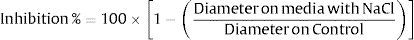Several fungal species represent a potential risk to embryos of Odontesthes bonariensis (Cuvier and Valenciennes, 1835), a euryhaline freshwater fish that lives in the Pampean inland waters and has potential economic relevance.
AimsTo identify two fungi isolated from O. bonariensis eggs exposed to saline conditions and to characterize their pathogenicity and tolerance to sodium chloride solutions.
MethodsThe isolates were identified by morphological features, and a preliminar phylogenetic analysis using sequences of translation elongation factor 1-alpha (EF-1α) and calmodulin (CAM) was performed. Koch's postulates were tested to identify the causative agent of fungal infection. The influence of NaCl on the fungal growth was evaluated in in vitro assays.
ResultsThe isolates LPSC 1001 and 1002 were identified as representatives of the genus Fusarium, and belonging to the Fusarium incarnatum-Fusarium equiseti species complex (FIESC) and the Fusarium solani species complex (FSSC), respectively. Histological observations on eggs exposed in vitro to both isolates in infectivity assays confirmed the ability of the fungal isolates to penetrate to egg's chorionic membrane, leading to the death of embryos. Increasing NaCl concentration in the culture medium reduced the growth of the isolates LPSC 1001 and 1002, being completely inhibited at 160 and 120g/l NaCl respectively.
ConclusionsThe isolates LPSC 1001 (FIESC) and 1002 (FSSC) were identified as fungal pathogens to O. bonariensis eggs. The use of NaCl solutions as antifungal treatment was not effective to control the infection with these strains.
Diversas especies de hongos pueden representar un riesgo importante para los embriones de Odontesthes bonariensis (Cuvier & Valenciennes 1835), un pez de agua dulce eurihalino que vive en las aguas interiores pampeanas y tiene una relevancia económica potencial.
ObjetivosIdentificar dos especies de hongos de huevos de O. bonariensis expuestos a condiciones salinas y caracterizar su patogenicidad y tolerancia a las soluciones de cloruro de sodio.
MétodosSe identificaron los aislamientos por sus características morfológicas, y se realizó un análisis filogenético preliminar utilizando secuencias de traslación del factor 1 alfa elongación (EF- 1α) y calmodulina (CAM). Se llevaron a cabo los postulados de Koch para identificar el agente causante de la infección fúngica. La influencia de NaCl sobre el crecimiento fúngico se evaluó en ensayos in vitro.
ResultadosSe identificaron los aislamientos LPSC 1001 y 1002 como representantes del género Fusarium, y pertenecientes al complejo de especies Fusarium incarnatum-Fusarium equiseti (FIESC) y al complejo de especies de Fusarium solani (FSSC), respectivamente. Las observaciones histológicas en los huevos expuestos in vitro a ambos aislamientos en los ensayos de infectividad confirmaron la capacidad de estos para penetrar en la membrana coriónica, lo que condujo a la muerte de los embriones. El aumento de la concentración de NaCl en el medio de cultivo redujo el crecimiento de los aislamientos LPSC 1001 y 1002, quedando completamente inhibidos a 160 y 120g/l de NaCl, respectivamente.
ConclusionesSe identificaron los aislamientos LPSC 1001 (FIESC) y 1002 (FSSC) como hongos patógenos para los huevos de O. bonariensis; el uso de soluciones de NaCl como tratamiento antifúngico no resultó eficaz para el control de la infección con estas cepas.
The Argentinian silverside, or “pejerrey”, Odontesthes bonariensis (Cuvier & Valenciennes 1835) is an atherinid fish that inhabits rivers and lakes located in the southern sector of La Plata basin. The silverside is considered one of the most promising fish species of Argentina for the development of its intensive culture.40 The great potential of this species leads to the need of having a better knowledge of its biology, as well as of its pathogens, for implementing an adequate management of intensive culture systems, which includes a control diseases program.
Fungal diseases can be a serious threat to culture fish species. Several fungal species and water molds can pose a substantial risk to fish embryos.35,45 In combination with environmental stressors, the pathogens can cause severe declines in fish populations. Infected eggs and larvae act as a dissemination source, and the infection spreads quickly. Spread of the diseases can occur via spores that are released to the aquatic environment adhering to healthy individuals, or by mycelial extension from infected to contiguous healthy embryos.35
Although several authors 21,45 point to the order of the Saprolegniales (Oomycetes) as the most important pathogens in freshwater fish and their eggs, Rand 35 highlights the role of anamorphic fungi, due to their wide distribution and adaptability to different environmental conditions. Fungal fish pathogens list include species of the genera Aureobasidium, Cladosporium, Exophiala, Hormoconis, Ochroconis, Paecilomyces, Penicillium, Phoma and Verticillium, which have been reported to cause mortality in fish of the orders Salmoniformes, Perciformes, Siluriformes, Squaliformes and Pleuronectiformes.9,11,13,20,39,42
Fusarium is a wide and taxonomically complex genus. Recent molecular phylogenetic studies have revealed that the most commonly reported pathogens are part of several species complexes.24,48 The wide distribution and diverse ecology of the Fusarium species reflect its metabolic diversity, since these fungi are abundant in soil, as saprophytes, mutualists, and parasites on plants and animals, in different habitats.2,18,36,38 In fish, Fusarium culmorum,14Fusarium oxysporum,13Fusarium solani5,29,39 and other Fusarium species2 have been found as cutaneous or visceral pathogens, capable of causing sometimes lethal infections.
The aim of the present work was to isolate and characterize by traditional morphology and molecular methodologies the fungal biota belonging to genus Fusarium associated with O. bonariensis eggs exposed to saline conditions. Furthermore, we prove the pathogenicity of the isolates by following Koch's postulates and examine the etiological agents’ tolerance to NaCl as potential control measure.
Materials and methodsFungal isolation and identificationEggs of O. bonariensis were obtained from the Chascomús Fisheries Station of the Ministerio de Agricultura de la Provincia de Buenos Aires, Argentina and incubated as described in Pacheco Marino & Salibián31 with the addition of NaCl (Anedra) in a range of 10–30g/l. During the incubation, the infected eggs were collected aseptically and washed with an antibiotic solution (chloramphenicol and streptomycin (Sigma) at a concentration, of 2.5 and 5‰ (w/v), respectively). Eggs were rinsed with sterile distilled water and then used as fungal inoculum in Petri plates containing 20ml of potato glucose agar (PGA),17 at a final concentration of 0.006 and 0.012‰ (w/v) of chloramphenicol and streptomycin.
The identification of the isolates was based on cultural and morphological features following the original taxonomic papers,2,7,18 including the culture on potato sucrose agar (PSA), carnation leaf agar (CLA) and oatmeal agar (OMA),16 and synthetic low nutrient agar (SNA),23 at 25°C and darkness during 2–5 weeks.
DNA isolation, sequencing and phylogenetic analysisFor the molecular identification genomic DNA was extracted using the CTAB method described by Stenglein and Balatti.41 PCRs were performed to amplify partial sequences of the elongation factor 1-alpha (EF-1α) and calmodulin (CAM) genes according to O’Donnell et al.26,27 Amplifications were carried out in a XP thermal cycler (Bioer Technology Co.). The successful amplifications were confirmed by gel electrophoresis. PCR products were purified with the aid of a PureLink PCR purification kit (Invitrogen). DNA sequencing from both the sense and antisense ends of the fragments was carried out using Big Dye Terminator version 3.1 Cycle Sequencing Ready Reaction Kit (Applied Biosystems) in an Applied Biosystems Sequencer (ABI/Hitachi Genetic Analyzer 3130). The sequences generated in this study were deposited into GenBank under the following accession numbers: JN614903.1, JN614904.1, HQ201752.1 and HQ201753.1.
For the phylogenetic reconstruction we used the dataset of O’Donnell et al.,27,28 though the Fusarium tricinctum (FTSC) and the Fusarium sambucinum (FSAMSC) complexes were excluded due to non-availability of sequences of calmoduline gene for these fungi (see Supplementary S1 Table 1: Data of sequences used in the phylogenetc analysis in Appendix A). All the dataset sequences for each single molecular marker were aligned using MAFFT v.6 (http://mafft.cbrc.jp/alignment/server/).15 The nexus files used in this analysis were uploaded to TreeBASE (study accession number 14120).
