Roseomonas is rarely involved in pathology but represents an important case of contamination when is associated to hematopoietic stem cells, having intrinsic resistance to multiple classes of antibiotics and the ability to gain new resistance factors. This is a report that follows a previous identification of contamination microorganisms in the cryopreserved Umbilical Cord Blood Units (UCBU) stored in the Cord Blood Bank (CBB) of the National Center of Blood Transfusion (NCBT) at Mexico City.
ObjectivePhenotypic and molecular characterization of Roseomonas genomospecies 5 isolated from UCBU.
Materials and methodsPhenotypic enzyme diffusion tests in solid phase (proteases, amylases, hemolysins and lipases detection) and antimicrobials (for Gram negative bacteria) resistance tests were performed to determine the potential virulence and resistance of the strain isolated of the Umbilical Cord Blood Unit 2191. Additional PCR assays were performed to determine the presence of genetic elements associated to antimicrobial resistance: bla genes and Class 1 integrons. Also, a 16S rRNA gene sequence analysis was done on microbial strains isolated from UCBU.
ResultsBroad-spectrum penicillins, third generation cephalosporins, and fluoroquinolones did not show inhibitory activity in the 2191 strain. We could only identify extracellular amylase activity. Gene detection by PCR of encoding antimicrobial resistance (β-lactamases) and Class 1 integrons revealed the presence of bla–HSV and bla–TEM variants in the 2191 strain. Phylogenetic analysis revealed the presence of Roseomonas genomospecies 5 in the 2191 UCBU (named 2191 strain).
ConclusionsThis is the first report on the isolation of Roseomonas genomospecies 5 in a UCBU for transplantation, an unusual bacteria isolated from umbilical cord blood, associated with a possible immunosuppression in the donor. Its presence in UCBU can be fatal in immunocompromised patients if it were used for transplantation of Hematopoietic Stem Cells (HSC), due to the potential virulence of the strains and the resistance to antimicrobials commonly used.
Roseomonas es poco común y normalmente no es patógena, sin embargo, representa un caso importante de contaminación cuando se asocia al transplante de Células Troncales Hematopoyéticas, debido a que presentan resistencia a múltiples clases de antibióticos y la habilidad de ganar nuevos factores de resistencia. Este reporte es la continuación de la idenficación de los microorganismos contaminantes en las Unidades de Sangre de Cordón Umbilical (USCU) criopreservadas en el Banco de Sangre de Cordón (BSC) del Centro Nacional de la Transfusión Sanguínea (CNTS) en la ciudad de México.
ObjetivoCaracterización fenotípica y molecular de Roseomonas genomospecies 5 aislada de USCU.
Materiales y métodosSe realizaron pruebas fenotípicas y de difusión enzimática en fase sólida (detección de proteasas, amilasas, lipasas, hemolisinas) y antimicrobianos (para bacterias Gram negativas) para determinar el potencial de virulencia y resistencia de la cepa aislada de la unidad de sangre de cordón umbilical 2191. Adicionalmente se realizaron ensayos de PCR para determinar la presencia de elementos genéticos asociados a la resistencia a antimicrobianos: genes bla e Integrones de Clase 1. También se realizó un análisis de la secuencias del gen 16S rRNA de las cepas aisladas de USCU.
ResultadosLas penicilinas de amplio espectro, cefalosporinas de tercera generación y las fluoroquinolonas no mostraron actividad inhibitoria en la cepa 2191. Sólo se identificó la actividad amilolítica extracelular. La detección de genes mediante PCR reveló la presencia de dos genes codificantes a β-lactamasas (variantes bla-HSV y bla-TEM) y no se detectaron elementos de Integrones de Clase 1 en el genoma de la cepa 2191. El estricto análisis filogenético reveló la presencia de Roseomonas genomospecies 5 en la USCU 2191 (nombrada cepa 2191).
ConclusionesEste es el primer reporte sobre el aislamiento de Roseomonas genomospecies 5 en USCU para el trasplante; una bacteria inusual aislada de USCU, asociada con una posibe inmunosupresión en el donante. Concluyendo que la presencia de Roseomonas genomospecies 5 en USCU puede ser fatal en pacientes inmunocomprometidos si fueran utilizadas para el trasplante de células troncales hematopoyéticas, debido a que esta cepa presenta potencial de virulencia y resistencia a los antimicrobianos utilizados comúnmente.
