The enzyme 3-hydroxy-3-methylglutaryl coenzyme A reductase (Hmgr) catalyzes the synthesis of mevalonate, a key compound for the synthesis of cholesterol in humans and ergosterol in fungi. Since the Hmgr enzymes of Saccharomyces cerevisiae, Schizosaccharomyces pombe and Candida glabrata are similar to the Hmgr enzymes of mammals, fungal Hmgr enzymes have been proposed as a model for studying antifungal agents.
AimsTo examine the correlation between inhibiting Um-Hmgr enzyme and the viability, sterols synthesis and mating in Ustilago maydis.
MethodsUsing in silico analysis, the ORF codifying for Um-Hmgr was identified and the protein characteristics were deduced. The effect of the competitive inhibitors of Um-Hmgr on the viability of this basidiomycota, the synthesis of its sterols, and its mating were evaluated.
ResultsThe Umhmgr gene (XP_011389590.1) identified putatively codifies a protein of 1443 aa (ca. MW=145.5kDa) that has a possible binding domain in the endoplasmic reticulum (ER) and high identity with the Hmgr catalytic domain of humans and other yeasts. The inhibition of Um-Hmgr caused a decrease of viability and synthesis of sterols, and also the inhibition of mating. The activity of Um-Hmgr is mainly located in the membrane fraction of the fungus.
ConclusionsGiven our results we believe U. maydis is a valid model for studying synthetic inhibitors with lipid-lowering or antifungal activity. Additionally, we propose the Hmgr enzyme as an alternative molecular target to develop compounds for treating both phytopathogenic and pathogenic human fungi.
La enzima 3-hidroxi-3-metilglutaril-coenzima A-reductasa (Hmgr) cataliza la síntesis de mevalonato, compuesto clave precursor en la biosíntesis del colesterol en el ser humano y en la del ergosterol en los hongos. Las enzimas Hmgr de Saccharomyces cerevisiae, Schizosaccharomyces pombe y Candida glabrata presentan similitud con la Hmgr de los mamíferos, motivo por el cual se han propuesto como modelo para el estudio de antifúngicos.
ObjetivosEstudiar la correlación que existe entre la inhibición de la enzima Um-Hmgr y la viabilidad, la síntesis de esteroles y el mating en Ustilago maydis.
MétodosPor medio de un análisis in silico se identificó el ORF de la Um-Hmgr, y se dedujeron las características de la proteína. Se evaluó el efecto de los inhibidores competitivos de la enzima Um-Hmgr en la viabilidad, la síntesis de esteroles y el mating.
ResultadosEl gen Umhmgr (XP_011389590.1) codifica una proteína putativa de 1.443aa (MW=145,5kDa), con un posible dominio de unión al retículo endoplásmico (RE) y una identidad alta con el dominio catalítico de la Hmgr humana y de otras levaduras. La inhibición de la Um-Hmgr ocasionó una disminución en la viabilidad y síntesis de esteroles del hongo, así como la inhibición del mating. La actividad de la Um-Hmgr está localizada principalmente en la fracción membranal del hongo.
ConclusionesLa enzima Um-Hmgr está anclada probablemente al RE y presenta una elevada homología con el dominio catalítico de otras Hmgr de eucariotas. La Um-Hmgr participa en la síntesis de esteroles de este basidiomiceto, y su inhibición provoca la pérdida de la viabilidad, la reducción de los niveles de esteroles y del mating del hongo.
Sterols regulate the fluidity of membranes and the function of some membrane proteins. 3-Hydroxy-3-methylglutaryl coenzyme A reductase (Hmgr) is the rate-limiting enzyme in the biosynthesis of sterols. It catalyzes the reduction of HMG-CoA in the presence of two molecules of NADPH to generate mevalonic acid, an important precursor in the synthesis of ergosterol (Erg) in fungi and cholesterol in mammals.8
Hmgr enzymes are classified in two groups, class I and class II. Class I includes the enzymes of eukaryotes and some archaea, and have a transmembrane sequence that allows them to bind to the endoplasmic reticulum (ER). Class II consists of the soluble enzymes of bacteria and other archaea.11