Phylogenetic analysis was conducted based on the aligned sequences for each of the data set using PAUP v. 4.0b10.44 All the alignment gaps were treated as missing data. Each data set was initially analyzed based on different optimality criteria: maximum parsimony (MP), neighbor-joining (NJ) and Bayesian Inference (BI). In the MP analysis the starting trees for branch swapping were obtained via a stepwise addition procedure. A heuristic search, with random sequence addition of 1000 replicates and tree-bisection-reconnection branch-swapping algorithm, was performed. The bootstrap consensus tree is inferred from 1000 replicates; branches corresponding to partitions reproduced in less than 50% bootstrap replicates are collapsed. Models of nucleotide substitution and parameters for phylogenetic reconstruction were selected by jModelTest version 0.1.112,33 and used to perform NJ and BI analysis (see models of nucleotide substitution and parameter related in Supplementary material S1B: Models of nucleotide substitution in Appendix A). Clade stability was assessed by 1000 bootstrap replicates with random sequence addition. Consistency index (CI) and retention index (RI) were calculated for the indication of the amount of homoplasy present in both MP and NJ analysis. Bayesian MCMC phylogenetic analysis using MrBayes 3.2.337 was performed using two simultaneous chains of 1.5×107 generations and a sample frequency of 300, for a total of 50,000 sample trees, and the prior parameters of the corresponding substitution model and distribution rate. To assess whether the tree topologies inferred from MP and BI analysis were more similar than expected by chance, the congruence index (Icong index) was used.6
Pathogenicity assaysA conidial suspension of each isolate was made according to Toledo et al. 46. Silverside eggs were exposed for 96h to 20ml of conidial suspensions of 1×106 to 1×109 conidia/ml of each fungal strain separately. Eggs not exposed to conidial suspensions were used as a control. Three trials with 10 individuals by replicate were carried out for each fungal isolate. Survival, mortality and infection rate (IR) were recorded at the end of the test, and median letal concentration (LC-50) was computed using the Trimmed Spearman-Karber 1.5 software program.
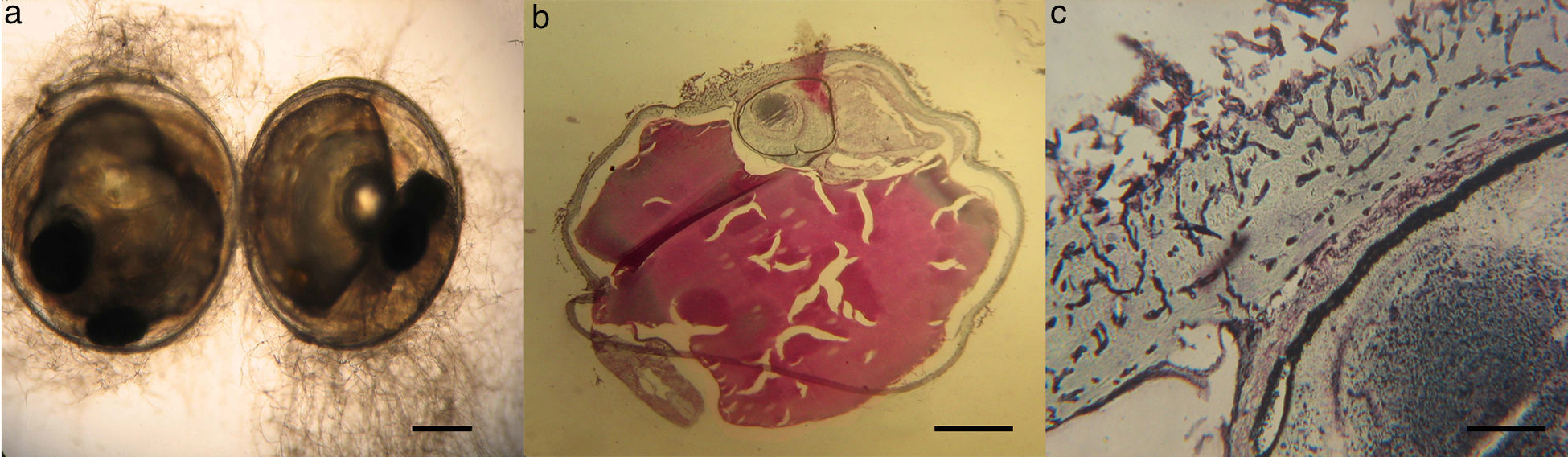
Histopathological examinationInfected and uninfected eggs were fixed by immersion in 10% phosphate-saline buffered formalin and were processed according to standard procedures. Sections (5μm) of each egg were stained with hematoxylin eosin (HE) and a silver methenamine staining (Grocott), as described by Prophet et al.,34 and examined by light microscopy.
Influence of NaCl on fungal growthThree sets of Petri dishes containing 20ml of Czapek's Dox medium17 with 6 different NaCl concentrations (0 – control–, 40, 80, 120, 160 and 200g/l) were used for each fungal strain. Plates were inoculated at the center, with a 3mm diameter plug cut from axenic and monosporic culture plates, and incubated at 25°C in darkness. Three days after the inoculation, the diameter of the colonies was measured every 24h in the same direction for two weeks, using a vernier caliper (accuracy=0.1mm); colonies were observed for an additional week to detect possible changes in the growth rate.
The inhibition rate was expressed as the percentage reduction in colony diameter compared with the control group (Czapek's Dox). The percentage of reduction was calculated as follows:
Statistical analysisIn the pathogenicity assays, the differences in survival and infection rates between treatments and the control group were analyzed by a Mann–Whitney U test. Growth rate was calculated from the linear regressions. Pearson's correlation coefficient was used to determine the linear equation. Variance was analyzed using Kruskal Wallis non-parametric tests to determine differences between growth rates of each species under different treatments. Comparisons between experimental and control values were performed by a posteriori Tukey multiple comparisons test. Statistical calculations were performed using the software program XL Stat7.5.