Roseomonas is a Gram-negative, pink pigmented, aerobic, slow-growing alpha-proteobacteria that has been isolated from aquatic and air environments, sludge and clinical samples such as blood, wound, urinary and respiratory specimens, peritoneal dialysis fluid, corneal scrapings and bones.1–4Roseomonas usually infects immunocompromised patients, such as those with leukemia, cancer chemotherapy, malignancy, sepsis, peritonitis, catheter associated infections and pulmonary tuberculosis.1,5,6 Some reports describe that the sources of infection are water and soil that infect patients with poor health practices.5,7,8 On the other hand, its virulence has been associated to the “mucoid phenotype” and it is well known that Roseomonas produces a biofilm on foreign materials inside the human body.9 Bacterial infection represents one of the most important risks associated to organ transplantation, including Hematopoietic Stem Cells (HSC) obtained from Umbilical Cord Blood (UCB).10,11 Patients with immunosuppression and onco-hematological disorders are susceptible to develop bacteremia by microorganisms that can be found in the UCBU. In order to ensure UCBU sterility, National Center of Blood Transfusion (NCBT) has a strict quality control, including a microbiological monitoring along the different procedures that involve UCB collection until its transplantation.3 It is important to point out that the units that are positive to microbiological culture are discarded for transplantation. The presence of bacterial contaminants in the Cord Blood (CB) might be due to (a) inadequate collection of CB in the operating room; (b) cross-contamination during CB processing (hematopoietic stem cells separation) and (c) by bacteremia or undetected infections in the donor.12,13 A disadvantage of microbiological controls (aerobic and anaerobic) from CB is the incubation temperature (37°C) of blood culture bottles, which restricts the growth of psychrophiles and facultative psycrophiles bacteria and limits the detection of contaminants such as fungi and yeast; another disadvantage is the culture medium composition that limits a great number of microorganisms. This generates a deviation in the detection of contaminants that may grow in the UCBU and produce post-transplantation bacteremia in the patient. Therefore, it is crucial to have an efficient contamination detection method, as well as pathogen-free donor. National Center of Blood Transfusion (NCBT) selects UCB donors reviewing their medical history and evaluating parameters such as history of isoimmunization, chronic diseases, drug addiction, congenital diseases, chromosomopaties, sexually transmitted diseases, risk activities, surgery, tattoos, perforations, transfusions, dental treatments, allergies, infectious and diseases, among others. In this paper we report the isolation, phenotypic and molecular characterization of Roseomonas genomospecies 5, an unusual bacterial contaminant associated with immunocompromised patients that was founded in UCBU (named 2191) in the CBB of the NCBT for transplantation.
Materials and methodsCollection and processing of 2191 UCBAn UCBU (2191) was collected in Morelos state, Mexico, after a vaginal delivery with placenta in situ by gravity in a closed bag system mixed with 20mL of Citrate Phosphate Dextrose (CPD) (Grifols, Spain). A nurse carried out the collection of UCB after Cord Umbilical (CU) antisepsis with 70% isopropanol, and 10% povidone iodine (the donor previously signed informed consent in order to use the donated umbilical cord blood for research purposes, if not used for transplantation). UCBU (2191) was analyzed for compliance with inclusion criteria such as minimum volume collected, Total Nucleated Cells count with the equipment ABX-Micros60 Horiba, CD34+ cell count by flow cytometry (FACS Calibur, BD Biosciences, USA), typing of HLA class I and II (One Lamba, E.U), blood group ABO and the Rh system. Infectious disease testing HBsAg, anti-HBc, anti-HIV, anti-HCV, anti-CMV-IgG and IgM, Chagas and Syphilis were done by chemiluminescence serology (Architect, Abbott Diagnostics, Germany). Using equipment Sepax-100 (Biosafe America Inc.) volume reduction to 20mL was performed and collected in a bag (Baxter Healthcare Corporation) adding 5mL of DMSO using automated equipment Coolmix-210 (biosafe America Inc.). The freeze bag was sealed and placed in an aluminum canister and cryopreserved in an automated tank (BioarchiveTM System TG 3626) to −196°C in liquid phase nitrogen.