The human Hmgr enzyme has an extreme COOH terminal that contains the catalytic region. This is separated from a linking sequence that anchors the other domain, the NH2-terminal region, which contains 339 amino acid residues that anchor to the membrane of the ER.15,20,21,28 The analysis of the proteins deduced from the sequences of the hmgr genes suggests that the Hmgr enzymes of fungi and yeasts have the same topology.2
Statins are the most studied human Hmgr (Hmgr-h) inhibitors. These semisynthetic or synthetic substances, which are derived from the metabolites of some species of fungi, have been utilized to inhibit the synthesis of cholesterol in mammals.9,16 Although statins and their derivatives are widely used, their slight hepatotoxicity, rhabdomyolysis and myopathy have been documented.18,32
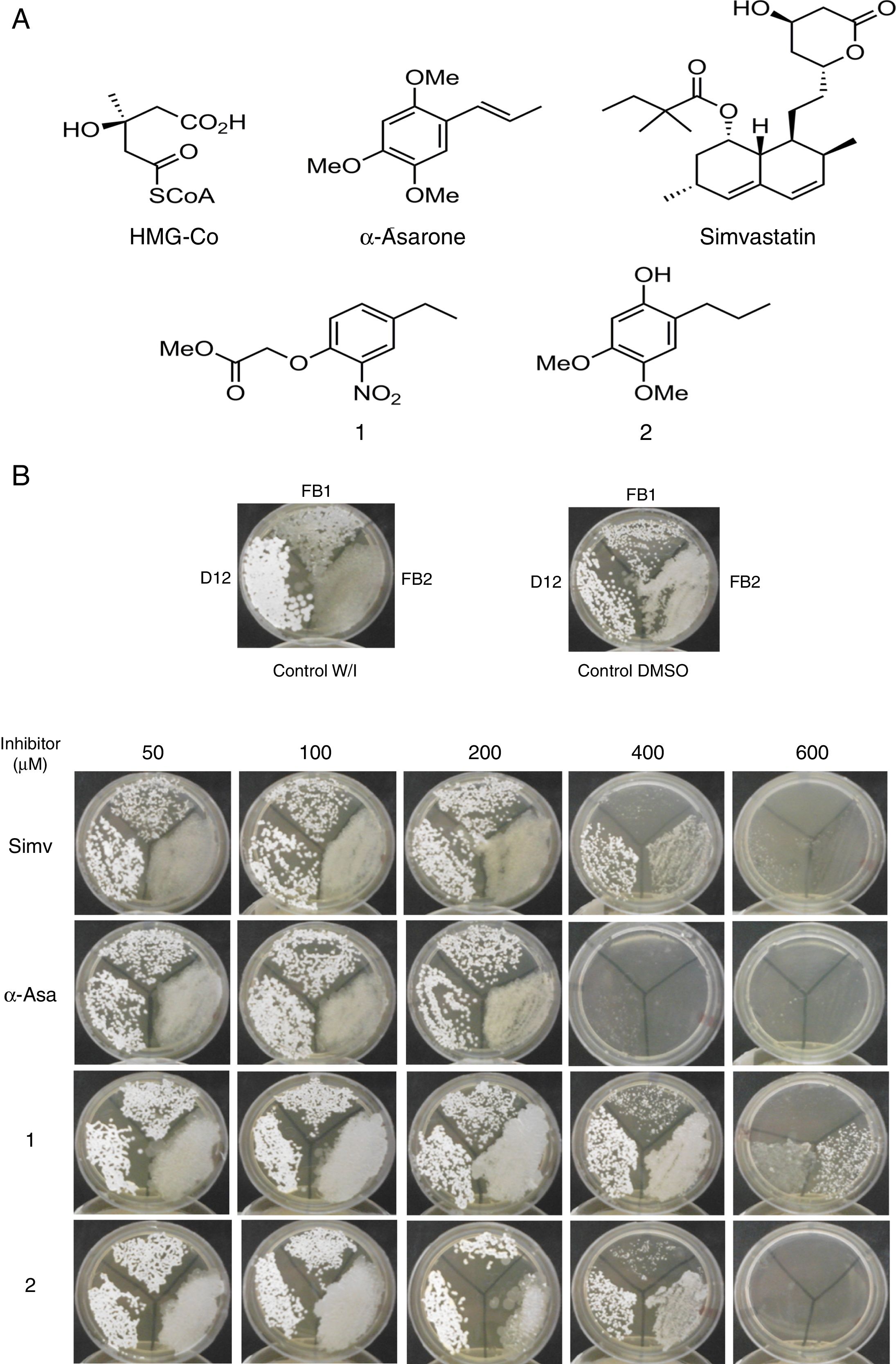
Currently, new compounds with lower toxicity that can inhibit Hmgr, as well or better than statins, are sought. In this sense, a series of structural analogs of α-asarone and fibrates, such as derivatives 1 and 2 were synthesized after being designed to maintain the essential pharmacological properties for inhibition of Hmgr, but with modifications to reduce their toxicity2,23 (Fig. 2). To test these inhibitors, Hmgr enzymes from yeasts have been proposed as a model for studying the regulation and inhibition of eukaryotic sterols.2,3
Ustilago maydis has been proposed as an alternative model for the study of basic cellular biology processes. Moreover, U. maydis genome has been already sequenced and a plethora of genetics and molecular biology techniques are available to be used in this organism.26 The Hmgr of U. maydis was presently evaluated as a possible model for testing Hmgr inhibitors. Therefore the idea is to establish an Um-Hmgr model for future research in relation to a given series of antihyperlipidemic (lipid-lowering) compounds with potential for medicinal application. It must be pointed out that the role of Hmgr of U. maydis in the metabolism or life cycle of this basidiomycota is unknown. However, statins have demonstrated to have antifungal activity. For instance, atorvastatin reportedly causes a decrease in the synthesis of sterols and in the viability of the opportunistic pathogen Candidaglabrata.31 The aim of the current study was to explore the effect of some inhibitors of Hmgr on the viability of U. maydis, its synthesis of sterols and mating.
Materials and methodsStrains and culture mediumWild-type U. maydis strains (FB1, FB2 and D12, kindly donated by Flora Banuett) were maintained at −70°C in 50% (v/v) anhydrous glycerol (J.T. Baker, Mexico), then recovered in liquid YPD medium (1% yeast extract (MCD Lab, Mexico), 2% casein peptone (triptone) (MCD Lab), 2% dextrose anhydrous powder (J.T. Baker)) at 28°C, shaken in a rotary shaker at 200rpm, and used as inoculum for subsequent experiments.14
Bioinformatic analysisThe nucleotide sequence of the Um-Hmgr was obtained by accessing the NCBI website http://www.ncbi.nlm.nih.gov/. The search was defined in the database in the drop-down menu located in the upper-left corner; the “protein” section was selected, the access number XP_011389590.1 was introduced, and the amino acid sequence was extracted from the FASTA format. Later the BLATp website was accessed https://blast.ncbi.nlm.nih.gov/Blast.cgi and the “protein BLAST” section was selected where the sequence obtained from the FASTA format was used for its analysis.
The U. maydis Um-Hmgr nucleotide sequence was identified through a BLASTp analysis with deduced amino acid sequences of other yeasts and filamentous fungi. The approximate isoelectric point (pI) was determined and molecular signatures predicted by using the ScanProsite database (http://psort.hgc.jp/form2.html).10 Subcellular location was identified with PSORTII (http://psort.hgc.jp/form2.html).27
Effect of inhibitors on the viability of U. maydisThe strains of U. maydis (FB1, FB2 and D12) were grown in liquid YPD medium at 28°C for 24h. An aliquot of each of the cultures (approximately 1×104 cells) was taken and inoculated in solid YPD medium having one of the inhibitors (simvastatin, α-asarone, compounds 1 or 2) at different concentrations (50, 100, 200, 400 and 600μM). Finally, the medium was incubated at 28°C for 24h and the growth of the fungus was observed.
