen sources. Nitrite was not assimilated by any of the strains. All strains were able to grow at 2% NaCl and at 2% MgCl2, but failed to grow at 5% NaCl and at 0.1% cycloheximide.
Carbon assimilation profiles for Apophysomyces species obtained with API 50 CH strips
| Species | ||||||||||||||||
| A. variabilis | A.elegans | A. trapeziformis | A. ossiformis | |||||||||||||
| Carbon source | CBS 658.93 | UTHSC 06-4222 | GMCH 211/09 | GMCH 480/07 | IMI 338332 | IMI 338333 | UTHSC 03-3644 | CBS 476.78 | CBS 477.78 | UTHSC 04-891 | UTHSC 06-2356 | UTHSC 08-1425 | UTHSC 08-2146 | UTHSC R-3841 | UTHSC 04-838 | UTHSC 07-204 |
| GLY (glycerol) | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + |
| ERY (erythritol) | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − |
| DARA (D-arabinose) | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − |
| LARA (L-arabinose) | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + |
| RIB (D-ribose) | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + |
| DXYL (D-xylose) | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + |
| LXYL (L-xylose) | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − |
| ADO (D-adonitol) | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + |
| MDX (methyl-ß-D-xylopyranoside) | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − |
| GAL (D-galactose) | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − |
| GLU (D-glucose) | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + |
| FRU (D-fructose) | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + |
| MNE (D-mannose) | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + |
| SBE (L-sorbose) | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − |
| RHA (L-rhamnose) | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − |
| DUL (dulcitol) | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − |
