Candidemia is one of the most frequent opportunistic mycoses worldwide. Limited epidemiological studies in Latin America indicate that incidence rates are higher in this region than in the Northern Hemisphere. Diagnosis is often made late in the infection, affecting the initiation of antifungal therapy. A more scientific approach, based on specific parameters, for diagnosis and management of candidemia in Latin America is warranted.
‘Recommendations for the diagnosis and management of candidemia’ are a series of manuscripts that have been developed by members of the Latin America Invasive Mycosis Network. They aim to provide a set of best-evidence recommendations for the diagnosis and management of candidemia.
This publication, ‘Recommendations for the diagnosis of candidemia in Latin America’, was written to provide guidance to healthcare professionals on the diagnosis of candidemia, as well as on the usefulness and application of susceptibility testing in patients who have a confirmed diagnosis of candidemia.
Computerized searches of existing literature were performed by PubMed. The data were extensively reviewed and analyzed by members of the group. The group also met on two occasions to pose questions, discuss conflicting views, and deliberate on a series of management recommendations.
‘Recommendations for the diagnosis of candidemia in Latin America’ includes diagnostic methods used to detect candidemia, Candida species identification, and susceptibility testing. The availability of methods, their costs and treatment settings are considered.
This manuscript is the first of this series that deals with diagnosis and treatment of invasive candidiasis. Other publications in this series include: ‘Recommendations for the management of candidemia in adults in Latin America’, ‘Recommendations for the management of candidemia in children in Latin America’, and ‘Recommendations for the management of candidemia in neonates in Latin America’.
This article is also published in Spanish in this issue. It can be found inhttp://dx.doi.org/10.1016/j.riam.2013.05.009
La candidemia es una de las micosis oportunistas más frecuentes en todo el mundo. El escaso número de estudios epidemiológicos llevados a cabo en América Latina indica que las tasas de incidencia en esta región son mayores que las descritas en el hemisferio norte. A menudo el diagnóstico de la infección se establece tardíamente, lo que afecta al inicio del tratamiento antimicótico. Por esta razón, para el diagnóstico y el manejo de la candidemia está justificada una estrategia más científica, basada en parámetros específicos.
Recomendaciones para el diagnóstico y manejo de la candidemia constituye una serie de artículos preparados por miembros del grupo Latin America Invasive Mycosis Network. Su objetivo es proporcionar las mejores evidencias disponibles para el diagnóstico y el manejo de la candidemia.
El presente artículo, Recomendaciones para el diagnóstico de la candidemia en América Latina, ha sido redactado con el objetivo de brindar asesoramiento a los profesionales de la salud en lo referente al diagnóstico de la candidemia en pacientes que la padecen o están en riesgo de padecerla.
Mediante la base de datos PubMed se emprendió una búsqueda informatizada de los estudios publicados. Los miembros del grupo revisaron y analizaron exhaustivamente los datos. El grupo también se reunió en 2 ocasiones para proponer preguntas, abordar los puntos de vista conflictivos y deliberar sobre las recomendaciones terapéuticas.
Recomendaciones para el diagnóstico de la candidemia en América Latina incluye diversas recomendaciones sobre aspectos relacionados con los métodos diagnósticos para la detección de la candidemia, la identificación de las especies de Candida y las pruebas de sensibilidad antifúngica. Se expone también la disponibilidad de los métodos, sus costes y el marco en el que se aplican los tratamientos.
Este manuscrito es el primero de los artículos de esta serie dedicada al diagnóstico y tratamiento de las candidiasis invasoras. Otras publicaciones de esta serie son Recomendaciones para el diagnóstico de la candidemia en adultos en América Latina, Recomendaciones para el manejo de la candidemia en niños en América Latina, y Recomendaciones para el manejo de la candidemia en neonatos en América Latina.
Este artículo está publicado en español en este mismo número. Puede encontrarlo enhttp://dx.doi.org/10.1016/j.riam.2013.05.009
The diagnosis of invasive candidiasis is often made late in the course of infection, causing a delay in the initiation of antifungal therapy. Late diagnosis can be the result of non-specific clinical signs and symptoms,18 the variable accuracy of the available diagnostic tests,27 delayed time to grow Candida,19 blood cultures that are not positive until late-stage infection,18,25 inadequate sample size for blood culture,25 and false-negative results due to the use of antifungal agents in prophylaxis.25
Overcoming the challenges of diagnosis can improve time-to-treatment of Candida infection. Here, we provide recommendations for the diagnosis of invasive candidiasis, including methods for detecting infection, Candida species identification, and susceptibility testing.