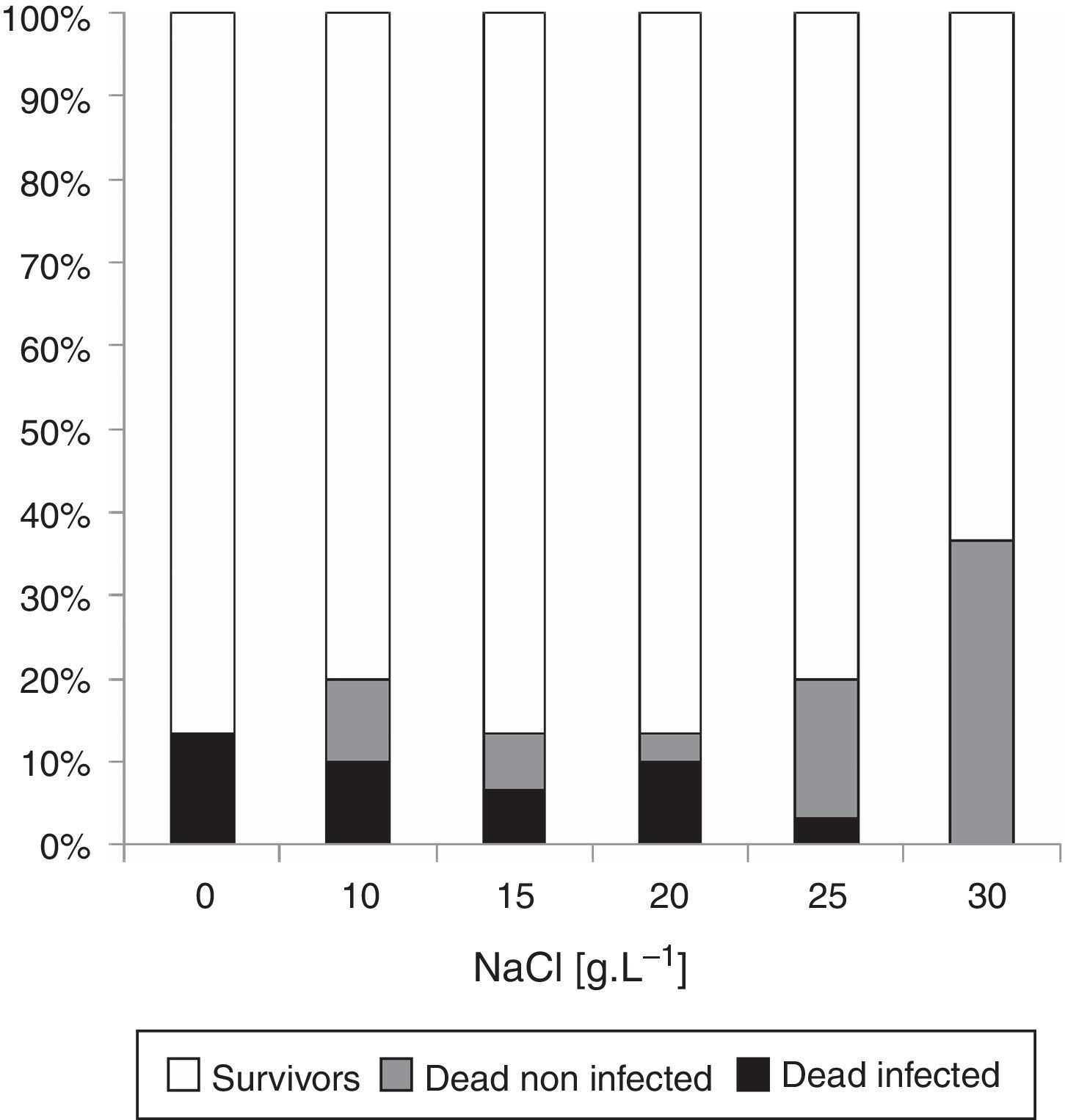
ResultsFungal isolation and morphological identificationA 13.3% of the non-exposed to NaCl eggs died due to fungal infections, and a mortality rate of 3.3–10% was found in eggs exposed to NaCl at a range of concentrations between 10 and 25g/l (Fig. 1). Two morphotypes isolated were morphologically identified as Fusarium incarnatum (Desm.) Sacc. (1986) and Fusarium solani (Mart.) Sacc. (see Supplementary S2A: Taxonomic descriptions, and S3: Illustrations in Appendix A) and were deposited in the strain-culture collection of Spegazzini Institute Herbarium (LPSC) with accession codes LPSC 1001 and 1002, respectively.
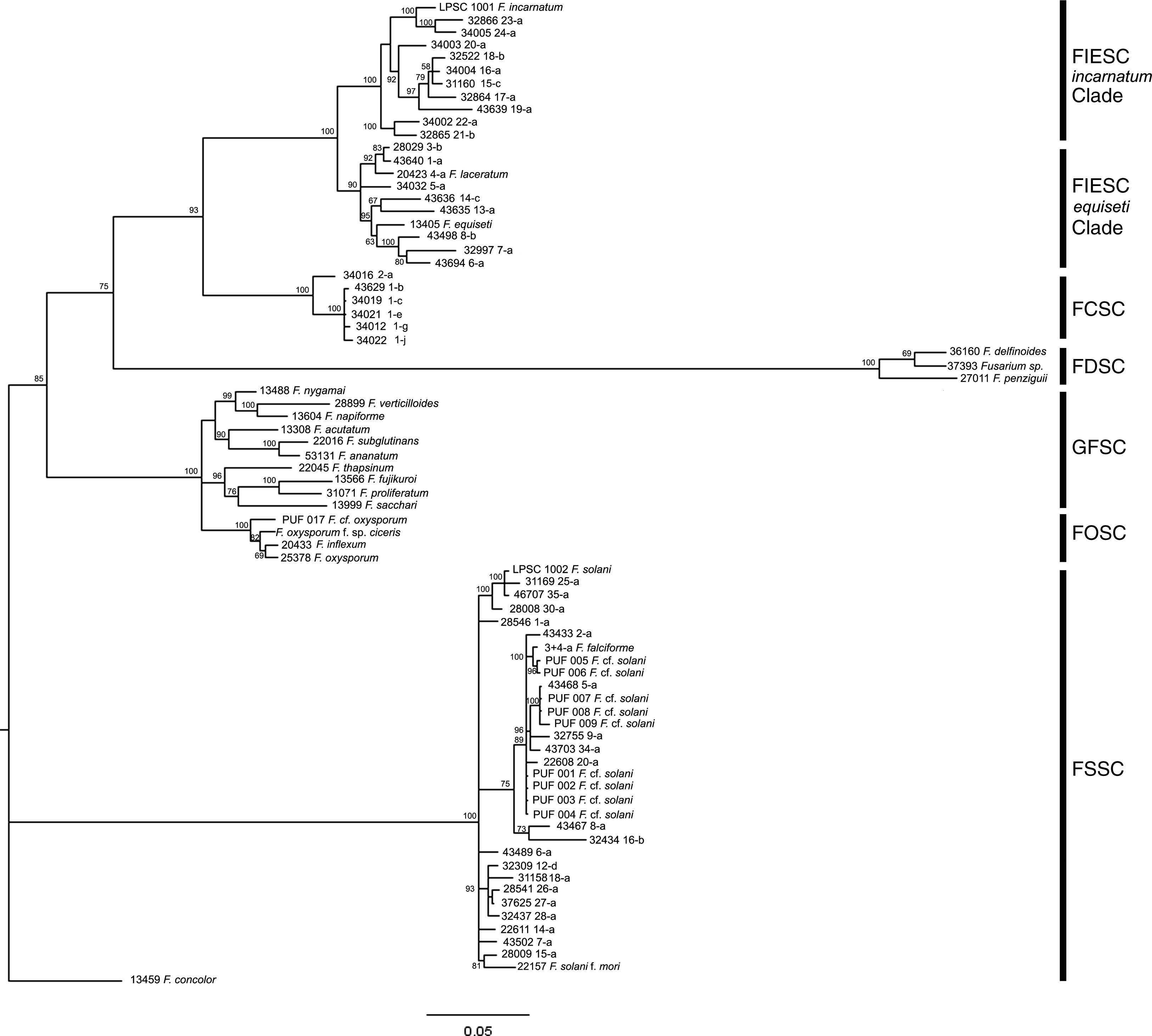
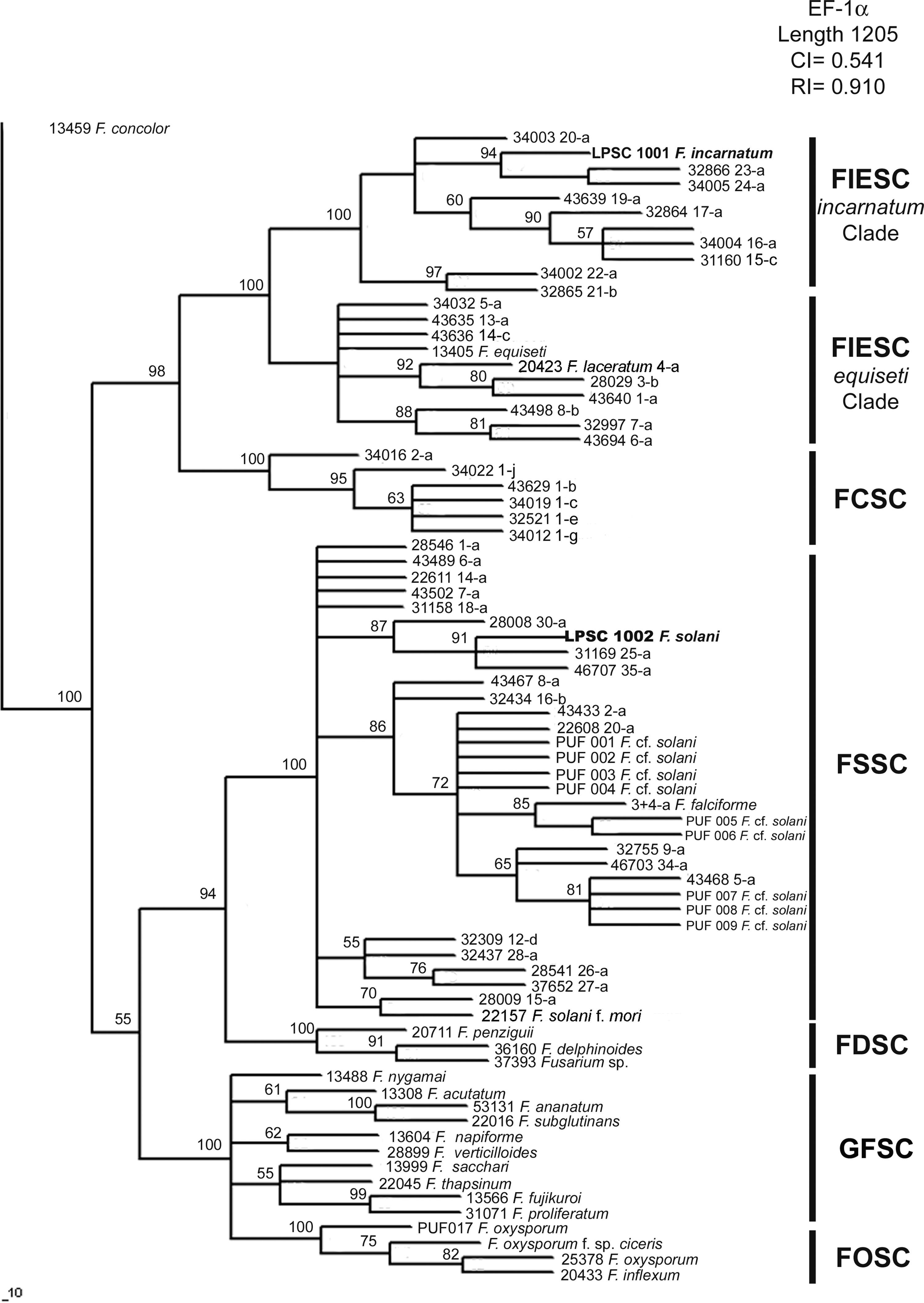
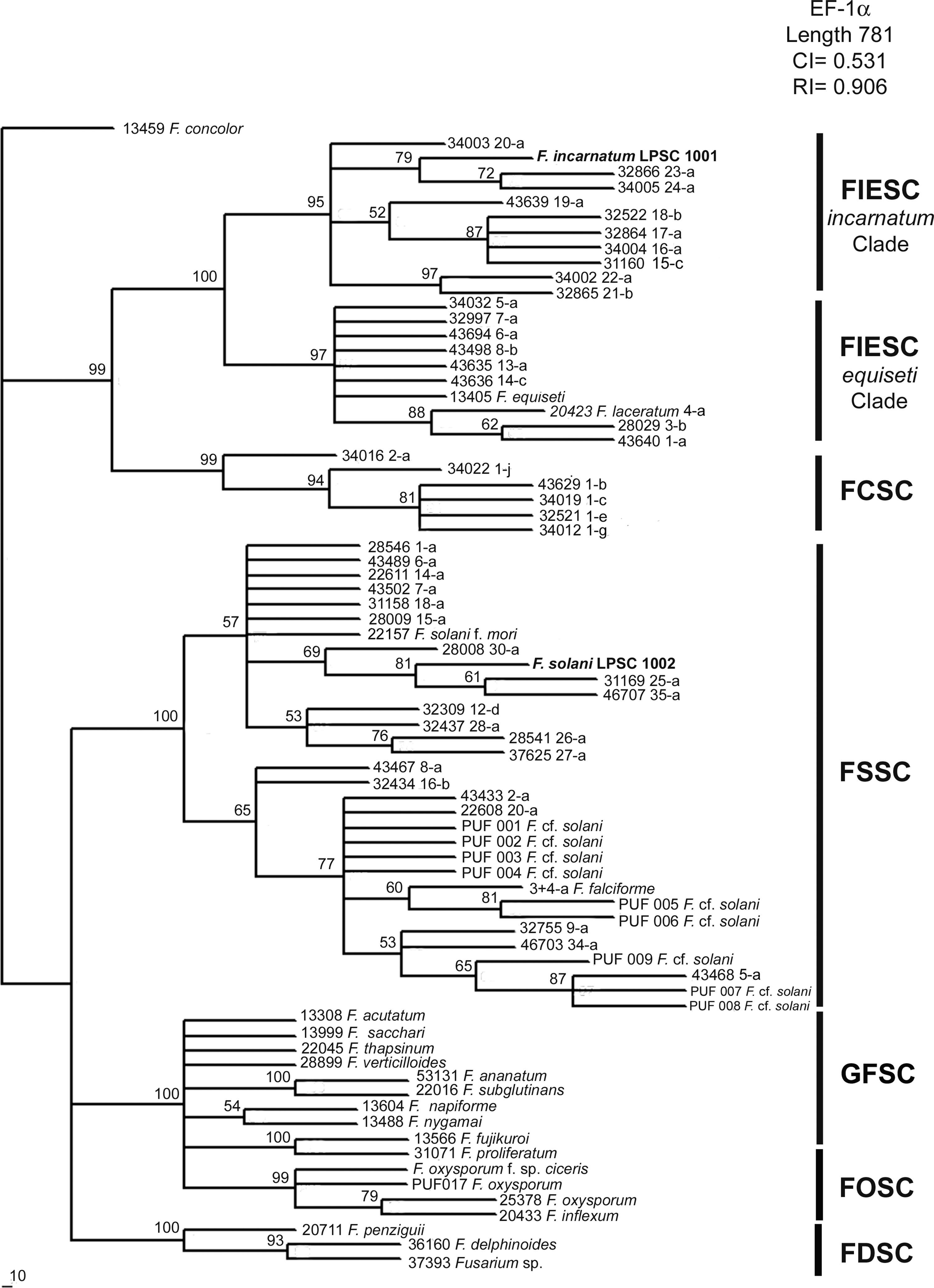
Molecular identification and phylogenetic analysisThe amplified products of isolates LPSC 1001 and 1002 had 699 and 653bp for the elongation factor 1-alpha (GenBank Accession Number: HQ201752-3), and 710 and 775bp for calmodulin (GenBank Accession Number: JN614903-4), respectively. A phylogenetic analysis based on MP, NJ and BI criteria was performed using separate data sets of the two loci and the Fusarium concolor strain 13459 as an outgroup. Bayesian analysis of EF-1α locus resolved six well supported clades (with a support value of 100%), and the distribution of the monophyletic clades was according