In vitro susceptibility of U. maydis to simvastatin, α-asarone and synthetic compounds 1 and 2 by agar disk diffusion assayIn vitro susceptibilities of U. maydis FB1, FB2 and D12 to simvastatin, α-asarone and synthetic compounds 1 and 2 were determined by agar disk diffusion assay using the Clinical and Laboratory Standards Institute (CLSI) M44-A guidelines. Briefly, Müller-Hinton agar medium (MHA) (0.3% meat extract (BD Bioxon, Mexico), 1.75% casein peptone (triptone) (MCD Lab), 0.15% starch (J.T. Baker), 1.5% bacteriological agar (MCD Lab) supplemented with glucose 2% dextrose anhydrous power (J.T. Baker)) was used as described by the CLSI; it was not necessary to add methylene blue, since growth inhibition halos were evident. Fluconazole was used as a control, as well as resistant and sensitive Candidaalbicans and Candida parapsilosis strains. The concentrations used are described by CLSI according to the solvent used: water for fluconazole and simvastatin, and DMSO for α-asarone and compounds 1 and 2.
Extraction and quantification of sterolsTotal intracellular sterols were extracted and ergosterol concentrations were assayed using a slight modification of a previously reported method.4 Briefly, U. maydis FB1 cells were grown in YPD medium and incubated at 28°C for 24h with agitation at 200rpm. The cell culture was obtained by adjusting it with YPD medium to a density of 0.2 (As600) in different flasks that contained 5ml of culture consisting of DMSO solvent (Sigma–Aldrich, USA) only as control, and 10, 50, 100, 200 or 300μM of inhibitor (simvastatin, α-asarone, compounds 1 or 2). The cultures were incubated at 28°C for 18h with agitation at 200rpm.
Cells were harvested by centrifugation and ergosterol was removed with 25% alcoholic KOH (25g KOH dissolved in 35ml of distilled water, and diluted to 100ml with ethanol) by methanolic KOH (Merck, Mexico) extraction. A second sterol extraction was performed by adding a mixture of 1ml of sterile distilled water and 3ml of n-heptane (Tsq, MX). The heptane layer was spectrophotometrically scanned between 230 and 300nm with a spectrophotometer (BioSpectrometer, Eppendorf). The presence of detectable ergosterol resulted in a characteristic 281.5 peak. Ergosterol content was calculated as a percentage of the wet weight of the cells, as previously described.4,7
The experiments and determinations were carried out in triplicate and data are presented as the mean of the inhibition of ergosterol synthesis (expressed as a percentage of the control). Equivalent quantities of cells were assayed, and the biomass concentration was determined by measuring dry cell weight, as previously described.13
Effect of inhibitors on sexual mating complementationU. maydis cells (FB1 + FB2) were grown in YPD liquid medium at 28°C for 24h (in the log phase of growth). The inhibitors (simvastatin, α-asarone, compounds 1 and 2) were added at distinct concentrations (50, 100, 200, 400 and 600μM). For this test two controls were used, one of them without any treatment (no addition of inhibitor) and another one with 10% DMSO. Growth under these conditions was assessed by placing a 10μl drop of each treatment in YPD solid medium (1% yeast extract (MCD Lab), 2% casein peptone (triptone) (MCD Lab), 2% dextrose anhydrous power (J.T. Baker), supplemented with 0.5% activated charcoal (Merck)). Plates were incubated at 28°C and the induction of the fuzz phenotype was monitored after 24h.
Preparation of soluble fractions and membrane fractionsThe U. maydis FB1, FB2 and D12 cells were harvested from YPD medium during early stationary phase by centrifugation at 5000×g at 4°C for 10min. Cells were fragmented in a FAST Prep-24 (MPTM) using glass beads (7.5g of glass beads, and 5g of cells) and pulses (3×20s at 6.5m/s with 2min intervals on ice). Broken cells were centrifuged at 5000×g at 4°C for 10min. The crude extract was carefully removed from the glass beads and centrifuged at 23,000×g at 4°C for 10min. The supernatant was removed and centrifuged at 100,000×g at 4°C for 1.5h using a Beckman ultracentrifuge. The corresponding soluble fraction or cell free extract and membrane fraction were used for enzyme assays and protein determination. 3
Enzymatic activity assayThe activity and inhibition assays were performed as previously reported.3 Briefly, the oxidation of NADPH (Sigma–Aldrich, USA) was spectrophotometrically monitored at 340nm in a BioSpectometer-Kinetic (Eppendorf). HMG-CoA reductase (Sigma-Aldrich) activity was assayed at least six times. The reaction mixture contained 0.13mM HMG-CoA, 1μl of Um-Hmgr with or without inhibitor, and 50mM Tris–HCl (IBI Scientific, USA), pH 7.5, to a final volume of 100μl. After 10min of incubation at 37°C, the reaction was started by adding 0.13mM NADPH and then monitored for 10min. For all reactions, 1 unit of enzymatic activity is defined as the amount of enzyme required to catalyze the oxidation of 1mmol of NADPH per min (1μU catalyzes 1μM of NADPH in 1min), using a modified version of a previously reported method.3,6
Inhibitory compoundsSimvastatin and α-asarone were purchased from Sigma–Aldrich while compounds 1 and 2 were synthesized as previously described.24,33
Statistical analysisFor all the measured parameters, averages and standard errors were considered. The differences between the means were analyzed by means of the Student's t test. The experimental data were considered significant with p<0.05 (*), being very significant with p<0.01 (**) and extremely significant with p<0.001 (***). The comparison of multiple groups was performed using the two-way ANOVA analysis of variance, followed by the Tukey test. The analysis and graph were made with the GraphPad Prism 6.0 software (GraphPad Software, San Diego, CA, USA).