Methods for detecting hematogenous Candida infectionDiagnosis can be achieved through various procedures, including conventional methods such as blood culture and serological assays for surrogate markers (1-3-β-d-glucan [BDG] and mannan antigen/antibody). Newer diagnostic methods include polymerase chain reaction (PCR) from blood or tissue biopsy, and enzyme-linked immunosorbent assay (ELISA) to detect an antigen produced by Candida.
Blood cultureBlood culture is by far the best method available for the diagnosis of candidemia; however, it is a lengthy procedure. This can affect timing of the initiation of antifungal therapy and contribute to increased morbidity and mortality.49 Time to detection can be influenced by Candida spp. In one retrospective study, mean times to positive culture for Candida albicans and Candida glabrata were 35.3±18.1h and 80.0±22.4h, respectively.19 The type of medium used for culture can also influence time to detection. A comparison of a selective fungal medium and a standard aerobic medium for the diagnosis of candidemia demonstrated a significant time saving when the fungal medium was used, with a mean time saving of 8.8h for C. albicans and 43.7h for C. glabrata.46
As mentioned previously, blood culture is the gold standard for the diagnosis of candidemia; however, its sensitivity is variable.51 In order to optimize this method, factors which are known to influence sensitivity (e.g. blood volume, number of cultures, and time to detection) should be carefully considered, as should the size of the inoculum, as well as type of culture bottles and media used. In one study with 15 Candida spp., higher inoculum sizes (10, 100, 1000 yeast cells per bottle) resulted in 70%, 73% and 79% detection of growth, respectively.27 A comparable rate of detection of Candida in aerobic and mycology bottles has been observed,8,27,45 which is higher than that in anaerobic bottles.8,27 The Working Group recommends the use of aerobic bottles for the diagnosis of candidemia by blood culture, as this is the standard procedure in every hospital.
Blood culture systemsA number of blood culture systems are available, which vary in sensitivity. These include conventional (manual), automated, and lysis-centrifugation methods. The use of biphasic media appears to be slightly better than or equivalent to conventional broth in yeast recovery.33 The sensitivity of automated blood culture systems, such as BacT/ALERT, BACTEC, and Isolator, is influenced by the type of culture bottle and media used.28,46 The lysis-centrifugation blood culture technique is the most sensitive method available. It has better diagnostic yields compared with conventional blood culture systems and reduces the time to result.38 However, lysis centrifugation is labor intensive, expensive, and has a high rate of contamination,18 and for this reason the Working Group considers automated blood culture systems the best culture option for the diagnosis of fungemia due to Candida spp.
SerologyA number of serological assays for the detection of candidemia are currently under development. Due to limited information on their diagnostic value, the Working Group cannot make a recommendation on their usage. These methods include the BDG assay,65 and mannan antigen and antibody assays.1,47 A recent systematic review of serological methods for the diagnosis of candidemia in critically ill patients showed a higher diagnostic sensitivity for the combined use of antigen and antibody.11
BDG is a structural component of Candida cell walls that shows promise as a biomarker for candidemia. Studies using the serum BDG assay for the diagnosis of candidemia reported assay sensitivities between 57% and 100% and specificities between 44% and 92%, much greater than blood culture methods.65 However, the BDG test is prone to false-positive results from β-glucan contamination by certain antibiotics and materials.25 Sources of β-glucan include dialysis membranes and filters made from cellulose, specific immunoglobulin products, cotton gauze, and sponges used in surgeries, as well as some drugs (including lentinan, crestin, scleroglucan, and schizophyllan).35,37,50,63 BDG has been extensively used in research and has been approved by regulatory agencies in both the USA and Europe for the diagnosis of Candida infection. However, owing to lack of availability and high costs, very few centers in Latin America are likely to be able to use this assay. Given its high negative predictive value, repeated negative BDG tests may help to rule out candidemia. Nevertheless, at present it is not clear how this test should be utilized at bedside, in terms of helping the clinician to decide whether or not an antifungal drug should be prescribed to a patient at risk of developing candidemia.