to the scheme proposed by O’Donnell et al.27 (Fig. 2). On the latter, the isolate LPSC 1001, morphologically identified as Fusarium incarnatum, was indeed located within the FIESC clade and specifically within the F. incarnatum subclade, with the haplotypes 23 and 24 (a support value of 100%). The isolate LPSC 1002 falls within FSSC clade, and was closely related to the haplotypes FSSC 25-a, 30-a, and 35-a (support value of 100%). We found similar topologies in the trees generated when this locus was analyzed under MP and NJ criteria (see MP and NJ trees of EF-1α locus in Supplementary S4 and S6, respectively, in Appendix A). The statistical support of this observation is provided by the Congruence Index (I cong) obtained from the comparative analysis of the tree topologies inferred from BI and MP analysis (4.04, p=2.96×10−28). Additionally, the MP analysis showed that 335 were parsimony informative on a total of 781.
Bayesian MCMC analysis of the single data set from EF1-α. Only Bayesian posterior-probabilities (100×) above 50 are shown. Fusarium concolor isolate (13459) was used as an outgroup for the phylogram. FIESC: Fusarium equiseti-incarnatum species complex, FCSC Fusarium chlamydosporum species complex, FDSC: Fusarium dimerum species complex, FFSC: Fusarium fujikuroi species complex, FOSC: Fusarium oxysporum species complex, FSSC: Fusarium solani species complex.