| INO (inositol) | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − |
| MAN (D-mannitol) | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + |
| SOR (D-sorbitol) | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + |
| MDM (methyl-D-mannopyranoside) | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − |
| MDG (methyl-D-glucopyranoside) | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − |
| NAG (N-acetyl-glucosamine) | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + |
| AMY (amygdalin) | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − |
| ARB (arbutin) | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − |
| ESC (esculin) | − | − | − | − | − | − | − | + | + | − | − | − | − | − | − | − |
| SAL (salicin) | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − |
| CEL (D-cellobiose) | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + |
| MAL (D-maltose) | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + |
| LAC (D-lactose) | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − |
| MEL (D-melibiose) | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − |
| SAC (D-saccharose) | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − |
| TRE (D-trehalose) | + | + | + | + | + | − | + | + | + | + | + | + | + | + | + | + |
| INU (inulin) | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − |
| MLZ (D-melezitose) | + | + | + | + | + | − | + | + | + | + | + | + | + | + | + | + |
| RAF (D-raffinose) | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − |
| AMD (amidon) | + | + | + | + | + | + | − | + | + | + | + | + | + | + | + | + |
| GLYG (glycogen) | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + |
| XLT (xylitol) | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + |
| GEN (gentiobiose) | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − |
| TUR (D-turanose) | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − |
| LYX (D-lyxose) | − | − | − | − | + | + | − | − | − | − | + | − | − | − | + | + |
| TAG (D-tagatose) | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − |
| DFUC (D-fucose) | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − |
| LFUC (L-fucose) | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − |
| DARL (D-arabitol) | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + |
| LARL (L-arabitol) | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + |
| GNT (potassium gluconate) | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − |
| 2KG (potassium 2-keto-gluconate) | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − |
| 5KG (potassium 5-keto-gluconate) | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − |
Nitrogen assimilation and tolerance to chemical compounds for the Apophysomyces species included in this study
| Species | ||||||||||||||||
| A. variabilis | A. elegans | A. trapeziformis | A. ossiformis | |||||||||||||
| Nitrogen source and other tests | CBS 658.93 | UTHSC 06-4222 | GMCH 211/09 | GMCH 480/07 | IMI 338332 | IMI 338333 | UTHSC 03-3644 | CBS 476.78 | CBS 477.78 | UTHSC 04-891 | UTHSC 06-2356 | UTHSC 08-1425 | UTHSC 08-2146 | UTHSC R-3841 | UTHSC 04-838 | UTHSC 07-204 |
| Cadaverine | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + |
| Creatine | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + |
| Creatinine | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + |
| L-lysine | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + |
| Nitrate | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + |
| Nitrite | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − |
| L-tryptophan | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + |
| L-proline | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + |
| L-leucine | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + |
| L-ornithine | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + |
| L-cysteine | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + |
| Arginine | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + |
| 2% NaCl | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + |
| 5% NaCl | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − |
| 7% NaCl | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − |
| 10% NaCl | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − |
| 2% MgCl2 | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + |
| Cycloheximide 0.1% | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − |
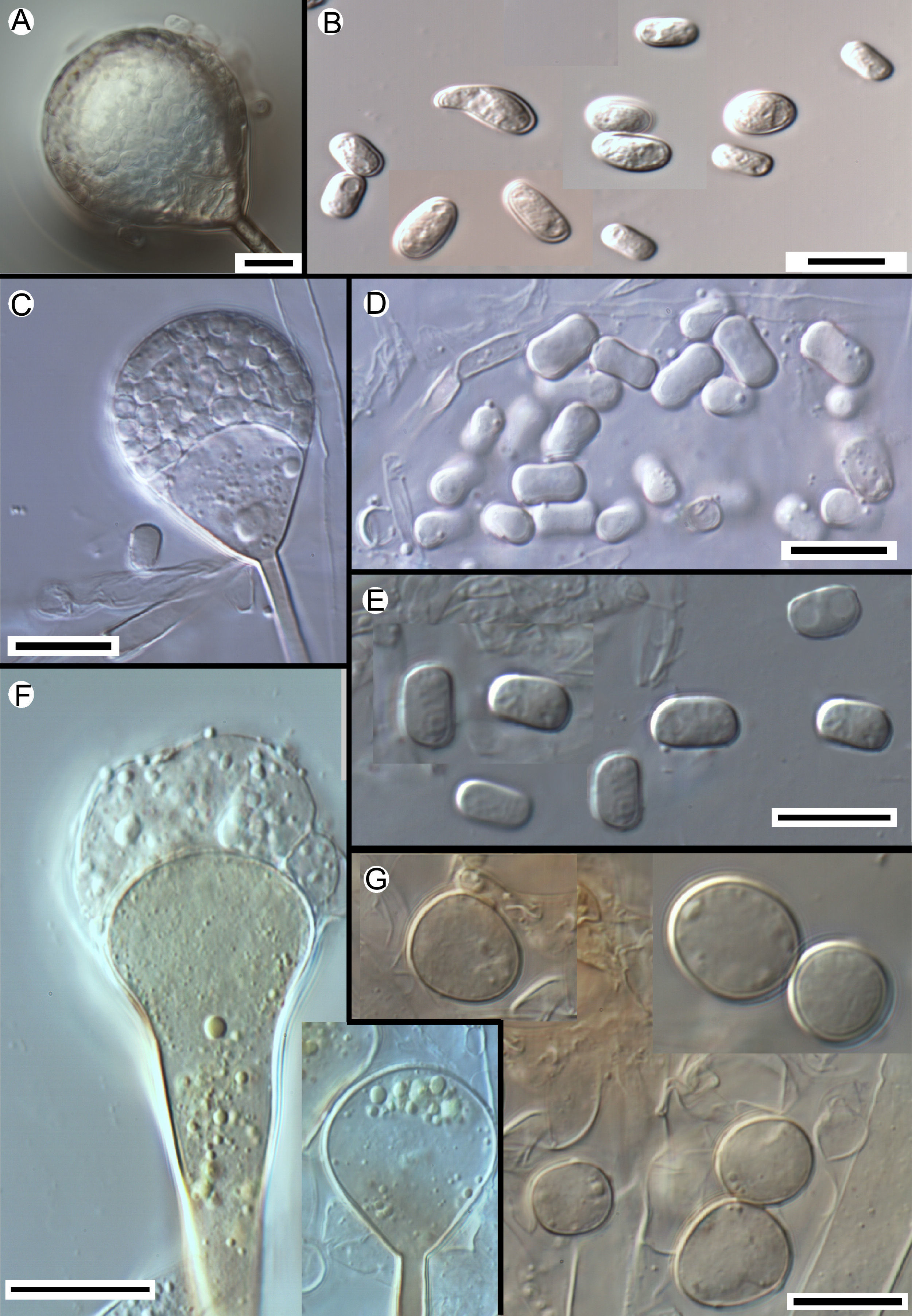
In general, all the strains examined displayed the typical features of the genus Apophysomyces described by Misra et al.18 However, a more detailed microscopic study of these fungi showed important and consistent differences, mainly in the morphology of sporangiophores and sporangiospores, which correlated with the different phylogenetic species. The strains included in clade 1 showed some morphological diversity. The sporangiospores ranged from broadly clavate to ellipsoidal, were flattened on one side, and measured 5–14×3–6μm. The strains included in clade 2, which comprises the type strain of A. elegans, showed ovoid, subspherical, broadly ellipsoidal to barrel shaped sporangiospores, although more irregularly shaped spores were also present, measured 6–12×5–8μm, and were the largest for the different species in the complex. The sporangiospores of the strains included in clade 3 were trapezoidal and smaller (5–8.5×3–5μm) while those of strains in clade 4 were thick-walled and clearly biconcave (bone-shaped) in side view, measuring 6–8×3–5.5μm. In addition to differences