Screening of Candida deep-seated infectionsImagingEchocardiography has demonstrated to be an effective tool for the diagnosis of Candida-induced endocarditis,15 and magnetic resonance imaging (MRI) and computed tomography (CT) have shown to be effective non-invasive tools for the identification of hepatosplenic fungal disease.26,43 Although MRI and CT are sensitive in detecting the fungal microabscesses associated with chronic disseminated candidiasis (CDC), the requirement for repeat images during the course of an infection limits their usage. Computer-assisted ultrasonography has also been shown to successfully detect microabscesses in the liver and/or spleen and can be repeated as often as required, making it a useful method for the detection of CDC and for the follow-up of patients with CDC.36
BiopsyThe diagnosis of infections due to Candida spp. from particular specimens can be complicated. These organisms commonly colonize human skin and mucous membranes and, therefore, culture from specimens (sputum, tracheal secretions, and urine) from these sites is not necessarily a proof of invasion.33 Deep-organ Candida infection may require tissue biopsy to establish diagnosis, as in the case of Candida-induced pneumonia; however, tissue samples may contain few organisms, resulting in negative culture.64 CDC is diagnosed through identification of fungal structures under the microscope, or fungal growth from biopsy materials, as blood cultures are positive in only <20% of CDC patients.43 However, Candida may not always be detected upon biopsy, and a biopsy may not be feasible in some patients owing to the risk of complications.43 In at-risk patients who develop skin lesions, biopsy is utilized for the confirmation of diagnosis of Candida pneumonia via the identification of fungal elements.
New perspectivesRecently an ELISA was developed for the detection of a 65kDa antigen produced by C. albicans, Candida tropicalis and Candida parapsilosis. In one study, this novel diagnostic test detected the 65kDa antigen in 80% of patients with candidemia.4 The advent of PCR for the detection of fungal nucleic acids has provided a faster and more accurate alternative to blood culture for the detection of Candida in blood samples.41 In addition, PCR has been used in the detection of Candida in tissue-biopsy specimens from patients with CDC.22,39
A recent meta-analysis covering 54 studies found PCR-positivity rates among patients with proven or possible candidemia to be 85% (78–91%), compared with just 38% (29–46%) for blood cultures.3 An improved sensitivity for PCR to detect candidemia has been achieved with the use of serum and plasma samples as compared with whole blood. In one study, Candida DNA was detected in 71% of serum samples and 75% of plasma samples, compared with 54% of whole blood samples.41 A limitation of this assay was the inability to detect fungal DNA in 25% of whole blood specimens drawn at the same time as positive blood culture samples.41
Reports of false-positive, as well as false-negative, results when PCR has been used for the detection of Candida in seeded clinical specimens such as blood, urine, and peritoneal fluid, have been described.7 In another study, a false-positive result was reported in two patients who had been treated with azole antifungal agents, which may indicate that PCR was able to detect non-viable organisms that could not be recovered by culture.21 Compared with antigen and antibody methods, PCR has a higher sensitivity and specificity for diagnosis.11 Despite this, no international standardization exists for this method, and there are no commercially available PCR kits for diagnosing Candida. Consequently, PCR is not considered a reliable tool for use in clinical laboratories to help clinicians to identify patients with candidemia early.
Recommendations summary for detecting hematogenousCandidainfection
- 1.
Blood culture is the gold-standard method for the diagnosis of invasive candidiasis.
- 2.
Aerobic bottles and automated blood culture systems are recommended to achieve optimum sensitivity.
- 3.
Following diagnosis, Candida isolates should be identified at species level.
Additional diagnostic method recommendations
- 1.
Imaging (e.g. echocardiography, MRI and CT) may play a part in the diagnosis of endocarditis and CDC.
- 2.
Histopathology and/or direct microscopy can be considered for the diagnosis of deep-seated Candida infections, as with CDC or pneumonia. In case of CDC, a negative microscopic examination does not indicate the absence of disease.