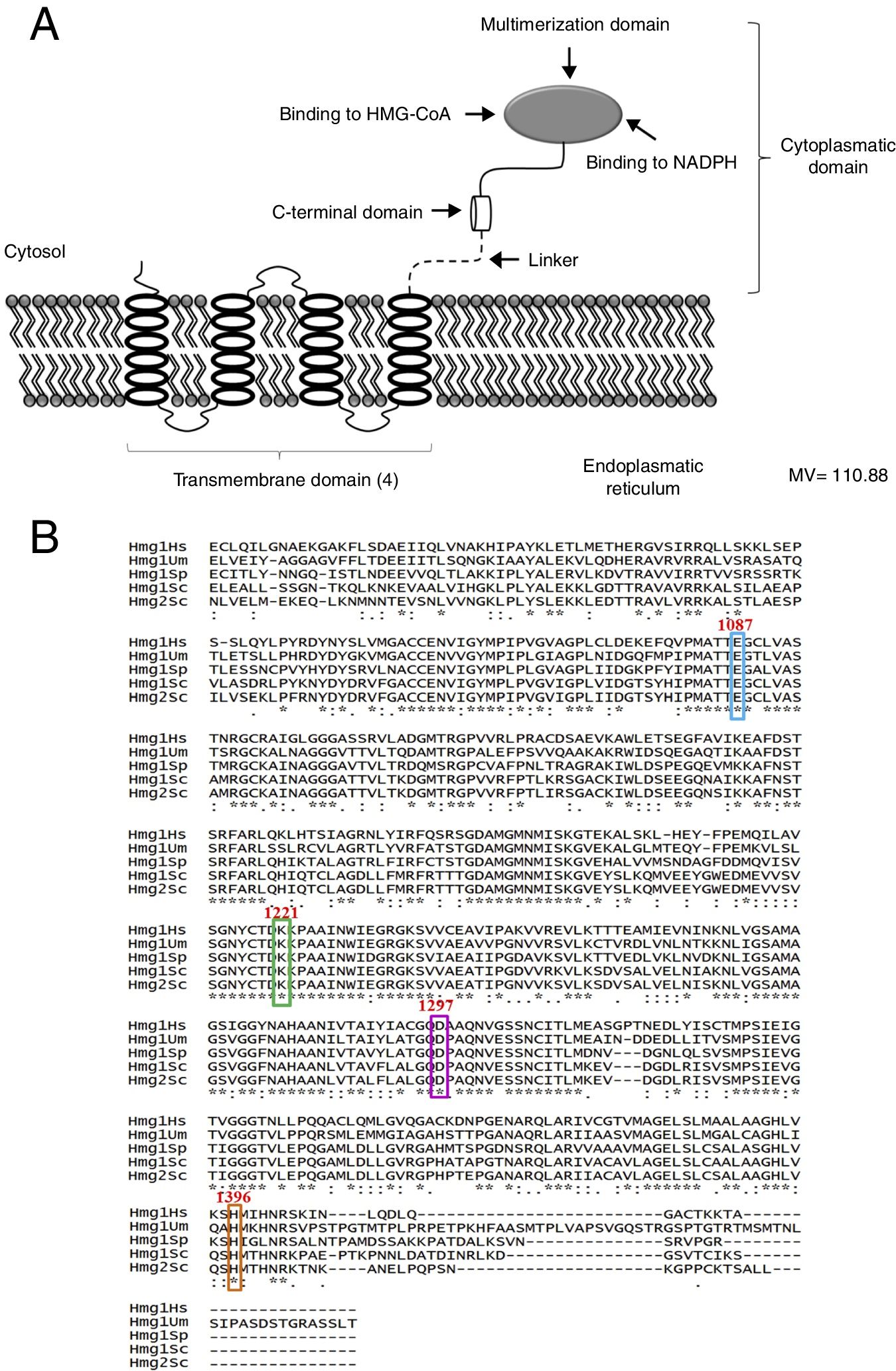
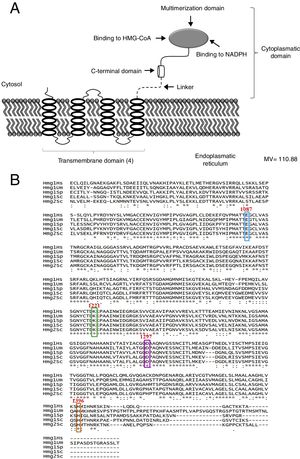
ResultsThe bioinformatic analysis allowed the identification of a 4332 pb ORF (UniProt, NCBI XP_011389590.1) in the U. maydis genome, with no introns, which codes for a protein of 1443 aa. This protein has a putative molecular weight of approximately 145.5kDa and an isoelectric point (pI) of 8.49. Our results suggest that Hmgr of U. maydis is likely located in the ER; this putative protein probably has a site sensitive to sterols (located in residues 335-532), and a catalytic domain (located in residues 968-1376). The topography of Hmgr of U. maydis (Um-Hmgr) showed four transmembrane segments, which are binding domains for the substrate of the dimerization cofactor. It is also likely that it has an amino extreme in the lumen of the ER and an extreme carboxylic acid in the cytoplasm, characteristics very similar to other eukaryotic Hmgr enzymes (Fig. 1A).
The Hmgr protein of Ustilago maydis, deduced from the nucleotide sequence. (A) Topography of Um-Hmgr in the endoplasmic reticulum. (B) Alignment of the sequences of amino acids of the catalytic domain of human, U. maydis, S. pombe and S. cerevisiae Hmgr enzymes. Glutamic acid1087, lysine1221, aspartic acid1297 and histidine1396.
The amino acid sequence of the Um-Hmgr was deduced from the nucleotide sequence and compared to the sequences of human, S.cerevisiae and Schizosaccharomyces pombe Hmgr enzymes. The amino acids located at the putative catalytic site are characteristic and consistent with the catalytic sites of other Hmgr enzymes. These residues are glutamic acid (E1087), lysine (K1221), aspartic acid (D1297) and histidine (H1396) (Fig. 1B).
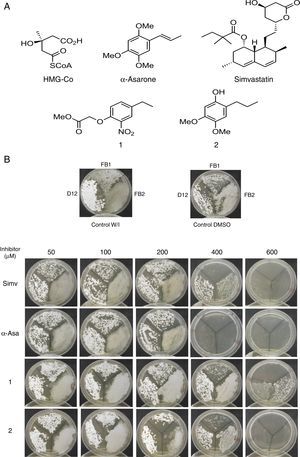
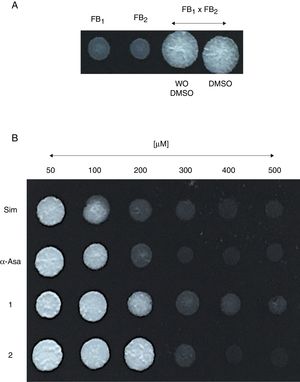
Um-Hmgr inhibitors diminished growthEvaluation of the effect of the Hmgr inhibitors (simvastatin, α-A asarone, compounds 1 and 2) (Fig. 2A) was made on the growth of the U. maydis FB1, FB2 and D12 strains in YPD solid medium. All the compounds inhibited the growth of the three strains. However, the U. maydis FB1 strain turned out to be more sensitive to all the compounds. α-asarone was more effective than simvastatin in growth inhibition (MIC=400 and 600μM, respectively). Compound 2 proved to be as effective as simvastatin for inhibiting the growth of the three U. maydis strains (Fig. 2B). MICs to the assayed compounds were determined according to the disk diffusion test described by CLSI, M44-A. Fluconazole and simvastatin were used as controls. U. maydis strain FB1 showed a MIC of 3.12μg to α-asarone and compounds 1 and 2. U. maydis strains FB2 and D12 MIC to compounds 1 and 2 was 40μg. U. maydis FB2 and D12 MICs to α-asarone were 10 and 20μg, respectively. The MICs to simvastatin were 8μg for the FB1 strain and 32μg for the FB2 and D12 strains. U. maydis FB1 growth was more sensitive to the α-asarone and to the synthetic compounds 1 and 2, than strains FB2 and D12. However, the three strains of U. maydis were more sensitive to fluconazole (MIC to fluconazole for strains FB1 and FB2 was 1μg, and 2μg for strain D12).