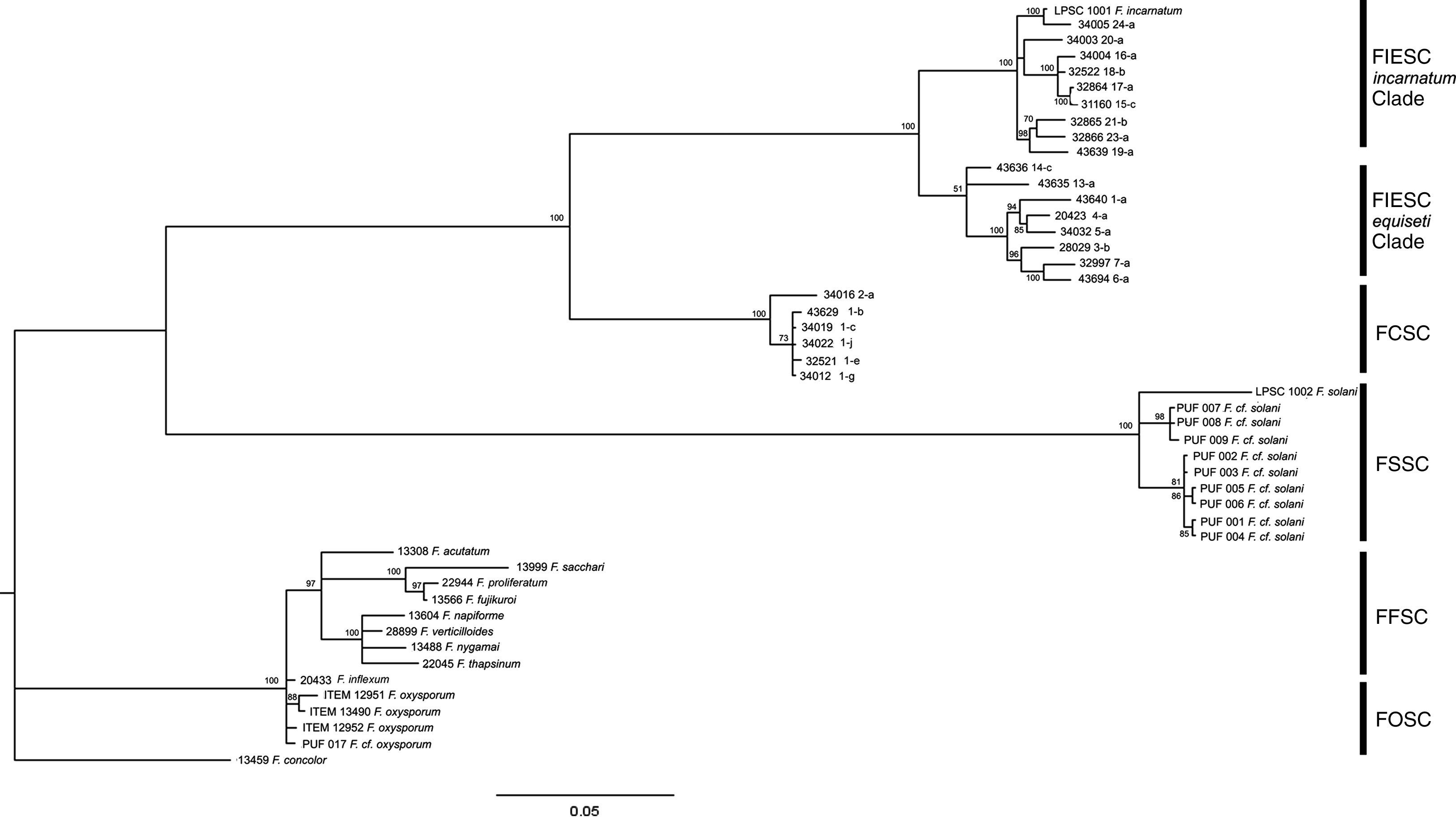
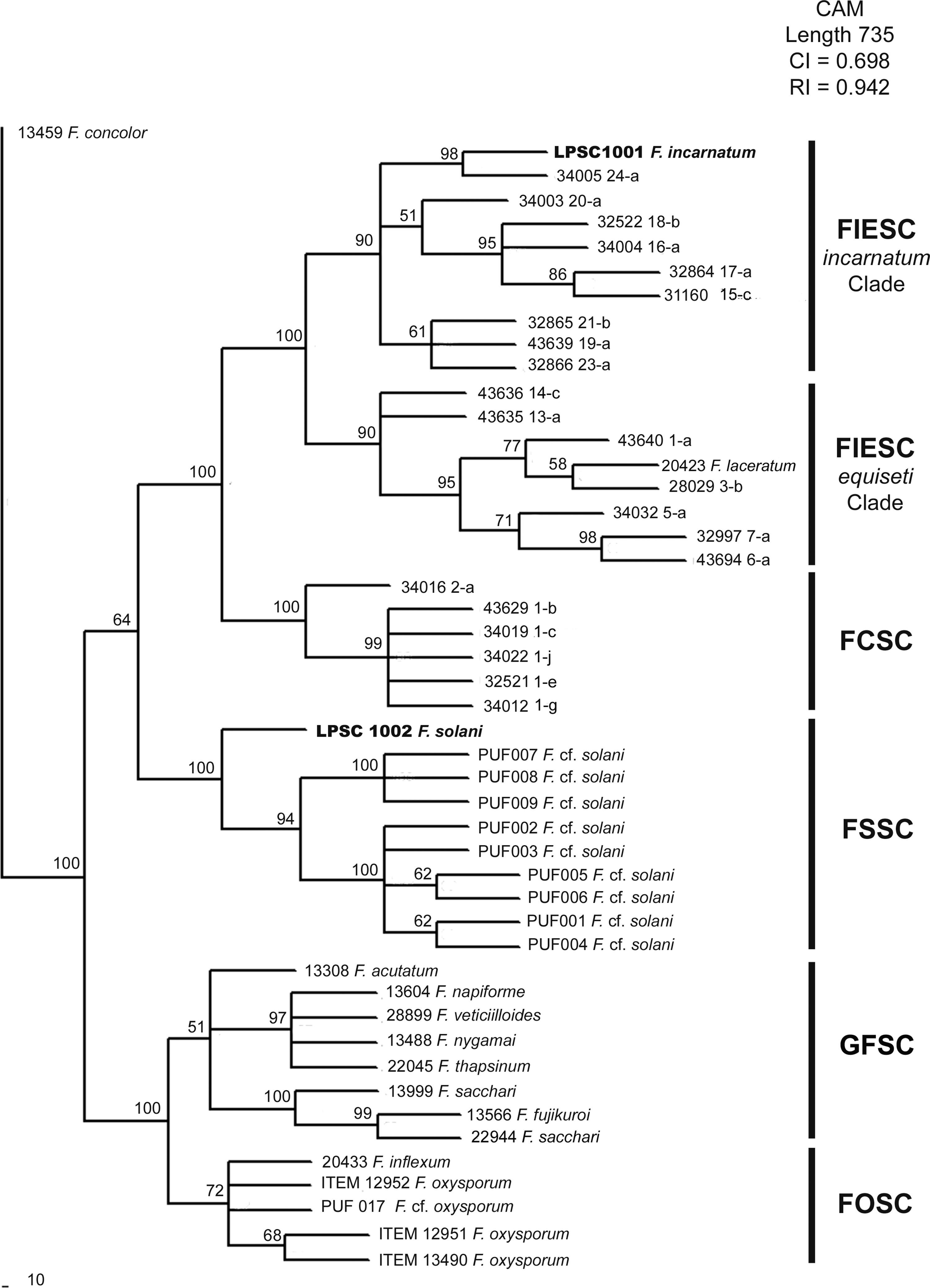
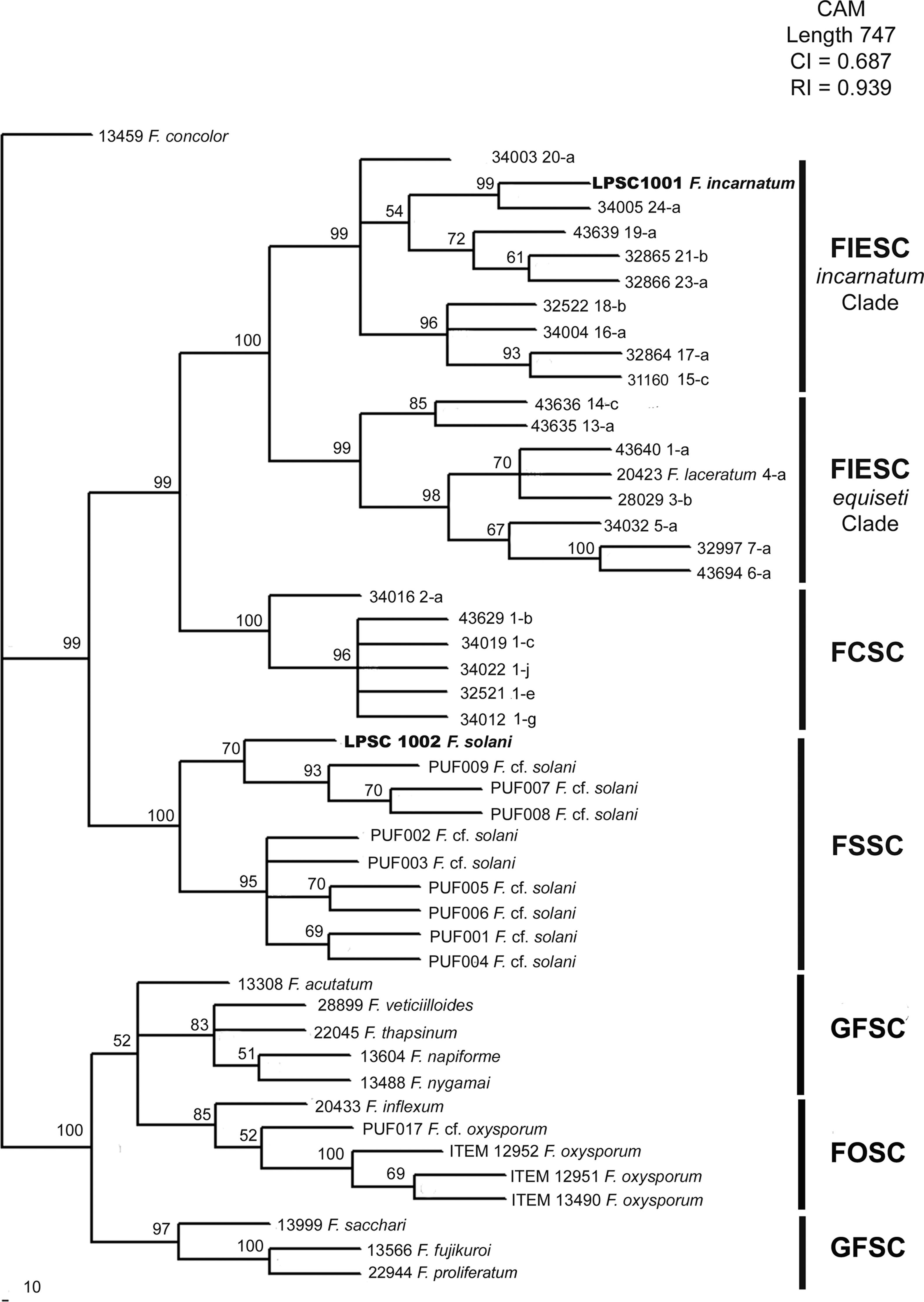
When the phylogenetic relationship using a BI analysis was based on available data from CAM locus, five clades well supported (a support value of 100%) were resolved (Fig. 3). In this case, LPSC 1001 was closely associated with haplotype 24 of Fusarium incarnatum-equiseti species complexes (FIESC) clade (incarnatum subclade; with a support value of 100%), and LPSC 1002 was related with the strains PUF 001 to 009 belonging to Fusarium solani species complex (FSSC) clade (a support value of 100%). Similar tree topologies were also found in the analysis performed under MP and NJ criteria (see MP and NJ trees of CAM locus in Supplementary S5 and S7, respectively, in Appendix A). The MP analysis revealed 301 out of 841 characters were parsimony informative, and the statistical comparison of the tree topologies obtained with MP and BI criteria revealed that these were more similar than expected by chance (I cong: 4.24; p=3.15×10−26). Finally, based on the