Following the detection of a pathogen, early species identification is important to initiate appropriate antifungal therapy or to modify existing antifungal therapy. Species identification can be achieved by one or more of the following methods: conventional direct examination and staining, screening (germ tubes or chromogenic media), the MicroScan Yeast Identification Panel, manual commercial systems (e.g. API 20C, and API 32C), automated commercial systems (e.g. VITEK 2) and molecular identification (PCR, matrix-assisted laser desorption ionization time-of-flight [MALDI-TOF] or peptide nucleic acid fluorescent in situ hybridization [PNA FISH]).
Conventional direct examination and stainingIn general, direct microscopy, using a number of different stains, is a quick and relatively cheap method to initially identify a pathogen as yeast or bacterium.17 In patients with deep-seated Candida infections, microscopic identification of fungal elements in biopsy material or fungal growth from such biopsies has been considered the gold standard for the diagnosis of CDC32 or Candida-induced pneumonia. It may also be useful for the diagnosis of hematogenous candidiasis in patients who develop skin lesions secondary to Candida dissemination.
The microbiological diagnosis of fungal infections via direct microscopy of tissue sections is generally performed using wet preparations (potassium hydroxide, calcofluor white) and Gram, Wright or Giemsa stains.48 Specimens should be examined for the presence of small, round-to-oval, thin-walled budding yeast cells in clusters and branching pseudohyphae62; however, other genera of fungi may also exhibit this morphology. For this reason, further examination may be necessary to exclude other genera. Following Gram staining, the capsule of Cryptococcus can be visualized as a faint red halo, and the presence of arthroconidia may indicate Trichosporon.53 It is important to note that a negative microscopic examination does not necessarily exclude infection.23,48
Peptide nucleic acid-fluorescent in situ hybridizationThe Yeast Traffic Light PNA FISH is a novel molecular method which may help the fast identification of the most clinically relevant species of Candida as soon as the culture is positive60. Following Gram stain to check for the presence of Candida, slides are prepared to generate fluorescence signals characteristic of individual Candida species. Despite being a useful test, the need of fluorescent microscopy and the cost of the reagents limit its use in clinical laboratories in Latin America.
Phenotypic culture examinationFast screeningA germ tube test (GTT) is a prompt and easy method for the screening of C. albicans.57
Chromogenic mediaIn Latin America, the Working Group recommends the use of CHROMagar media for species identification as it is more accurate, although less expensive chromogenic media are also available (e.g. Oxoid media). In one study, CHROMagar Candida medium allowed the isolates from 93% of clinical specimens to be identified at the species level, with up to 92% accuracy, within 48h. The cost of this technique was comparable with conventional methods, making it an affordable testing method for laboratories from countries with limited resources.30 A restriction of the CHROMagar method is the lack of a wide range of colors, which confines identification to a limited number of species. These include C. albicans (indistinguishable by this method from Candida dubliniensis), C. tropicalis and Candida krusei. The isolates of C. glabrata, C. parapsilosis, Candida guilliermondii, Candida kefyr and Candida lusitaniae have various tones on CHROMagar from off-white to pink, which are difficult to define because they are similar pastel tones and not typical for any species.66 This limitation highlights the requirement for alternative methods for extended Candida spp. identification.5 When culturing biological materials or yeast colonies in chromogenic media, it is important to check for mixed cultures and infection by more than one fungal pathogen.
CHROMagar is prepared according to the manufacturer's instructions and poured into sterile Petri dishes or tubes and allowed to gel and dry. Prepared media plates can be kept for 24h at ambient temperature or for up to one month if refrigerated and protected from light and dehydration.9 If refrigerated, plates should be warmed to room temperature prior to inoculation via streaking the sample onto the plate and incubation at 30–37°C for 48h.9 Identification of the species is based on colony color (Table 1).29,52
Colony colors of yeast isolates incubated for 2 days on CHROMagar Candida medium at 37°C.
| Species | Color and morphology52 |
| Candida albicans | Green |
| Candida tropicalis | Dark blue to blue-gray (with dark halo in agar) |
| Candida krusei | Pale pink to purple (rough with spreading pale edges) |
It is essential that all yeasts causing invasive fungal infections in humans are identified to species level. Species identification can be determined by the following methods.