Microbiological techniquesFor microbiological quality control, 5mL of the final UCBU (2191) was inoculated into the FA (FAN aerobic) and FN (anaerobic FAN) bottles (bioMérieux, Germany), respectively. Over seven days incubation, bottles that exhibited a positive signal of contamination by the unit monitor of the BacT/ALERT 3D system were included in this study. Bottles with positive signs were subcultured in solid media: 5% sheep blood agar, chocolate agar, Eosin Methylene Blue agar, mannitol salt agar, Pseudomonas agar, Trypticase Soy Agar (TSA) and Sabouraud Dextrose Agar (SDA). The plates were incubated aerobically and anaerobically (gas-pack system “BD GasPakTM EZ Gas Generating System”) at 37°C and 30°C for 24–48h and 28°C for 24–72h (only for SDA plates). Subsequently, 2191 UCBU strain was purified in the TSA for future tests. A Gram stain test was performed followed by the biochemical reactions, primarily in VITEK-2® (36 substrates tested) (Biomeriux, France) and results were read after 48-h post-incubation.
Antibiotic resistance and virulence factorsResistance of 2191 strain to antibiotics was determined using disk diffusion method on Mueller–Hilton agar according to recommendations of “the clinical and laboratory standards institute (CLSI)”.5Pseudomonas aeruginosa ATCC 25923, Escherichia coli ATCC 25922 and Staphylococcus aureus ATCC 25923 were used as controls. Results were interpreted as susceptible or resistant by measuring the diameter of inhibition zone according to the criteria stipulated by the CLSI.14 For the detection of virulence factors, 2191 strain was grown on TSA and two colonies were suspended in 2mL of LB-broth. The density of cellular suspension was adjusted to 1×109CFU/mL in a spectrophotometer 3000 [SmartSpecTM Plus (BIO-RAD)] at 600nm. Cell suspension was placed on various substrates by dripping for the phenotypic determination of virulence factors (proteases, amylases, nucleases, hemolysins and lipases). All strains were tested by triplicate.15
Molecular biology assaysPolymerase Chain Reaction (PCR) of the 16S rRNA gene were performed with universal primers 27F (5′-AGA GTT TGA TCM TGG CTC AG-3′) and 1492R (5′-TAC GGY TAC CTT GTT ACG ACT T-3′) using conditions recommended by De Santis et al.16 The bla genes (TEM, SHV, CTX-M, FOX and ACC) and Class 1 integrons (IntI-dfrA16-qacEΔ1/sulI) were detected by PCR as previously reported.17–20Aeromonas salmonicida (pRAS1) was used as the control for Class 1 integrons. 16S rRNA amplicon was sequenced by the Biology Institute of “Universidad Nacional Autónoma de México” using an ABI PRISM® 310 Genetic Analyzer sequencer (Applied Biosystems, USA). The nearly complete 16S rRNA gene sequence (1361bp) was determined as described by Kwon et al.21 Phylogenetic analysis was carried out by maximum-likelihood after a multiple alignments of data by Thompson et al.22 Phylogenetic tree was built based on maximum-likelihood phylogenetic analysis of 16S rRNA gene sequences available from GenBank. Finally, the sequence obtained was deposited in the GenBank.
ResultsCollection and processing of 2191 UCBUThe donor was a Mexican female, 32 years, clinically healthy, with three gestations and two childbirths without a history of isoimmunization, chronic diseases, drug addiction, congenital diseases, chromosomopaties, sexually transmitted diseases, risk activities, surgery, tattoos, perforations, transfusions, dental treatments, allergies or infectious diseases. The donor gave birth to a clinically healthy male by normal delivery. A final volume of 156.2mL of CB was collected successfully. The bag containing the CB was transported at room temperature to CBB from the NCBT in a Styrofoam container for processing. The following cell parameters were identified from the 2191 UCBU: white blood cells 13.6×108 and 7.6×108 initial and final, respectively. Resulting in a recovery percentage of 55.8% and 4.03×106 cells CD34+. These cell parameters are considered suitable for cryopreservation and future transplantation. Infectious markers were negative for infectious serology and agglutination in maternal and cord plasma.