Inhibition of Ustilago maydis growth with Hmgr inhibitors. (A) The chemical structure of the competitive inhibitors utilized herein is compared to the substrate of the HMG-CoA enzyme. (B) Inhibition of U. maydis strains by simvastatin, α-asarone, and compounds 1 and 2. As controls, the growth of U. maydis strains was tested without any treatment (W/I) and with the DMSO solvent only.
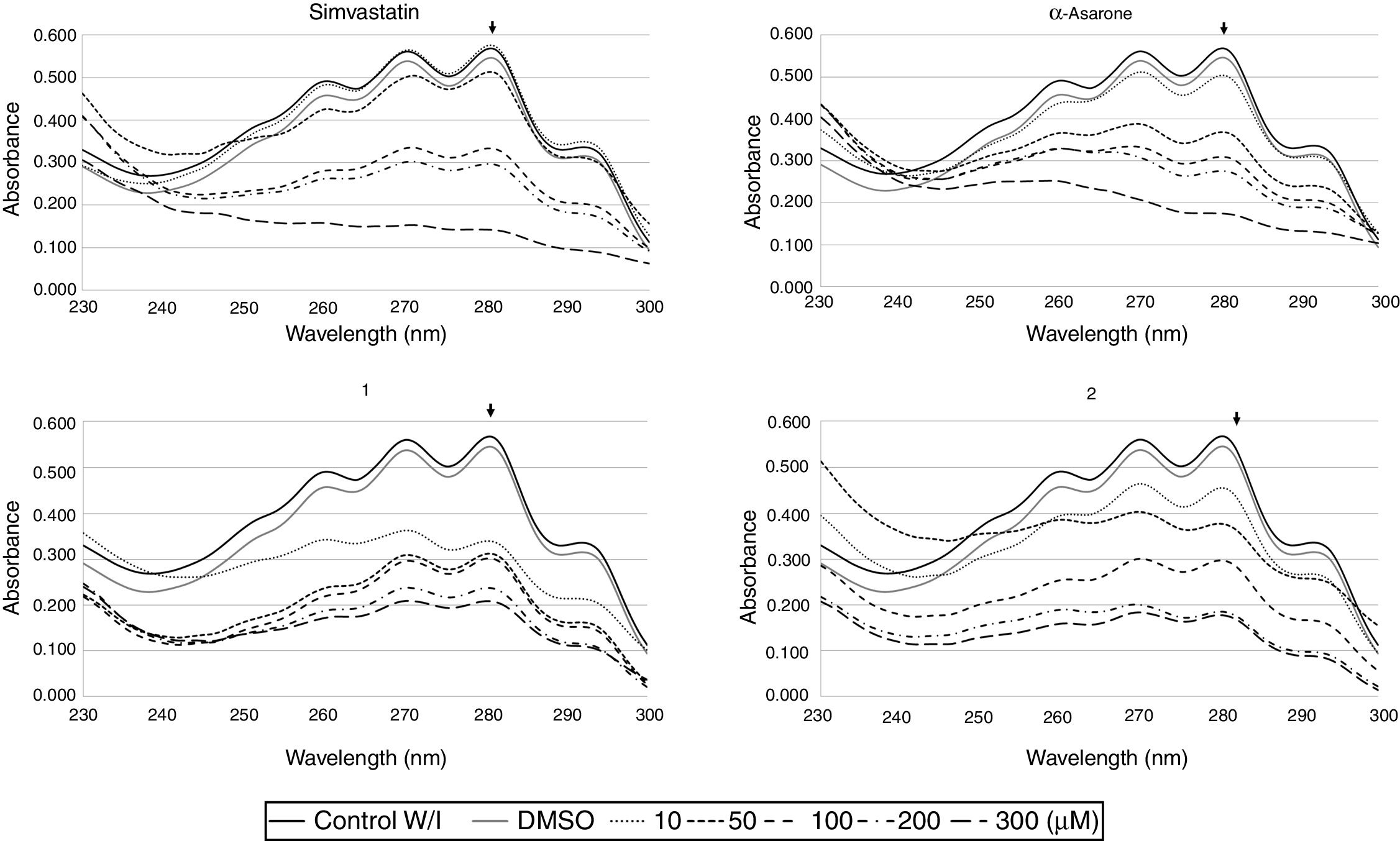
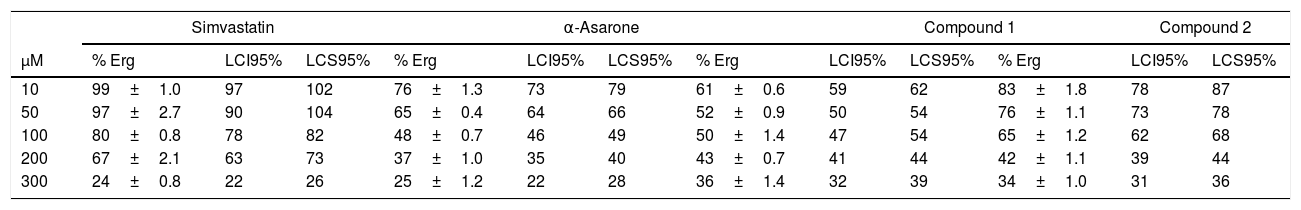
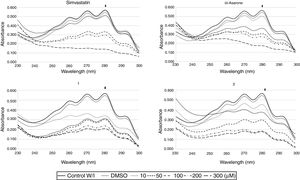
To explore the possible association between the loss of viability of U. maydis and the inhibition of sterol synthesis, the level of ergosterol in U. maydis cells was measured after 18h of treatment with the inhibitors. The compounds employed to inhibit Um-Hmgr activity caused also a reduction in the level of sterols of this fungus. Absorption spectra obtained after using different concentrations of the inhibitors (Fig. 3) showed the characteristic four peaks of sterols. The decrease in the height of these peaks indicates lower levels of sterols. The levels of ergosterol were quantified based on the peak corresponding to As at 281.5nm (Table 1). The percentage of inhibition of ergosterol synthesis varied according to the different inhibitors and the distinct concentrations. The percentage of inhibition of ergosterol synthesis in U. maydis FB1 varied according to the different inhibitors and the distinct concentrations. As shown in Table 1, all tested compounds resulted in decreased levels of ergosterol; however, the IC50, concentration of the inhibitor causing a decrease in ergosterol levels by 50%, were slightly different: 112μM for compound 1, 188μM for compound 2, 133μM for α-asarone and 224μM for simvastatin.