in spore morphology, the strains in clade 2 also showed two types of sporangiophores: (i) large (up to 540μm), bearing vase- or bell-shaped apophyses and (ii) shorter (up to 400μm), bearing funnel-shaped apophyses. The sporangiophores in strains of clades 1, 3, and 4 are similar to the short ones of clade 2.
Mating testZygospore formation was not observed after 6 months of incubation in all the mating tests assayed.
Based on the described morphological and physiological differences, which correlated with the molecular data, we concluded that clades 1, 3, and 4 represent three species of Apophysomyces, different from A. elegans (clade 2), which are proposed here as new species.
Apophysomyces variabilis Alvarez, Stchigel, Cano, D.A. Sutton et Guarro, sp. nov. (Figs. 2A, B; 3H, I).
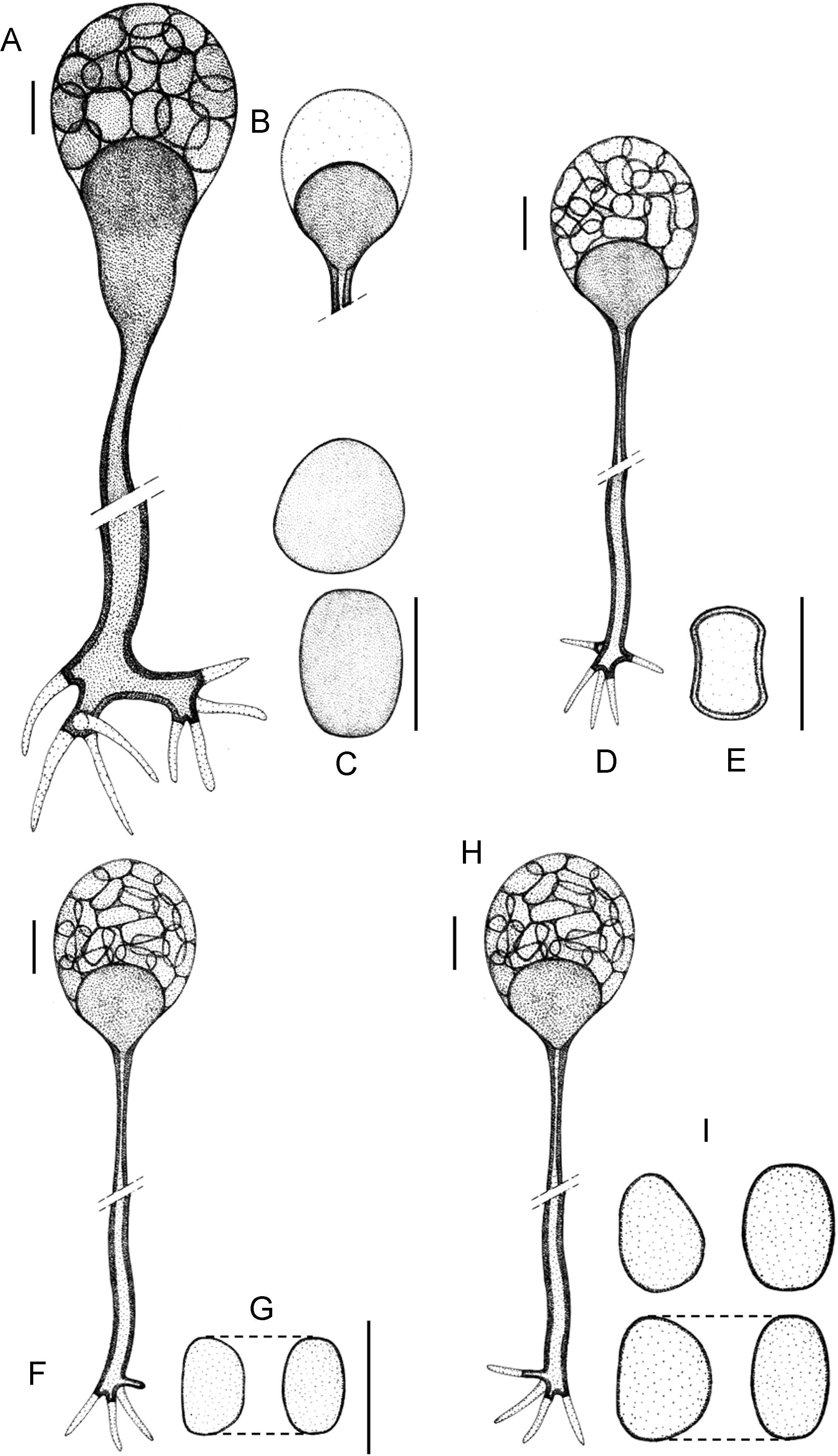
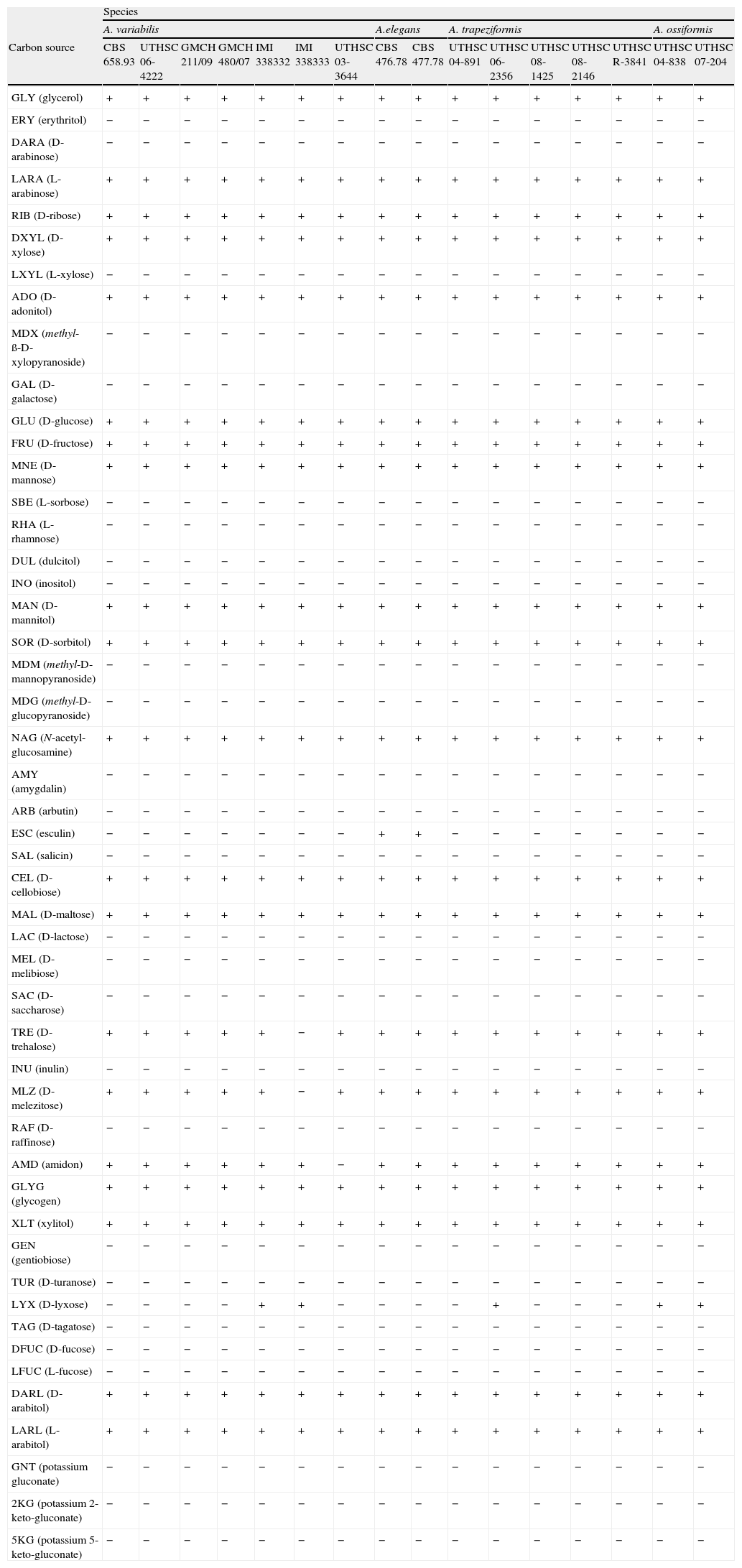
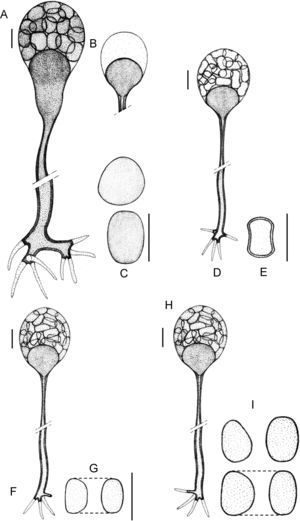
Morphology of the apophyses and sporangiospores of Apophysomyces: (A, B) A. variabilis CBS 658.93 (A, apophyses funnel-shaped; B, sporangiospores). (C, D) A. ossiformis UTHSC 04-838 (C, apophyses funnel-shaped; D, bone-like shaped sporangiospores). (E) A. trapeziformis UTHSC 08-1425 (E, trapezoid-shaped sporangiospores). (F, G) A. elegans CBS 476.78 (F, apophyses bell- and funnel-shaped; (G) subspherical to broadly ellipsoidal sporangiospores). All bars=10μm.
Coloniae in CZA ad 37°C rapide crescentes, albae, sed sparsis, inmersis pro parte maxima compositae. Sporangiophora erecta, plerumque simplicia, 100–400μm longa, 2–3.5μm lata, brunnea, cum sporangio apohysati. Apophyses plerumque infundibuliformes, 15–20×15–20μm2. Sporangiosporae variabiles in forma et magnitudine, trapezoides, ellipsoideae, subtriangulares vel claviformis, 5–14×3–6μm2. Holotypus, CBS H-658.93, ex osteomielitis (cultura viva FMR 10381, CBS 658.93).