rate between parsimony informative characters (PIC) and bp, the EF-1α locus was the most phylogenetically informative since it showed the greatest nucleotide diversity (PIC/bp=0.4289) in relation to that obtained for CAM locus (0.3579).
Bayesian MCMC analysis of the single data set from CAM. Only Bayesian posterior-probabilities (100×) above 50 are shown. Fusarium concolor isolate (13459) was used as an outgroup for the phylogram. FIESC: Fusarium equiseti-incarnatum species complex, FCSC Fusarium chlamydosporum species complex, FSSC: Fusarium solani species complex, FFSC: Fusarium fujikuroi species complex, FOSC: Fusarium oxysporum species complex.
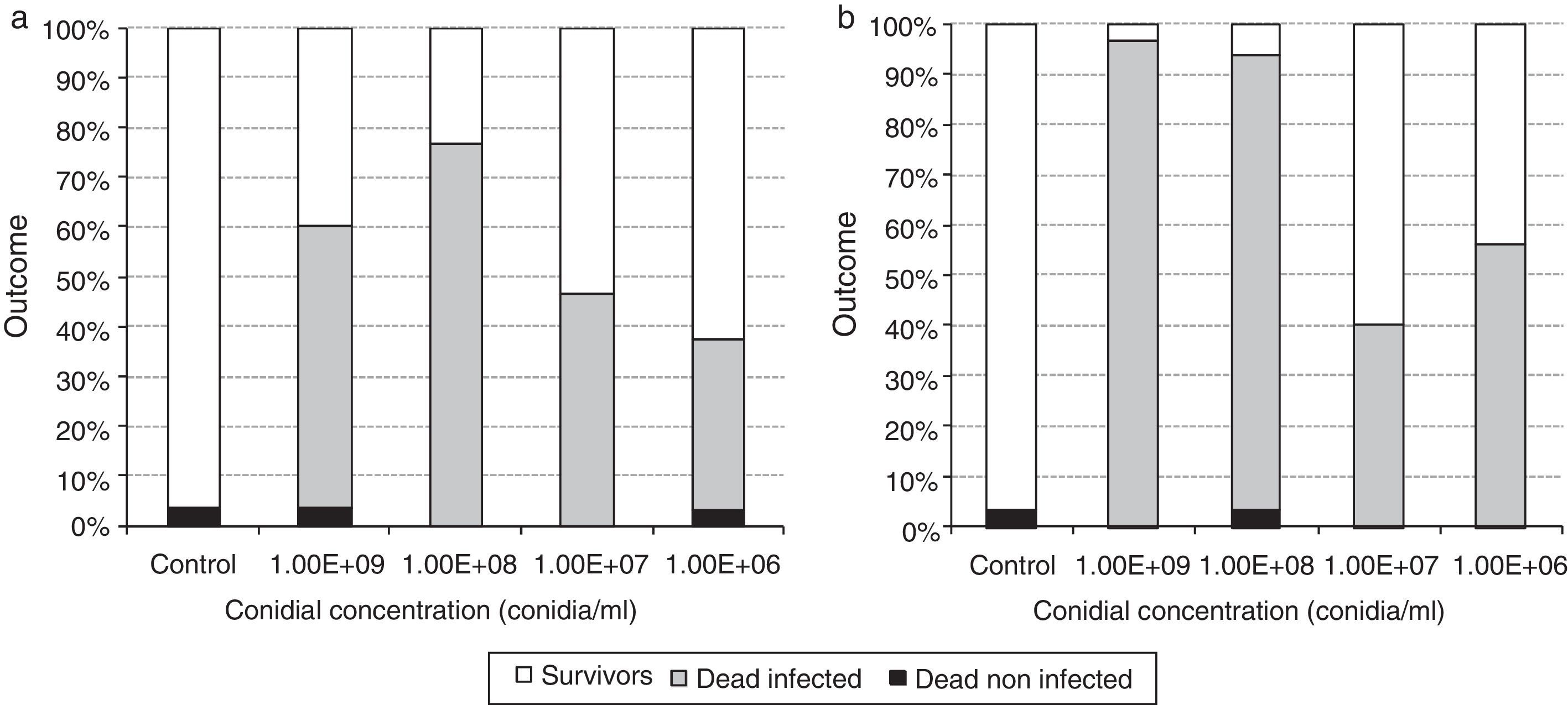
Both isolates were able to infect silverside eggs. Not fungal infections were recorded in the control groups, and mortality rates of non-infected eggs were lower than 3.3% for both strains under treatment as well as in the control groups. In the eggs exposed to 1×107 and 1×106 conidia/ml of LPSC 1001, and 1×107conidia/ml of LPSC 1002 the infection rate (IR) was lower than 50% (Fig. 4).