Isolation and presumptive identification of the 2191 strainThe microbiological control was positive in both bottles (aerobic and anaerobic). The time of detection of the initial positive culture was 60.96h for aerobic bottles and 87.84h for post-incubation anaerobic bottles. Regular microbiology techniques were performed to obtain pure cultures for biochemical and genetic identification of 2191 isolate. The UCBU (2191) strain exhibited a colonial morphology similar to the previously reported for genus Roseomonas: 4–5mm in diameter, pink pigmentation non-diffusible (on chocolate agar plate), bright, creamy, humid appearance, smooth edge and convex colonies. Gram stain was gram-negative, coccoid and sometimes chaining. The biochemical reactions were performed in VITEK-2® system which revealed that only 7 substrates out of 36 were assimilated by the organism with a 91% probability assignment (l-proline-arylamidase, tyrosine arylamidase, urease production, use citrate, lactate and succinate alkalinization and production of sulfhydryl “ELLMAN test”). The analysis of biochemical tests by the automated system assigned the level Roseomonas to UCBU 2191 strain. Therefore, the analysis of 16s rRNA gene sequence was done.
Antibiotic resistance and virulence factors of the 2191 strainThe UCBU 2191 strain was susceptible to gentamicin, amikacin, netilmicin, trimethoprim/sulfamethoxazole, nitrofurantoine and chloramphenicol but resistant to ampicillin, cephalotin, norfloxacin, ciprofloxacin and cefotaxime. The determination of virulence factors showed that UCBU (2191) strain is only capable of starch hydrolyzing due to the production of extracellular amylase.
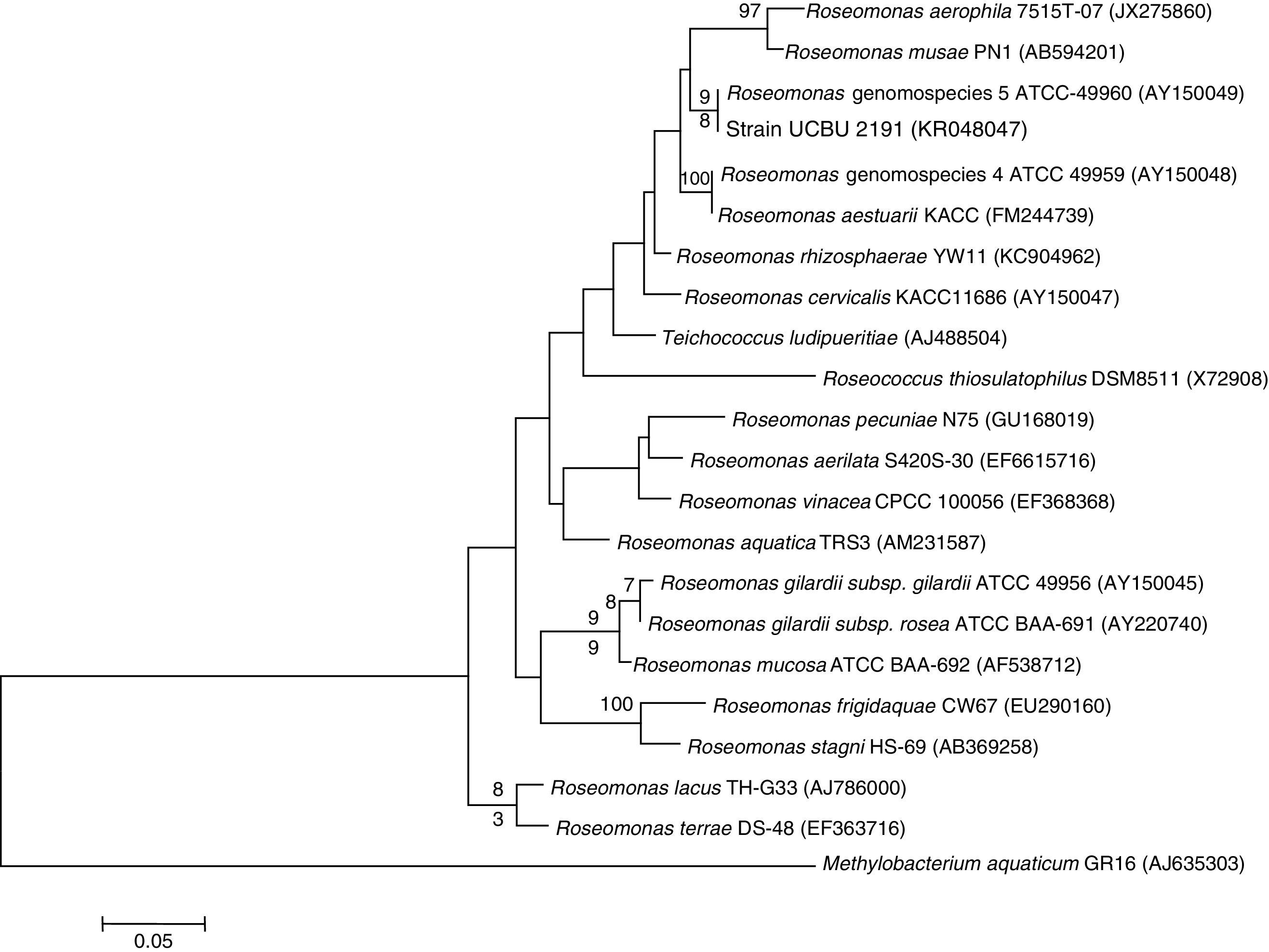