Etymology: the epithet refers to the variable morphology of the sporangiospores.
Colonies attaining a diameter of 90mm after 4 days of incubation at 37°C on CZA, whitish, with scarce aerial mycelium; hyphae branched, hyaline, smooth-walled, 3–5.5μm in diameter; reverse concolorous. Sporangiophores erect, generally arising singly, at first hyaline, soon becoming light greyish brown, generally straight, slightly tapered towards the apex, unbranched, 100–400μm long, 2–3.5μm wide, and smooth-walled. Sporangia apophysate, terminal, pyriform, multispored, white at first, becoming light greyish brown when mature, and 15–50μm in diameter. Apophyses short, funnel-shaped, and 15–20×15–20μm. Sporangiospores variable in shape, trapezoid, ellipsoid, subtriangular or claviform, hyaline to light brown in mass, smooth- and thin-walled, and 5–14×3–6μm. Not able to assimilate esculin.
Colonies on SA, PDA, and MEA showed similar features than on CZA, but they were more floccose, white, and with less sporulation. The optimum growth temperature was 35–42°C and the minimum temperature of growth was 15°C. The fungus did not grow at 50°C.
Apophysomyces ossiformis Alvarez, Stchigel, Cano, D.A. Sutton et Guarro sp. nov. (Figs. 2C, D; 3D, E).
Coloniae in CZA ad 37°C rapide crescentes, albae, sed sparsis, inmersis pro parte maxima compositae. Sporangiophora erecta, plerumque simplicia, 100–400μm longa, 2–3.5μm lata, brunnea, cum sporangio apohysati. Apophyses plerumque infundibuliformes, 15–20×15–20μm. Sporangiosporae ossiformis, 6–8×3–5.5μm. Holotypus, CBS H-20328, ex cellulitis cruris vulnus hominis (cultura viva FMR 9913, UTHSC 04-838).
Etymology: the epithet refers to the bone-like shape of the sporangiospores.
Colonies attaining a diameter of 90mm after 4 days of incubation at 37°C on CZA, whitish, with scarce aerial mycelium, branched, hyaline, smooth-walled, and 3–5.5μm in diameter; reverse concolorous. Sporangiophores erect, generally arising singly, at first hyaline soon becoming light greyish brown, generally straight, slightly tapered towards the apex, unbranched, 100–400μm long, 2–3.5μm wide, and smooth-walled. Sporangia apophysate, terminal, pyriform, multispored, white at first, becoming light greyish brown when mature, 15–50μm in diameter. Apophyses short, funnel-shaped, 15–20×15–20μm. Sporangiospores mostly bone-like shaped, hyaline to light brown in mass, smooth- and thick-walled, and 6–8×3–5.5μm. Not able to assimilate esculin.
Colonies on SA, PDA, and MEA showed similar features than on CZA, but they were more floccose, white, and with less sporulation. The optimum growth temperature was 35–42°C and the minimum 15°C. The fungus did not grow at 50°C.
Apophysomyces trapeziformis Alvarez, Cano, Stchigel, D.A. Sutton et Guarro sp. nov. (Figs. 2E; 3F, G).
Coloniae in CZA ad 37°C rapide crescentes, albae, sed sparsis, inmersis pro parte maxima compositae. Sporangiophora erecta, plerumque simplicia, usque ad 400μm longa, 2–3.5μm lata, brunnei, cum sporangio apohysati. Apophyses plerumque infundibuliformes, 15–20×15–20μm. Sporangiosporae trapezoids vel ellipsoideae, 5–8.5×3–5μm.
Holotypus, CBS H-20329, ex abscessus abdominis humanus (cultura viva FMR 10456, UTHSC 08-1425).
Etymology: the epithet refers to the trapezoid shape of the sporangiospores in side view.