“In-house” classical methodIt is possible to verify the biochemical profile of yeast colonies by using “in-house” sugar assimilation and fermentation tests. Several laboratories in Latin America prepare their own biochemical test panels, which include 7–12 different sugars for assimilation and fermentation. Results from such assays can then be compared against existing databases (in textbooks) to verify species.
Manual commercial systemsThe biochemical profile of yeasts under identification may be checked by commercially available tests. Following inoculation with yeast, a number of commercially available systems provide different parameters associated with the growth of colonies and a particular substrate. Some detect change in turbidity compared with a control well (e.g. API 20C AUX, and API 32C), whereas others use color generation in a series of wells (e.g. API Candida, Auxacolor, and Uni-Yeast-Tek).18 These tests provide accurate results for the commonest Candida species; however, they are not as reliable for the identification of uncommon species of Candida. Accuracy of results is highly dependent on the number of tests available in each commercial system as well as their database (i.e. limited or broad).
Automated commercial systemsVITEK 2 is a fully automated instrument which uses a fluorescent-based technology for identification of yeasts and yeast-like organisms within 15h.24 The fluorometric card has been replaced by a colorimetric card to broaden the database.2 The MicroScan Yeast Identification Panel is a 4-hour microdilution system for the identification of yeast isolates. It has a 94% identification success rate for 22 species of Candida.40,59
Molecular identificationAs mentioned previously, PCR can be used for the detection of fungal nucleic acids. It can also serve to identify Candida in blood samples, with species identification time reduced from a mean of 3.5 days by routine phenotypic methods to 7h.58 Furthermore, PCR is able to detect the presence of more than one species of Candida in the same patient, whereas blood cultures may yield only one species. The ability to detect and identify more than one species of Candida may affect therapy and outcome, particularly when one of the species is known to be resistant to azole compounds.14
New perspectives (proteomics)Mass spectrometry (MALDI-TOF system) has been used for the identification of Candida species in less than 30min with a 91% success rate.20 MALDI-TOF has been shown to successfully identify 100% of Candida isolates, compared with 92% identification using existing biochemical methods (VITEK 2 and API C AUX).42
The availability of specific diagnostic methods for the detection and identification of candidemia is dependent on the clinical setting. For small hospitals that do not provide care for transplant patients or treat many hematological and immunocompromised patients, the minimal requirements suggested by the Working Group for yeast identification are the capability to evaluate the micromorphology of the colonies complemented by a screening of the main relevant species of Candida by means of CHROMagar Candida medium and/or any in-house or a commercial kit to conduct biochemical tests. For tertiary hospitals, in addition to micromorphology and CHROMagar, species identification should be determined by manual commercial systems (API 20C, API 32C), automated commercial systems (VITEK 2) and/or molecular methods. Molecular methods should be considered in the identification of emerging pathogens where conventional tools usually generate inconsistent identification and when facing outbreaks of fungal infections.
Recommendations summary forCandidaspecies identification
- 1.
The Working Group recommends, as the minimum requirement, colony micromorphology observation complemented by macromorphology using CHROMagar Candida medium.
- 2.
For secondary hospitals, species identification may be determined using one or more of the following methods:
- (a)
Colony micromorphology.
- (b)
Colony macromorphology (CHROMagar Candida medium).
- (c)
Biochemical tests.
- I.
In-house conventional methods.
- II.
Manual commercial systems with a limited database (e.g. Auxacolour and Uni-Yeast-Tek).
- I.
- (a)
- 3.
For tertiary care hospitals that provide care for transplant patients or treat many hematological and immunocompromised patients, the Working Group recommends, as minimum requirement:
- (a)
Micromorphology observation complemented by biochemical tests (API 20C, API 32C, VITEK 2 or MicroScan Yeast Identification Panel).
- (b)
Molecular methods in specific situations.
- (a)
- 4.
Molecular methods (PCR and MALDI-TOF) should be considered in the identification of emerging pathogens and when investigating outbreaks.
It is important to note that yeast identification should always take priority over susceptibility testing. Knowledge of the susceptibilities of local clinical isolates to individual antifungal agents can influence treatment choice. The Working Group encourages laboratories to collaborate with each other when performing susceptibility testing as this will ultimately reduce costs and increase accuracy.