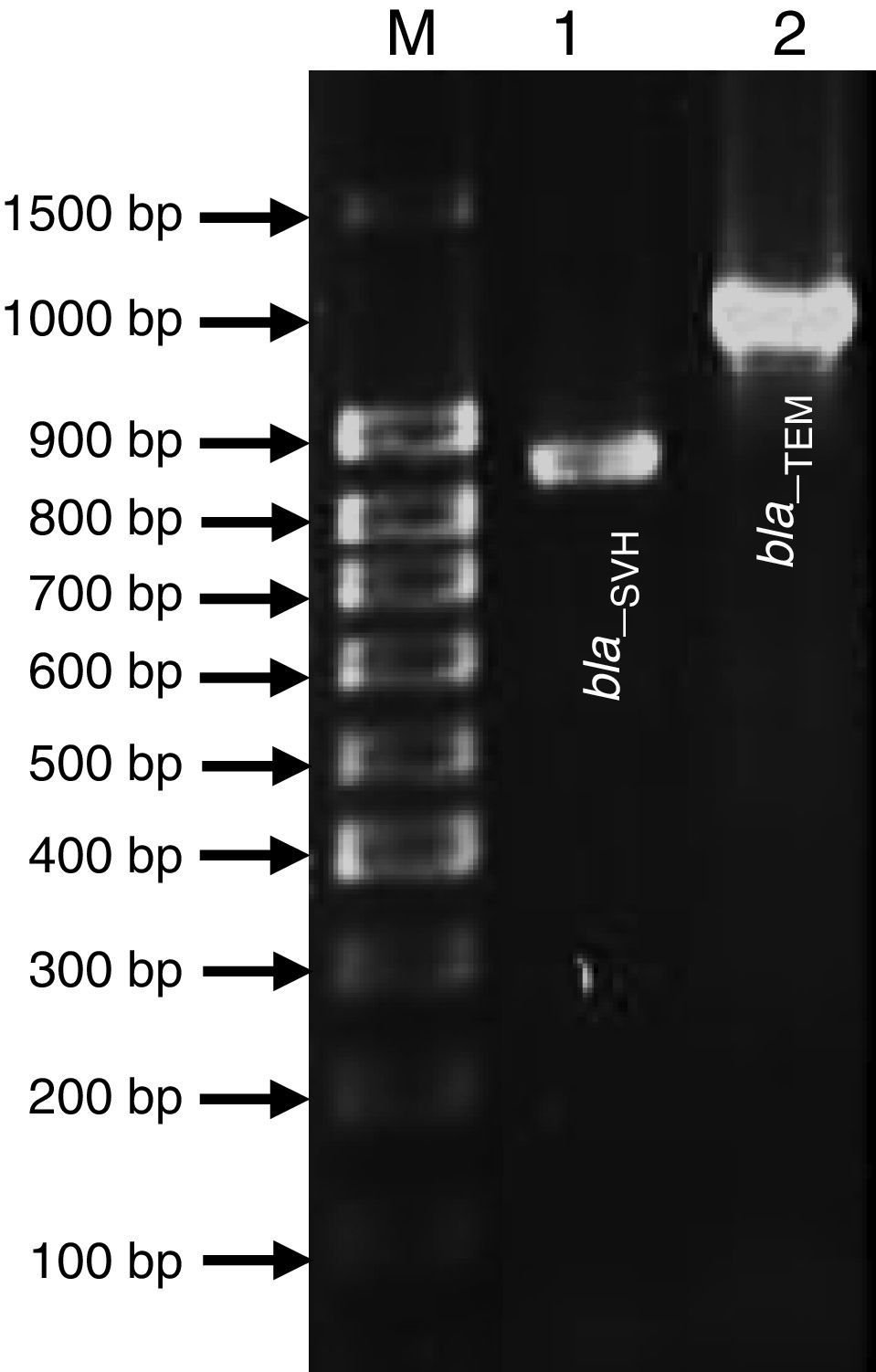
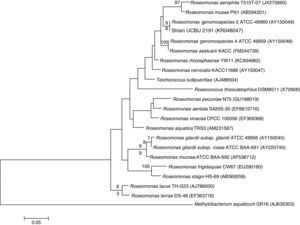
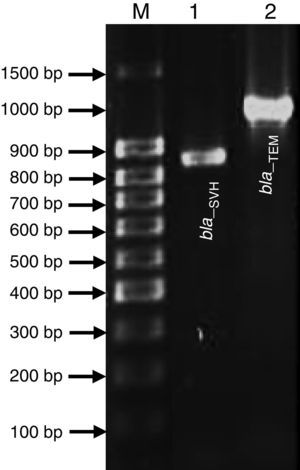
Genetic Identification, phylogenetic analysis and gene detection of 2191 strainPhylogenetic analysis based on the 16S rRNA gene and maximum-likelihood indicated that UCBU 2191 was closely related to Roseomonas genomospecies 5 ATCC 49960 (AY150049) (Fig. 1), with 99.99% of similarity. Based on these results, it is concluded that 2191 strain belongs to the Roseomonas genomospecies 5. The assigned accession number in GenBank for 16S rRNA sequence of 2191 strain was KR048047.1. PCR assays revealed the only presence of bla–TEM and bla–SHV in the genome of 2191 strain. Amplicons molecular sizes 1018bp and 858bp were detected for bla–SHV variants and bla–TEM, respectively (Fig. 2).
Phylogenetic tree of Roseomonas genomospecies 5 2191 UCBU and related taxa based on maximum-likelihood phylogenetic analysis of 16S rRNA gene sequences available from GenBank (accession numbers in parentheses). Bootstrap values based on 1000 replications are listed as percentages at the branching points.
Bacterial contamination can affect any blood component including CB and their detection in CB becomes important due to its therapeutic use (transplant). Therefore, it must be ensured that any component of blood, including CB, is free of contaminants. An important aspect is that cord blood units that show evidence of positive microbiology are excluded for purposes of transplantation. Prior to UCBU transplantation, the patient receives immunosuppression treatment to prevent graft versus host disease. However, this immunosuppression can generate sepsis by opportunistic