Colonies attaining a diameter of 90mm after 4 days of incubation at 37°C on CZA, whitish, with scarce aerial mycelium, branched, hyaline, smooth-walled, 3–5.5μm in diameter; reverse concolorous. Sporangiophores erect, generally arising singly, at first hyaline soon becoming light greyish brown, generally straight, slightly tapered towards the apex, unbranched, up to 400μm long, 2–3.5μm wide, and smooth-walled. Sporangia apophysate, terminal, pyriform, multispored, white at first, becoming light greyish brown when mature, and 15–50μm in diameter. Apophyses short, funnel-shaped, and 15–20×15–20μm. Sporangiospores mostly trapezoid-shaped in side view, more or less cylindrical in front view, flattened at one side and broadly convex on opposite side, hyaline to light brown in mass, smooth- and thin-walled, and 5–8.5×3–5μm. Not able to assimilate esculin.
Colonies on SA, PDA, and MEA showed similar features than on CZA, but they were more floccose, white, and with less sporulation. The optimum growth temperature was 35–42°C and the minimum 15°C. The fungus did not grow at 50°C.
Based on our morphologic and physiologic studies, the type species of Apophysomyces is redefined as follows:
A. elegans Misra, Srivastava, and Lata (Figs. 2F,G; 3A–C).
(A)–(C) Apophysomyces elegans (A, vessel-shaped sporangiophore; B, funnel-shaped sporangiophore; C, sporangiospores). (D, E) Apophysomyces ossiformis (D, sporangiophore; E, sporangiospore, frontal and side views). (F, G) Apophysomyces trapeziformis (F, sporangiophore; G, sporangiospore). (H, I) Apophysomyces variabilis (H, sporangiophore; I, sporangiospores). Bars: 10μm.
Colonies attaining a diameter of 90mm after 4 days at 37°C on CZA, whitish at first, becoming brownish grey, with scarce aerial mycelium; reverse concolorous. Sporangiophores generally arising singly, emerging from aerial hyphae, straight or curved, mainly unbranched or some times branched at the apex, light greyish brown, with two types of morphology, i.e. (i) those that were large (up to 540μm), bearing vase- or bell-shaped apophyses (15–46×11–40μm) and (ii) those that were shorter (up to 400μm) and bore funnel-shaped apophyses (15–20×15–20μm), of 4–7.5μm wide, and smooth-walled. Sporangia produced terminally, pyriform, with distinct apophyses, and 20–60μm in diameter. Sporangiospores ovoid, subspherical, broadly ellipsoidal to barrel-shaped, frequently irregularly shaped, subhyaline, smooth- and thin-walled, and 6–12×5–8μm. The strains analyzed were able to assimilate esculin.
Similar colonies features as described on CZA were observed on AS, PDA, and MEA, with the exception of lesser production of mycelium in CZA. The optimum growth temperature was 35–42°C and the minimum 15°C. The fungus did not grow at 50°C.
Antifungal susceptibility testsThe results of antifungal susceptibility testing for Apophysomyces strains are shown in Table 4. Amphotericin B and posaconazole were the most active antifungal agents. Itraconazole and ravuconazole were more active than voriconazole, and caspofungin and anidulafungin were inactive against all strains.