Only when the eggs were exposed to 1×109conidia/ml, the IR of the isolate LPSC 1002 (96.7% of IR) was higher than LPSC 1001 (56.7%) (p=0.043). No significant differences were found. When comparing the IR of different doses of isolate LPSC 1001; however, the IR obtained in eggs exposed to 1×109conidia/ml of the isolate LPSC 1002 was significantly higher than the ones recorded in 1×108 (p=0.002) 1×107 (p=0.043) and 1×106 (p=0.002) conidia/ml. For both isolates (LPSC 1001 and 1002), the IR of the eggs exposed to 1×108 was higher than in those exposed to 1×107 (p=0.008 and 0.012). On the other hand, the mean lethal concentrations were 1.25×107 (CL 95%= 3.98×106–3.95×107) and 1.09×107 (CL 95%= 5.50×106–2.16×107) to LPSC 1001 and 1002 respectively.
Histopathological studyMicroscopical observations of infected eggs from pathogenicity tests were done. A tiny and inconspicuous mycelial development was found on infected eggs (Fig. 5a). Using Grocott-stained histological sections of these eggs, septate hyphae in the corionic membrane were also detected (Figs. 5b and c).
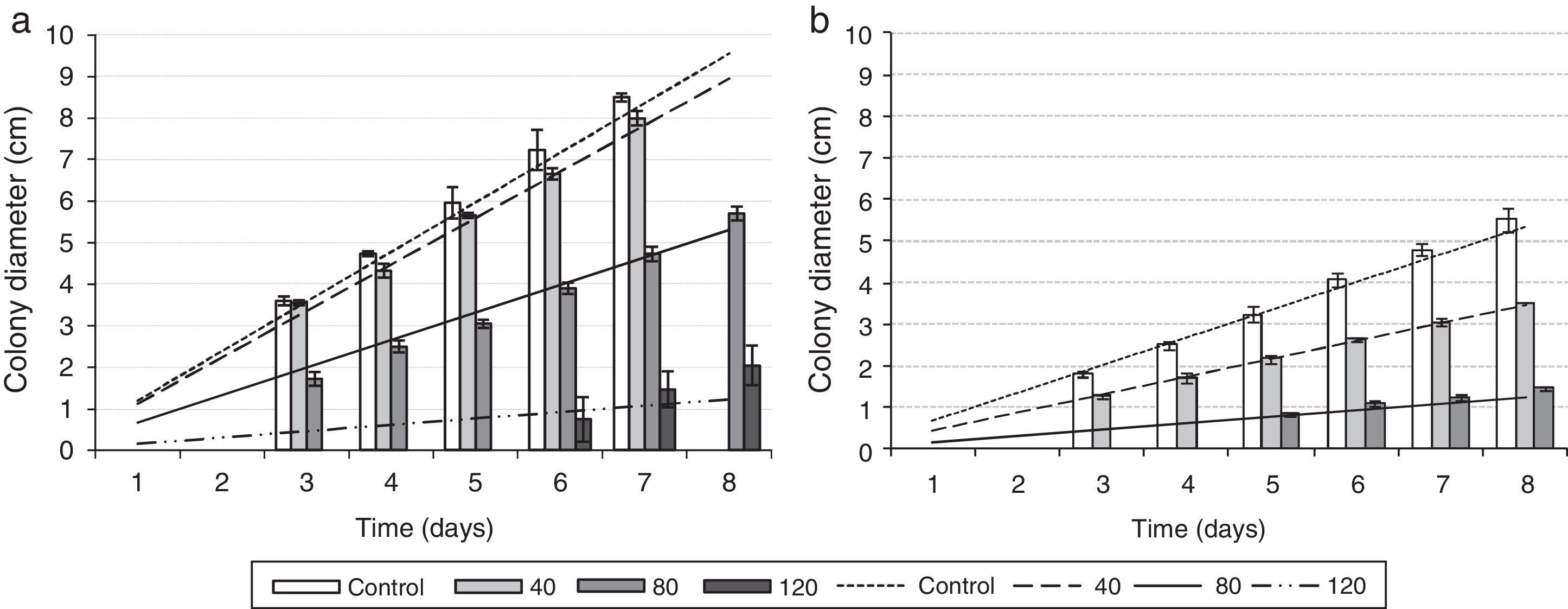
Influence of NaCl on the in vitro fungal growthThe effect of NaCl on the growth of both isolates is shown in Fig. 6. The growth rate of LPSC 1001 is greater than that of LPSC 1002 on basal medium, while the latter is more susceptible to NaCl. Growth was completely inhibited at 120g/l and 160g/l for both LPSC 1001 and LPSC 1002, respectively.
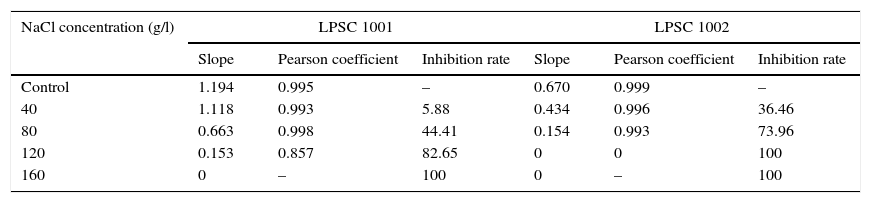
The slope values obtained in the linear regression, the corresponding Pearson's coefficient, and the inhibition rate for Fusarium isolates are shown in Supplementary material S8 in Appendix A (Table 1). In general, the higher NaCl concentrations exhibited inhibitory effect on the growth rates. The analysis of variance showed, compared with the control, significant differences in the growth rate of the isolate LPSC 1001 when exposed to 80 and 120g/l NaCl. Significant differences were also found in the isolate LPSC 1002 growth rate between treated and control groups.
Slope of the linear regression of the colony diameters (growth rate), Pearson correlation coefficient values and growth inhibition rate for the isolates LPSC 1001 and LPSC 1002 exposed to different concentrations of NaCl for 8 days. Data correspond to the mean values of Fig.6.