bacteria or potentially virulent pathogens. In a previous work,3 the most frequently isolated strains in Umbilical Cord Blood Units were Enterococcus faecium, followed by Staphylococcus epidermidis, E. coli, Enterococcus faecalis, Staphylococcus haemoliticus, Klebsiella pneumoniae, Enterococcus durans, Lactobacillus helveticus, Enterococcus hiriae.3
The contaminants identified were mainly skin microflora, fecal and vaginal microflora of the donor. Roseomonas genomospecies 5 (a taxonomic group within the genus Roseomonas) was identified in UCBU 2191.15 Although Roseomonas is not considered a primary human pathogen, it has been isolated as commensal bacteria in immunosuppressed patients and young adults with sexually transmitted diseases, catheters, respiratory problems, wound infection or bone disease, peritonitis, enteritis and/or abdominal pain, kidney transplant infection, eye infection and middle-age women with one of several underlying conditions, including cancer and diabetes.24–27
This is the first work reporting the identification and phenotypic and molecular characterization of Roseomonas genomospecies 5, strain isolated from UCBU for transplantation. In a previous work, the acquisition of bacterial sepsis associated with the allogenic transplantation of hematopoietic stem cell in a patient with lymphoblastic lymphoma was reported.28 Therefore, Roseomonas has been recognized as the causative agent of sepsis associated with the transplantation of UCBU in an immunosuppressed patient. To our knowledge, this is the first report of early detection of Roseomonas genomospecies 5 in a UCBU as part of quality control in the manufacture of UCBU for transplantation.