Um-Hmgr inhibitors (simvastatin, α-asarone, and compounds 1 and 2) caused a reduction in the level of ergosterol. Ustilago maydis was grown in YPD medium and treated with different concentrations (10, 50, 100 and 300μM) of the inhibitors. The control was YPD without any treatment or with the vehicle (DMSO) only. The extracted sterols were analyzed with an absorption spectrum between 230 and 300nm. The quantities examined correspond to the same amount of biomass. The arrow in each graph indicates the absorbance at 281.5nm, corresponding to the peak at which ergosterol absorbs.
Percentage of ergosterol present in Ustilago maydis FB1 cells treated with inhibitors of the enzyme Hmgr.
| Simvastatin | α-Asarone | Compound 1 | Compound 2 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| μM | % Erg | LCI95% | LCS95% | % Erg | LCI95% | LCS95% | % Erg | LCI95% | LCS95% | % Erg | LCI95% | LCS95% |
| 10 | 99±1.0 | 97 | 102 | 76±1.3 | 73 | 79 | 61±0.6 | 59 | 62 | 83±1.8 | 78 | 87 |
| 50 | 97±2.7 | 90 | 104 | 65±0.4 | 64 | 66 | 52±0.9 | 50 | 54 | 76±1.1 | 73 | 78 |
| 100 | 80±0.8 | 78 | 82 | 48±0.7 | 46 | 49 | 50±1.4 | 47 | 54 | 65±1.2 | 62 | 68 |
| 200 | 67±2.1 | 63 | 73 | 37±1.0 | 35 | 40 | 43±0.7 | 41 | 44 | 42±1.1 | 39 | 44 |
| 300 | 24±0.8 | 22 | 26 | 25±1.2 | 22 | 28 | 36±1.4 | 32 | 39 | 34±1.0 | 31 | 36 |
| Control W/I | Control DMSO |
|---|---|
| % Erg | % Erg |
| 100 | 100 |
The presence of detectable ergosterol resulted in a characteristic 281.5nm peak of absorbance. Ergosterol content was calculated as a percentage of the wet weight of the cells, as described by Arthington-Skaggs et al. (1999).4,7U. maydis was grown in YPD medium and treated with different concentrations (10, 50, 100 and 300μM) of the inhibitors: simvastatin, α-asarone, and compounds 1 and 2. For the controls, YPD was used without any treatment or with DMSO only. Each treatment was run in triplicate in independent assays. Values are expressed as the mean. Control W/I (without inhibitor), Control DMSO (dimethyl sulfoxide, solvent of the synthetic compounds). Statistic analysis: The differences between the means analyzed by t-Student and p < 0.001 were considered statistically significant.
The possible relationship between the decrease in the level of sterols and the inhibition of Um-Hmgr was evaluated. An enzymatic extract was obtained from the soluble and membrane fractions of this fungus. Um-Hmgr activity was mainly detected in the membrane fraction (8.0 mU/mg vs. 0.2 mU/mg in the soluble fraction) of U. maydis FB1. The Um-Hmgr membrane activity was inhibited with all four tested compounds by 50% (IC50) with a concentration of approximately 50μM. A similar activity was observed in the membrane fraction (7.2 and 7.8 mU/mg) and in the soluble fraction (0.2 and 0.15 mU/mg) of cells of FB2 and D12U. maydis strains, respectively. Moreover, the IC50 for all four tested compounds was also very close to 50 μM.