If susceptibility testing is performed, it should be noted that there are still some controversies in the definition of clinical breakpoints for echinocandins and amphotericin B. Screening tests for fluconazole resistance provide the most reliable results compared with other antifungal agents. A number of methods exist for Candida susceptibility testing. These include reference broth-based methods (e.g. standard M27-A of the Clinical and Laboratory Standards Institute [CLSI] and standard EDef 7.1 of the European Committee on Antimicrobial Susceptibility Testing [EUCAST]), disk-based reference methods (e.g. CLSI standard M44-A), commercially available systems (including Fungitest, Sensititre YeastOne, Etest and Neo-sensitabs) and other methods (including flow cytometry and VITEK 2).
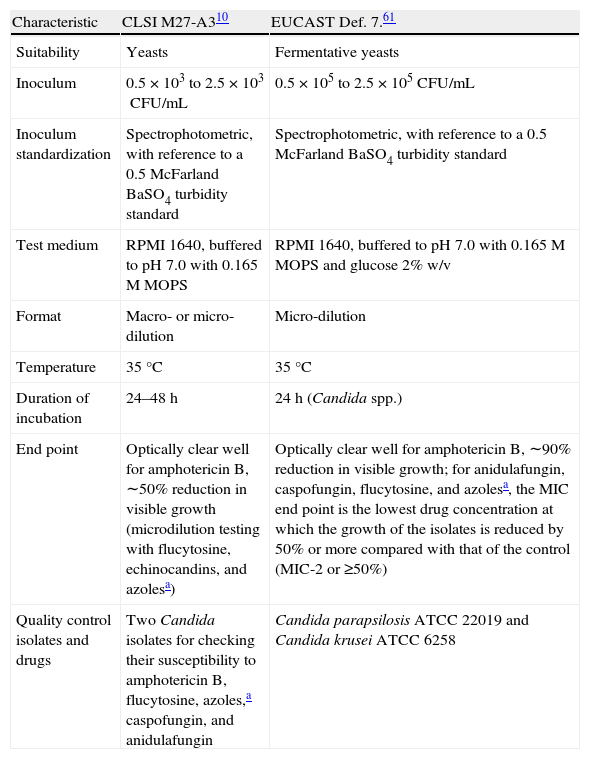
Candida susceptibility testing reference methodsThe reference broth-based method (M27-A) became a National Committee for Clinical Laboratory Standards Institute approved method for antifungal-susceptibility testing in 1997.54 Since the Institute's change of name in 2005, it is now known as the CLSI method.10 The EUCAST61 reference broth-based method is known as EDef 7.1 Both CLSI M27-A and EUCAST EDef 7.1 specify the size of the inoculum, test media, incubation temperature and duration, and end-point readings for the test drugs (Table 2).
CLSI and EUCAST conditions for antifungal susceptibility testing.
| Characteristic | CLSI M27-A310 | EUCAST Def. 7.61 |
| Suitability | Yeasts | Fermentative yeasts |
| Inoculum | 0.5×103 to 2.5×103CFU/mL | 0.5×105 to 2.5×105CFU/mL |
| Inoculum standardization | Spectrophotometric, with reference to a 0.5 McFarland BaSO4 turbidity standard | Spectrophotometric, with reference to a 0.5 McFarland BaSO4 turbidity standard |
| Test medium | RPMI 1640, buffered to pH 7.0 with 0.165M MOPS | RPMI 1640, buffered to pH 7.0 with 0.165M MOPS and glucose 2% w/v |
| Format | Macro- or micro-dilution | Micro-dilution |
| Temperature | 35°C | 35°C |
| Duration of incubation | 24–48h | 24h (Candida spp.) |
| End point | Optically clear well for amphotericin B, ∼50% reduction in visible growth (microdilution testing with flucytosine, echinocandins, and azolesa) | Optically clear well for amphotericin B, ∼90% reduction in visible growth; for anidulafungin, caspofungin, flucytosine, and azolesa, the MIC end point is the lowest drug concentration at which the growth of the isolates is reduced by 50% or more compared with that of the control (MIC-2 or ≥50%) |
| Quality control isolates and drugs | Two Candida isolates for checking their susceptibility to amphotericin B, flucytosine, azoles,a caspofungin, and anidulafungin | Candida parapsilosis ATCC 22019 and Candida krusei ATCC 6258 |
Adapted from Ref.55
CFU=colony-forming unit; CLSI=Clinical and Laboratory Standards Institute; EUCAST=European Committee for Antimicrobial Susceptibility Testing; RPMI=Roswell Park Memorial Institute; MIC=minimum inhibitory concentration; MOPS=3-(N-morpholino)propanesulfonic acid; w/v=weight/volume.