This microorganism belongs to the Acetobacteriaceae family, a Gram-negative coccobacillus that develops pink-pigmented colonies.23 We presume that Roseomonas presence in immunocompromised patients is due to a process of immunosuppression that allowed this unusual pathogen to entry. Hence, it has been assumed that the organism had colonized her vaginal or enterical flora thus contaminating the CB at the time of delivery during collection. A non-thorough disinfection procedure could be also considered. Therefore, we consider the possible potential sources of contamination of the umbilical cord blood are surgical equipment, laboratory equipment during manufacture of the unit, and the possibility of maternal contamination during the collection of umbilical cord blood.
A previous work reported Roseomonas genomospecies 5 isolated as a commensal from urogenital specimens in young adults.29 Although serological infectious markers against viral and bacterial pathogens were negative in the donor, who could be indicative of a process of immunosuppression, it is known that during the gestation period, a process of suppression of the immune systems is present, which may favor the acquisition of opportunistic infections; this could be the case.
Rihs et al. divided the genus into six Roseomonas groups based on biochemical and DNA hybridization techniques.29 A consistent feature of this organism is its slow-growth properties on culture; it often takes 4–5 days before any growth is seen. Another common characteristic of Roseomonas is its antibiotic susceptibility pattern, especially in regard to its behavior with cephalosporins, which appears ineffective against any of the Roseomonas species.23 However, with the recent increase in horizontal transfer of DNA between bacterial strains and evidence of the presence of bla genes of enterobacterial origin in Roseomonas genomospecies 5 (2191 strain), the feature of antibiotic resistance is not absolute for Roseomonas. Recent studies demonstrate this phenomenon.17 Previous researchers have studied bacteria of the genus Roseomonas, focusing on the genomospecies 1, 2, 3, 4 and 6.
These strains have been assigned to species according to the origin of their isolation.3 Future research are needed for the assignment of species to taxonomic group 5 (genomospecies 5) as found in this experiment. Tests such as determinations of fatty acids, percentage of G+C, electronic microscopy are needed for the proposal of a new species to Roseomonas genomospecies 5. In spite of the strict quality control in the manufacture of UCBU, it is well known that some bacteria such as P. aeruginosa and Roseomonas itself require special culture characteristics. These include temperature (28–30°C) and incubation time, critical for their detection. Current automated equipment for the detection of contaminants in blood, blood components and CB are exclusively restricted to mesophilic microorganisms, excluding those that require lower temperatures for growth as in the case of Roseomonas.3,26,30,31
Bacteria possess virulence factors that are related to the invasion, replication, adhesion and evasion of host immune system, causing injuries during the pathogenesis of the disease. The amylolytic activity detected in Roseomonas genomospecies 5 can play an important role in the virulence of the pathogen. As an extracellular enzyme, Roseomonas genomospecies 5 can promote invasion of host tissues, breaking connective tissue and intercellular junctions, due to chemical compounds of this tissue like glycogen, starch, and complex sugars, as it has been observed in other pathogens.32 Several researchers have reported different virulence factors in P. aeruginosa and Aeromonas hydrophila such as extracellular amylases.15 The CBB of NCBT is governed by international standards in manufacturing UCBU for transplantation.
However, it is necessary to have scientific evidence that demonstrates the presence of microorganisms that are difficult to detect in CB, the latter in order to implement methods to identify molecular level genus and species of bacteria that hardly grow like Roseomonas, mainly to inadequate selection of the donor as well as emergence of pathogens associated with transplant UCBU that could be associated to wrong collection of CB in the operating room.
It is necessary to conduct a strict scrutiny to differ donors with a history of chronic diseases in which the immune system is depressed taking into account the medical history of the donor and its immune status. These actions will avoid UCBU contamination during collection by bacteria that are hard to detect.
Ethical disclosuresProtection of human and animal subjectsThe authors declare that no experiments were performed on humans or animals for this study.
Confidentiality of dataThe authors declare that no patient data appear in this article.
Right to privacy and informed consentThe authors declare that no patient data appear in this article.
Conflict of interestThe authors declare that they have no conflict of interests.