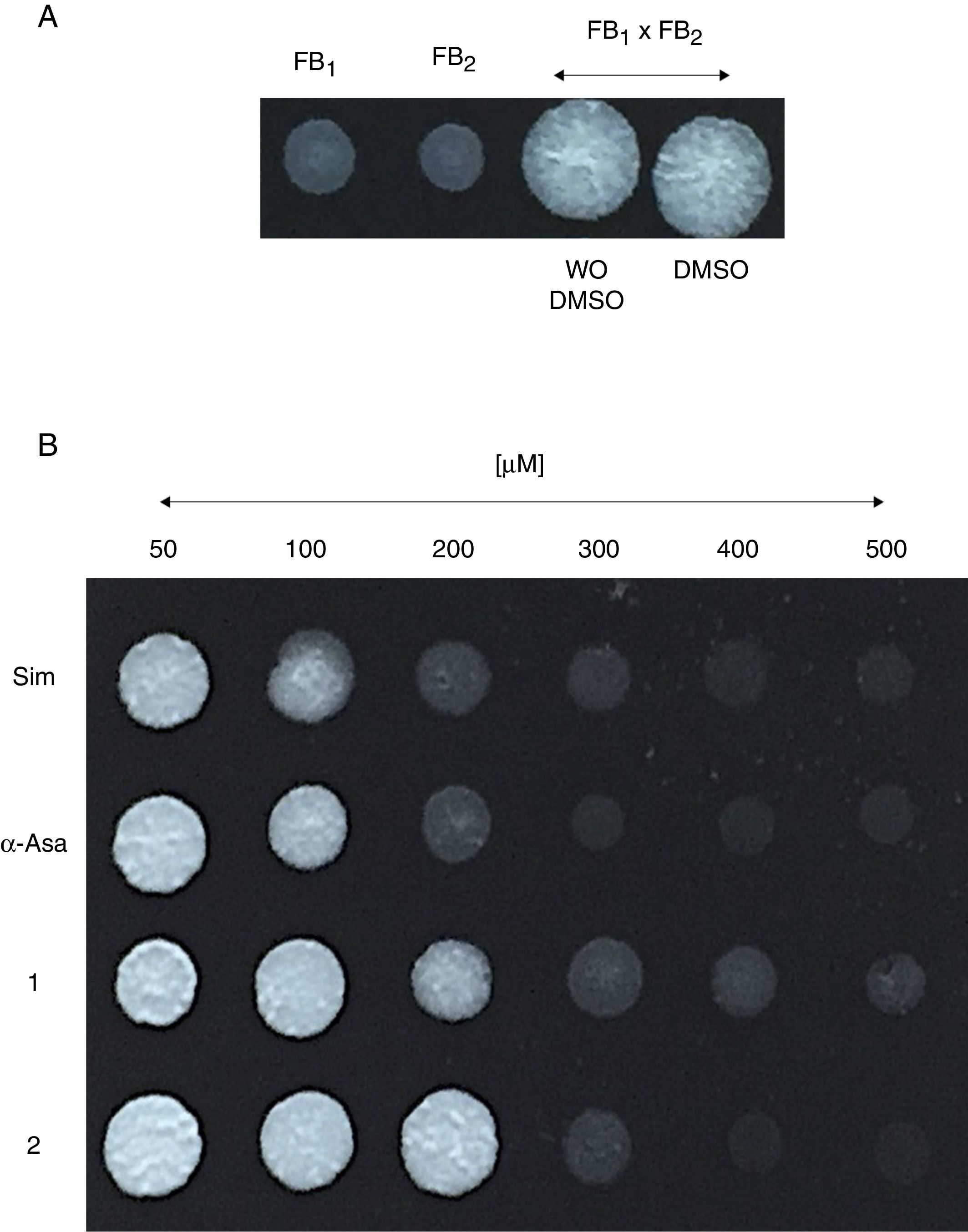
Um-Hmgr inhibitors affect matingMating of U. maydis is a relevant event in the life cycle of the fungus, as only dikaryon or diploid cells are pathogenic and, therefore, able to infect corn. Moreover, the sexual fusion of complementary cells is a requirement for the formation of the dikaryon. The sexual complementation of U. maydis was evaluated with or without one of the inhibitors present and without an inhibitor (Fig. 4). When tested at different concentrations (50, 100, 200, 400 and 600μM), the inhibitors (simvastatin, α-asarone, 1 and 2) were capable of impeding mating between the complementary strains, FB1 and FB2, of U. maydis.
Effect of the compounds on sexual complementation of Ustilago maydis, which was grown in YPD. (A) Without any treatment or with DMSO only (control groups). (B) With the inhibitors (simvastatin, α-asarone, and compounds 1 and 2) at different concentrations (50, 100, 200, 300, 400 and 500μM). All treatments were assayed at 28°C for 24h. For mating, a mixture of FB1 and FB2 strains was made. Mating was evidenced by the formation of a white button (FB1×FB2).
Due to the importance of being able to lower lipid levels in patients, inhibitors were designed for this purpose previously.3,24 Statins, the first of such inhibitors administered to patients, were based on the strategy of lowering cholesterol synthesis. Because of the toxicity of statins, novel strategies are sought. Nowadays, there is an effort to find Hmgr-h inhibitors that are less toxic and more efficient than statins.3,24 Therefore, alternative models have been proposed.
In general, there is an advantage of using yeast models to study Hmgr inhibitors that may eventually be administered to patients with high cholesterol concentrations. Yeast cells are easily cultivated, the culture media is well defined, and generation times are short. Moreover, there are high yields and easy accessibility to techniques of biochemistry, genetics, and molecular biology. In this sense, the inhibition of the Hmgr of S. pombe by some synthetic compounds has been demonstrated.3
Among different yeasts, U. maydis and Um-Hmgr could offer an attractive model for examining Hmgr inhibitors. The proteome of U. maydis is more similar to the one of Homo sapiens than that of S. cerevisiae, the yeast used as the model par excellance.26 For this reason, U. maydis has been proposed as an alternative model for the preliminary study of cellular biology of mammals in general.
The current bioinformatic analysis of U. maydis Hmgr revealed an ORF of 4332 pb that likely codifies for a protein of 1443 aa and a MW of approximately 145.5kDa. Um-Hmgr topology suggests that this protein is bound through its amino domain to the membrane of the ER with four segments of anchoring. Contrarily, other Hmgr enzymes, such as those of S. cerevisiae and C. glabrata, have 7–8 anchoring domains.1 This difference in the hydrophobic size of the amino domain, and therefore in the number of transmembrane segments, does not differentiate ascomycetes from basidiomycetes. Studies carried out by our group shown that the Hmgr of another basidiomycota, Cryptococcus neoformans, has 10 transmembrane segments,2 while Ganoderma lucidum has 8 segments.30 Mammalian Hmgr is a key enzyme in the synthesis of sterols, so it is highly regulated at multiple levels: transcription, translation, post-translational modifications and protein degradation (sterol-dependent-degradation). The transmembrane sequences of Hmgr are involved in the degradation of the enzyme: when the sterol levels are high, Insig protein binds to the membrane domain of Hmgr, which undergoes ubiquitination and then is degraded via the proteasome.8 The study and characterization of the transmembrane sequences of Um-Hmgr will allow in the future to understand the regulation of the levels of this enzyme by a possible path of degradation due to a sterol-dependent-degradation, in a model such as U. maydis. On the other hand, the study of Um-Hmgr transmembrane domain would shed light on the possible differences or similarities in the regulation of the levels of this enzyme compared to what happens in other yeasts such as S. cerevisiae, S. pombe and Cryptococcus neoformans.8
Regarding the proteins deduced from the nucleotide sequences of diverse genes codifying for Um-enzymes, previous analyses demonstrated that the active site of these enzymes is highly conserved.2,11 Given the bioinformatics data, Um-Hmgr is classified as class I enzyme that is probably located in the ER of this basidiomycete.