| NaCl concentration (g/l) | LPSC 1001 | LPSC 1002 | ||||
|---|---|---|---|---|---|---|
| Slope | Pearson coefficient | Inhibition rate | Slope | Pearson coefficient | Inhibition rate | |
| Control | 1.194 | 0.995 | – | 0.670 | 0.999 | – |
| 40 | 1.118 | 0.993 | 5.88 | 0.434 | 0.996 | 36.46 |
| 80 | 0.663 | 0.998 | 44.41 | 0.154 | 0.993 | 73.96 |
| 120 | 0.153 | 0.857 | 82.65 | 0 | 0 | 100 |
| 160 | 0 | – | 100 | 0 | – | 100 |
Two Fusarium isolates obtained from O. bonariensis eggs (LPSC 1001 and 1002) were characterized morphologically, physiologically and with molecular biology. The morphology of isolates LPSC 1001 and 1002 was similar to that of F. incarnatum (formerly F. semitectum) and F. solani respectively, as described by Booth,2 Leslie and Summerell,18 Nelson et al.,22 and Sutton et al.43 Traditionally the systematic of the Fusarium genus was based on morphological species concept,49 but in recent years molecular phylogenetic analyses have been applied to examine the taxonomy of this genus has and have led to a new taxonomic system based on the phylogenetic species concept.24,48 Studies based on genealogical concordance of multilocus DNA sequence data revealed an extensive cryptic speciation across the phylogenetic breadth of the Fusarium genus, especially within the F. solani species complex (FSSC) and F. incarnatum-F. equiseti species complexes (FIESC).27,28 Recently, O’Donell et al.28 published an Internet-accessible database for identifying novel etiologic agents of fusarioses within a precise phylogenetic framework. This is based on the multilocus analysis of partial sequences from three nuclear genes: translation elongation factor (EF-1α), the largest subunit of RNA polymerase (RPB1), and the second largest subunit of RNA polymerase (RPB2). Their use for identifying isolates to the species level using a single DNA marker was anticipated by their authors.28
Since molecular identification of LPSC 1001 and 1002 was based in the phylogenetic analysis of two genetic markers, EF-1α and CAM, our analysis was done using additional sequences of Fusarium species not included in O’Donnell's dataset. The phylogenetic analysis of EF-1α sequences under NJ, MP and BI criteria resolved six well supported clades with a similar topology to that reported by O’Donnell et al.28 In MP analysis the statistics values of more parsimonious tree were similar to that reported by O’Donnell et al.,28 being the MP tree length of 1205 and 1183 steps; the rate PIC/bp=0.45 and 0.43; the consistency index CI=0.54 and 0.47; and the retention indexes 0.91 and 0.87, respectively. Finally, all phylogenetic criteria showed that the isolates LPSC 1001 and 1002 are placed in the monophyletic groups FIESC and FSSC respectively. The phylogenetic analysis of CAM sequences resolved five monophyletic clades well supported, with a similar distribution than that observed in EF-1α analysis. The most parsimonious tree was 1183 steps in length, other parameters as rate PIC/bp=0.36, IC=0.70 and IR=0.94 indicated that the data set used in this analysis were more informative than those used by O’Donnell et al. 27. Finally, the topological comparison of trees obtained in BI and MP phylogenetic analyses of each single data set exhibited high level of topological concordance with similar bootstrapping levels of clade support. Considering that the phylogenetic analysis shows that isolates LPSC 1001 and 1002 shared a strong relationship with clinical isolates belonging to FIESC and FSSC respectively, and since both are considered the two main groups of pathogenic representatives of Fusarium genus, a potential risk of zoonoses cannot be discarded.25,43
Previously, Fusarium strains have been described as etiological agents of infections in carp (Cyprinus carpio) and red sea bream (Pagrus sp.).18,19 Ferguson10 isolated a F. solani strain from a mycetoma of desert pupfish (Cyprinodon macularius). Furthermore, fusariosis in sharks in marine habitats has been reported by Rand,2 Smith et al.,39 Crow et al.5 and Marancik et al.20 One case of infection by F. incarnatum was reported by Khoa et al. 16 in a black tiger shrimp (Penaeus monodon). Although these reports show the diversity of hosts and environments from which they were isolated, these data only correspond to diseases in adult hosts; therefore, our data extend the host range of fusariosis to teleost fish eggs and gives evidence that pathogenic ability of some fungi might be not suppressed only by increasing osmotic pressure. As Fusarium species are commonly observed in soil and in organic matter from fresh and brackish water, it is highly probable that the isolates LPSC 1001 and 1002 are part of the aquatic mycobiota associated to culture systems of O. bonariensis. Environmental conditions such as the presence of suspended organic matter, the increase of temperature, and overcrowding can contribute to fungal proliferation and therefore increase the infective burden in aquatic environment, making possible the colonization of a susceptible host. However, such events could be overlooked because the infections caused by these fungi generally have moderate mortality rates and their growth on eggs is inconspicuous compared to that caused by Saprolegniales.30
Sodium chloride is widely used in aquaculture as a therapeutic chemical to control infectious diseases. Application of salt baths (1–3%) in freshwater fish and amphibians is extensively used to control and treat ectoparasites and other pathogens as Ichthyophthirius multifiliis, Edwardsiella spp., and the causative agent of saprolegniasis.47 Since O. bonariensis is a euryhaline species able to live in habitat with up to approximately 21g/l salinity,40 the use of sodium chloride as an efficient microbicide is feasible; however, we found that O. bonariensis eggs exposed to a range of NaCl concentrations of 10–25g/l were infected by two strains of the Fusarium genus. A decrease in the mortality by the fungal infection was observed when increasing the NaCl concentration, until the mortality observed was only due to a deleterious effect by NaCl on eggs in concentrations up 20g/l; similar findings were reported by Pacheco Marino and Salibián.31
Many fungi isolated from extreme environments possess diverse mechanisms that allow them to counterbalance the effects of NaCl and other salts, though their growth may also be reduced when the salt concentration in the medium increases, reason why the NaCl may be applied as a fungistatic agent.3,47 Deleterious effects of the increase in the salinity can have a direct response in mycelia, causing osmotic stress or acting as a toxic component interfering with membrane stability and/or enzymatic activity.1 Although only a few fungi are considered as obligate halophiles, some others may grow better in the presence of salt, thus having a competitive advantage over less salt-tolerant species.50 The capacity to adapt metabolically to environments with low osmotic potentials has been observed in some members of Fusarium genus isolated from saline environments.32 Other studies on Fusarium spp. showed that conidial germination in F. solani and F. equiseti was positively affected by the decrease of osmotic potential of the aqueous medium.32 El-Abyad et al.8 also found that the addition of salt to the media increased mycelial growth and/or conidial germination of F. avenaceum, F. culmorum, F. diversisporum, F. equiseti and F. oxysporum f. sp.betae, but reduced also the growth rate in F. verticillioides. In contrast to our results, typical water molds that commonly infect eggs and adults of fish such as the members of the Saprolegniaceae family are not tolerant to saline conditions.4
In order to corroborate the halotolerance of the isolates LPSC 1001 and LPSC 1002, their response to a range of 40–160g/l NaCl was evaluated. The inhibiting NaCl concentrations for the in vitro growth of isolates LPSC 1001 and LPSC 1002 was 160 and 120g/l NaCl, respectively. The mean percentage of colony diameter reduction for LPCS 1001 was 82.65% with 120g/l of NaCl; in the case of LPSC 1002, with 80g/l of NaCl, this value was 73.96%. Similarly F. solani and other species of the Fusarium genus, recovered from hypersaline habitats (up to 5% NaCl), exhibited an increase (up 25%) of their in vitro tolerance to NaCl.19 This might be due, at least in part, to the availability of osmolytes synthesized by the fungus that alleviate the saline stress and, therefore, increases its osmotolerance.4In vitro assays performed by Palmero et al.32 showed that the colony diameter of F. equiseti exposed to NaCl concentrations was similar to that observed in LPSC 1002, but lesser than that from LPSC1001. In higher NaCl concentrations the growth of LPSC 1002 was higher than those reported for F. equiseti.
In conclusion, isolates LPSC 1001 and LPSC 1002 that were recovered from infected eggs of O. bonariensis were identified as representatives of the genus Fusarium based on classical (morphological) and molecular taxonomy. In this sense, our phylogenetic analysis confirmed the morphological identification, clustering the isolate LPSC 1001 in FIESC clade and the isolate LPSC 1002 in FSSC clade, belonging to the two main groups pathogens involved in human infections, within the Fusarium genus, according to O’Donnell et al.28 Pathogenicity assays showed that both isolates are capable to infect O. bonariensis eggs, while LPSC 1002 is more pathogenic than LPSC 1001. Furthermore, the response of the isolates LPSC 1001 and 1002 to NaCl, a chemical agent that could be used to control the occurrence of fungal infections, showed that a concentration of 160 and 120g/l of NaCl inhibits in vitro growth of each isolate respectively. Such information is key for the prevention and control of fusariosis in fish farms, although further studies are necessary to understand the physiological mechanisms of salt tolerance in both isolates and how their pathogenicity might be affected by NaCl concentration.
Conflict of interestThe authors declare no conflict of interest.
We thank the Fisheries Office of the Ministry of Production of the Buenos Aires Province for supplying test organisms. This research was partially supported by grants from Agencia Nacional de Promoción Científica y Técnica (PRH32/PICT 110) and from Comisión de Investigación Cientificas de la Provincia de Buenos Aires (CICPBA). Stenglein S.A. and Saparrat M.C.N. are researches from CONICET; Cabello M.N. and Salibián A. are researches from CICPBA. Dinolfo M.I. and Pacheco Marino are fellows from CONICET.