In vitro antifungal susceptibility data for Apophysomyces species
| Species (♯ of strains tested) | Antifungal agent | MIC or MEC (μg/mL) 24h | |
| GM | Range | ||
| A. variabilis (7) | Amphotericin B | 1.0 | 1 |
| Posaconazole | 1.1 | 1–2 | |
| Voriconazole | 29.0 | 16–>16 | |
| Itraconazole | 1.0 | 0.5–2 | |
| Ravuconazole | 1.0 | 0.5–2 | |
| Anidulafungin | 7.2 | 4–8 | |
| Caspofungin | 19.5 | 4–>16 | |
| A. elegans (2) | Amphotericin B | 0.5 | 0.5 |
| Posaconazole | 0.5 | 0.5 | |
| Voriconazole | 8.0 | 8 | |
| Itraconazole | 1.0 | 0.5–2 | |
| Ravuconazole | 1.0 | 0.5–2 | |
| Anidulafungin | 8.0 | 8 | |
| Caspofungin | 32.0 | >16 | |
| A. trapeziformis (5) | Amphotericin B | 0.8 | 0.5–1 |
| Posaconazole | 0.8 | 0.5–1 | |
| Voriconazole | 16.0 | 8–>16 | |
| Itraconazole | 0.9 | 0.5–2 | |
| Ravuconazole | 0.9 | 0.5–2 | |
| Anidulafungin | 6.1 | 4–8 | |
| Caspofungin | 27.9 | 16–>16 | |
| A. ossiformis (2) | Amphotericin B | 1.4 | 1–2 |
| Posaconazole | 0.7 | 0.5–1 | |
| Voriconazole | 5.7 | 4–8 | |
| Itraconazole | 1.4 | 1–2 | |
| Ravuconazole | 2.8 | 2–4 | |
| Anidulafungin | 5.7 | 4–8 | |
| Caspofungin | 32.0 | <16 | |
Apophysomyces has been traditionally considered a monotypic genus. However, on the basis of genetic, physiological, and morphological data, we have demonstrated here that the genus constitutes a complex of species. DNA sequences from three different loci were analyzed to infer phylogenetic relationships and species boundaries within strains morphologically identified as A. elegans. The informations provided by the three loci evaluated were similar, and proved to be useful markers for species level differentiation in Apophysomyces. Although our study included strains from very diverse origins, the number of isolates we could obtain was small, and we anticipate even greater diversity as more strains become available. Given this limitation we were, however, able to recognize at least four phylogenetically, morphologically, and physiologically different species. The shape and size of the sporangiospores, the type of the sporangiophore, and the shape of the apophyses were the most useful characters for this purpose.
As carbon assimilation profiles can be useful for differentiation of human pathogenic mucoralean genera,23 we tested the assimilation of numerous carbon sources (Table 2). Low interspecific variability within Apophysomyces was noted, with the only exception of esculin assimilation, which was positive for A. elegans, and negative for the other species in the complex. In this study, Apophysomyces strains also showed negative results for D-galactose, amygdalyn, arbutin, salicin, and gentiobiose assimilation, while in the study of Schwarz et al.23 these same tests were positive for the members of six other pathogenic genera, i.e. Cunninghamella, Lichtheimia (Absidia), Mucor, Rhizopus, Rhizomucor, and Syncephalastrum. In contrast, carbon sources such as L-sorbose, L-rhamnose, dulcitol, inositol, erythritol, D-arabinose, methyl-ß-D-xylopiranoside, methyl-D-mannopyranoside, methyl-D-glucopyranoside, D-tagatose, D-fucose, L-fucose, and inulin were all negative for both the Apophysomyces strains and the other six mentioned genera.23 Other carbon sources such as glycerol were assimilated by Apophysomyces, Rhizopus, and Cunninghamella but not by Lichtheimia, Rhizomucor, Mucor, and Syncephalastrum;D-ribose was assimilated by Apophysomyces, Rhizopus, and Mucor and not by Cunninghamella, Lichtheimia, Rhizomucor, and Syncephalastrum; L-xylose was assimilated only by Mucor and some strains of Rhizopus, and D-lactose was assimilated only by Lichtheimia, Rhizomucor, and Syncephalastrum, although it was species dependent in Rhizomucor. The nitrogen assimilation profiles and tolerance to various chemical agents for the Apophysomyces strains tested in this study were non-discriminatory (Table 3).
From a clinical point of view it is also worth mentioning that none of the clinical strains included in this study belonged to clade 2, which contains the type strain, A. elegans, and which has previously been considered a pathogenic species. Clade 2 included only two environmental strains isolated from Indian soils.
The in vitro activity of the antifungal drugs tested appeared to corroborate data from previous studies.1,2,7,25
This work was supported by the Spanish Ministerio de Ciencia y Tecnología Grants CGL 2007-65669/BOS and CGL 2009-08698/BOS.