Both methods produce similar minimum inhibitory concentrations (MICs [although EDef 7.1 results usually produce slightly lower MICs]),16,55 which indicates that methodology does not pose any obstacle to obtaining uniform standards in antifungal-susceptibility testing of yeasts. However, these methods are expensive and can be very laborious. Therefore, their actual usage in hospital laboratories is limited; many hospital laboratories prefer to use commercially available products, which claim to be faster and more accurate than the M27-A method.
The disk-based susceptibility testing method (CLSI M44-A) provides a zone of inhibition, a measurement that can be correlated with the MIC value. A good correlation between disk-based susceptibility testing and the M27-A reference method has been shown for fluconazole.44,54 This method is not widely used in Latin America, as fluconazole and voriconazole disks have a limited distribution.
Commercially available susceptibility testsIt is important to note that some of the commercially available products presented in this section may not currently be available in Latin America. Commercial broth-based MIC systems on the market include Candifast, Integral Systems Yeasts, and Fungitest. However, such methods show a limited correlation with the M27-A reference method.54
The Sensititre YeastOne generates a colorimetric response. This method has ≥85% rate of concordance with the M27-A reference method,54 and a 92% rate of concordance with the EDef 7.1 reference method.13 A more recent paper presented high concordance between Sensititre Yeast One and EUCAST, with an essential agreement rate of ≥95.5% (dependent on antifungal agent tested).12
E-test is an antimicrobial-susceptibility test which has been adapted for using with fungal agents. This simple method involves surface inoculation of an agar plate followed by the application of a plastic strip, which contains a concentration gradient of the antifungal agent. Following incubation of a susceptible yeast, a zone of inhibition can be seen and the MIC is read at the point at which the zone intersects the strip.31 The correlation between the E-test method and the reference methods M27-A and EDef 7.1 has been described as acceptable for most species of Candida and azole-based antifungal agents.13,54
Neo-Sensitabs is a commercial disk-based system. It has been reported that the results from this system do not correlate well with the M27-A and EDef 7.1 reference methods.13,54
Other susceptibility testing methodsThe use of flow cytometry as a potential tool for antifungal-susceptibility testing has long been recognized and it is reported to provide rapid results which compare well with standard reference methods.34,56 The VITEK 2 system, which is used for the identification of yeasts, can also be used for susceptibility testing. A comparative evaluation study found an excellent categorical agreement between VITEK 2 and the M27-A reference methods for determining the MIC values of antifungal agents (92–98.2%); however, discrepancies were observed within species.6
Conflict of interestsA.L. Colombo has received research grants from Pfizer, MSD, United Medical and Luminex, medical education grants from Pfizer, MSD, United Medical and Astellas. Moreover, he has also been a consultant for MSD, Pfizer and Gilead. J.A. Cortes has received research grants and support to attend educational meetings from Pfizer and MSD. M. Nucci has received research grants from Pfizer and MSD, and has acted as a consultant and speaker for Pfizer, MSD, Astellas and Gilead. F. de Queiroz Telles has participated in Continuing Education activities in laboratories for Astellas, MSD, Pfizer and United Medical, and in research activities in laboratories for Astellas, MSD and Pfizer. I.N Tiraboschi has been a speaker for Pfizer and Gilead. J. Zurita has been advisory board member and consultant for Pfizer, and has received research grants from Wyeth and MSD for participating in the SMART study.
Editorial support in the form of assistance with the first draft, collating author comments and editorial suggestions to draft versions of this manuscript was provided by Jacqueline Adam, PhD, of Choice Healthcare Solutions and funded by Pfizer. Responsibility for opinions, conclusions and recommendations lies with the authors.