The cellular fractionation of U. maydis FB1 yeasts allowed the detection of specific activity that was 40-fold higher in the membrane fraction than in the soluble fraction. This result supports the idea that this enzyme may be located in the ER.8,11 In order to corroborate in silico predictions and these preliminary results it is necessary to study more in-depth this finding and verify the subcellular location of the Um-Hmgr. With regard to the sensitivity of the enzymatic activity of Um-Hmgr to the inhibitors used in this study in the different U. maydis strains (the haploids FB1 and FB2, and the diploid D12) we observed there was practically no difference. However, as previously mentioned, the growth of these strains was affected differently, since the growth of the FB1 strain was more sensitive to the inhibitors than that observed in strains FB2 and D12. It is possible that the accessibility of these compounds for strains FB1, FB2 and D12 is different. It will be necessary to perform other studies to explain the observed differences.
U. maydis was selected as a model for the study of the Hmgr enzyme for an additional reason: its genome has a single codifying sequence for the enzyme. Hence, biochemical assays carried out with Um-Hmgr will correspond to one single type of protein instead of a mixture of isoenzymes, even if the protein has not been purified. This characteristic is not possible with the prototype yeast S. cerevisiae, which has two genes codifying for Hmgr enzymatic activity that are expressed under distinct conditions.8,11
Previous reports indicate that the levels of sterols diminished after applying simvastatin and atorvastatin to C. glabrata.3,24 In the present study the cells of U. maydis cultivated in YPD medium and harvested during the log growth phase were exposed to the four inhibitors herein tested, all designed and synthesized to competitively inhibit the Hmgr-h enzyme. There was a dose–response effect, observing lower levels of sterols with a greater concentration of each inhibitor. It was demonstrated quantitatively that the level of ergosterol also declined.
Other studies have shown that the inhibition of the P-450-α-14-sterol-demethylase enzyme diminishes the level of sterols. These assays were based on the technique of thin layer chromatography (TLC) coupled with high resolution chromatography.12,29 To our knowledge, the current contribution represents the first time a reduction in the level of sterols is detected after inhibiting the Um-Hmgr enzyme. This result was observed with the standard compounds (simvastatin and α-asarone) as well as the new test compounds 1 and 2. The decrease in sterols was especially notable in relation to ergosterol.
The use of Hmgr inhibitors led to an evident decline in the synthesis of sterols in C. glabrata and Aspergillus fumigatus.22,31 The reduced level of ergosterol can alter the activity of membrane enzymes and sterols (as demonstrated with statins), as well as affect the mitochondrial function (production of coenzyme Q) and diminishes the level of prenylated proteins, including those that prompt cell death.25
The life cycle of U. maydis involves a stage of complementation between two sexually compatible strains.5 Here we have shown that this process of mating was affected by the use of Um-inhibitors, evidenced by a clear fuzz reaction (white colonies were shown in contrast to the activated carbon of the medium). Compared to compound 1, compound 2 apparently caused increased inhibition of this process as well as reduced viability of U. maydis and its synthesis of sterols.
The infectious phase of this fungus requires the fusion of two haploid yeast cells of complementary genders, a process that is regulated by the interaction of the α and β mating loci. This leads to the formation of the dikaryotic mycelium form, which the plant requires for its growth. It has been described that the depletion of ergosterol in S. cerevisiae affects two important aspects of yeast mating: signaling by pheromones and the fusion of plasmatic membranes. However, this subject has not been studied in relation to basidiomycota.17
The compounds studied in this work could be used as antifungals for both phytopathogenic fungi and human fungal infections. The results of this work suggest that the Hmgr enzyme may be an alternative molecular target to those already described.2 In the case of a maize phytopathogenic fungi, such as U. maydis, molecular modeling studies could be carried out to allow the design of new compounds that are specific against the Hmgr of this fungus. The Hmgr enzyme of maize, involved in the synthesis of terpene compounds, is a clear example of the expansion of genes and seven genes HMGR have been found.19 The recombinant Um-Hmgr protein expressed in a heterologous system will allow for more detailed studies of inhibition with the compounds. The Hmgr of human pathogenic fungi has been proposed as an alternative molecular target that could help to solve the problem of strains resistant to conventional antifungals.2,31 In this regard, very good results have been obtained with the recombinant protein of Candida glabrata rec-Cg-Hmgr, which showed higher activity levels and stable storage conditions at −20°C. The catalytic domain of Cg-Hmgr was expressed in Escherichia coli, which showed that inhibitors of human Hmgr can inhibit rec-Cg-Hmgr, in addition to inhibiting sterol synthesis and yeast growth.1 Therefore, the compounds studied in this work could also be proposed as antifungals for yeasts of the genus Candida, especially for C. glabrata, and help in some way to solve the problem of resistance that this species presents to conventional antifungals.
The present results establish the foundation for future research on the synthesis of sterols in U. maydis and their role in growth, as well as on the regulation of Um-hmgr1 gene expression. For example, the heterologous expression of this gene codifies for the recombinant protein, which could be utilized to test synthetic inhibitors such as lipid-lowering and antifungal agents. The inhibition of sterol synthesis clearly reduced the viability and mating of U. maydis. In conclusion, we propose U. maydis as a valid model for studying sterol synthesis inhibitors and as a target for antifungals. As mentioned before, the need to uncover drugs for both instances is urgent and being able to test them in a non-mammal background as a first approach will favor the possibility of taking those Um-Hmgr inhibiting drugs to the next level of study.
Conflict of interestThe authors declare that they have no conflict of interests
This work was supported by grants from the Instituto Politécnico Nacional [20195606, 20195228, 20180198, 20170902, 20181873, 20171553, 20171714, 20161403, 20161245, 2016791] and CONACyT CB-283225 y CONACyT-178319. BRA was the recipient of fellowships from CONACyT and BEIFI-IPN. JAI, JT, CHR and LVT received support from COFAA-IPN, EDI-IPN and SNI CONACyT. JAI and LVT were hired through the “Programa Institucional de Contratación de Personal Académico de Excelencia IPN”.