Among the risks associated with hematopoietic stem cells transplantation of umbilical cord blood are bacterial infections. This becomes dangerous when bacterial sepsis is related to resistance to antibiotics commonly used to eliminate of this type infections. Among the widely used antibiotics are those of the β-lactam family. Among the genes responsible for the resistance are designated “bla genes” which encode for broad-spectrum lactamases. Its detection in the laboratory shown the importance in the manufacture of cord blood units.
ObjectiveTo correlate the antibiotic resistance in bacterial contaminants isolated from Umbilical Cord Blood Units (UCBU), with detection of genes encoding resistance of β-lactamics antibiotics.
Materials and methodsTest of resistance to antibiotics was determined in 107 bacterial strains using disk diffusion method according to the recommendations of “The Clinical and Laboratory Standards Institute (CLSI)” and multiplex amplification by PCR of genes blaTEM, blaSHV and blaCTX-M were performed in order to detect genetic elements associated to antimicrobial resistance tested.
ResultsBroad-spectrum penicillins showed lower inhibitory activity on the tested strains. Gene detection by multiplex PCR of encoding antimicrobial resistance (β-lactamases) revealed the presence of bla–HSV, blaCTX-M and bla–TEM genes in most strains tested even when these genes have been reported in Enterobacteriaceae family members.
ConclusionsThe genotype detection encoding antibiotic resistance in isolates of UCBU clearly indicates the potential risk of sepsis difficult to eradicate in the patient, if present in a UCBU available for transplant. Additionally, the existence of these genes reveals horizontal transfer events of genetic material between bacteria. So the importance to realize this studies before transplant.
Entre los riesgos asociados con el trasplante de células progenitoras hematopoyéticas de sangre de cordón umbilical son las infecciones bacterianas. Esto se convierte en peligroso cuando la sepsis está relacionada con la resistencia a antibióticos comúnmente utilizados para combatir infecciones. Entre los antibióticos ampliamente usados son los de la familia β-lactámicos. Los genes responsables de la resistencia a estos antibióticos se designan “genes bla” codificantes para lactamasas de amplio espectro. Su detección en el laboratorio demuestra la importancia del control en la manufactura de unidades de sangre de cordón.
ObjetivoCorrelacionar la resistencia a los antibióticos en bacterias contaminantes aisladas de Unidades de Sangre de Cordón Umbilical (USCU), con la detección de genes codificantes a antibioticos β -lactámicos.
Materiales y métodosSe realizaron ensayos de resistencia a antibióticos en 107 cepas por el método de difusión en disco de acuerdo a las recomendaciones de “El Instituto de Estándares Clínicos y de Laboratorio (CLSI)” y amplificación por PCR multiplex de genes blaTEM, blaSHV y blaCTX-M.
ResultadosLas penicilinas de amplio espectro mostraron menor actividad inhibitoria en las cepas probadas. La detección de genes que codifican resistencia a antibióticos mediada por β-lactamasas reveló la presencia de variantes blaTEM, blaSHV y blaCTX-M en la mayoría de las cepas, incluso cuando estos han sido reportados en miembros de la familia Enterobacteriaceae.
ConclusionesLa detección del genotipo de resistencia a antibióticos en aislamientos de USCU indica claramente el riesgo potencial de sepsis de difícil erradicación en el paciente, si estuviesen presentes en USCU disponibles para trasplante. La existencia de genes bla revela eventos de transferencia horizontal de ADN entre bacterias. De allí la importancia de realizar estos estudios antes del transplante.
Transplantation of Umbilical Cord Blood (UCB) is one of the major technologies used to treat oncohematologic and non-oncohematologic disorders, since it is an invaluable source of Hematopoietic Stem Cells (HSC).1,2 The HSC transplantation has been used to treat diseases such as myeloblastic and lymphoblastic leukemia, primary immunodeficiencies, aplastic anemia, severe combined immunodeficiency, genetic and/or metabolic inherited diseases as mucopolysaccharidosis and leukodystrophies.3–5 However, the use and success of this type of transplantation is limited by several obstacles, mainly related to the donor-recipient compatibility, therefore the Human Leukocyte Antigen (HLA) typing becomes very important.6 It is well known that antigens of HLA Class I and Class II are the major immunological barrier to perform transplantation of HSC,7 therefore patients included in transplant protocols are submitted to immunosuppression processes in order to prevent graft rejection; a dangerous situation for the patient who is exposed to a broad range of opportunistic infections by bacteria that normally would be considered innocuous.8,9 The procedure of collecting UCB must be performed under conditions that ensure its complete sterility. The risk of bacterial contamination is present in any stage, beginning with the collection, processing and cryopreservation.10,11 Therefore, in Cord Blood Banks (CBB) that collect and process UCB, the use of aerobic and anaerobic microbiological controls (preferably automated) is mandatory at the end of the process in order to ensure the safety of the final product. Antimicrobial treatment in immunocompromised patients is essential for the recovery after transplantation of HSC. In case of bacterial contamination of the UCBU, there is a potential risk of sepsis when bacteria are resistant to antibiotics, making it difficult to erradicate with antimicrobial therapy. The presence of mobile genetic elements such as plasmids and transposons are associated directly with the acquisition of resistance to antibiotics in the nature.12 A predominance of negative and Gram-positive bacteria such as Escherichia coli, Klebsiella pneumoniae, Staphylococcus aureus, Enterococcus faecalis and non-fermentative bacilli such as Pseudomonas aeruginosa have been recognized as contaminats,10 and producers of β-lactamases, generating resistance to penicillins, cephalosporins and monobactams.13,14 Antibiotics of the β-lactamic family have been widely used in the treatment of bacterial infections. Due to the irrational and indiscriminate use of antibiotics in nosocomial institutions, selection pressure results in emergence of multiresistant strains.15 Strains producing β-lactamases are responsible of nosocomial outbreaks, especially in intensive care units, complicating treatment and increasing morbidity and mortality.16 In a previous study, bacterial contaminants isolated from UCBU of CBB from National Center of Blood Transfusion for transplant were identified at molecular level; additionally the profile of antibiotic resistance and virulence factors were detected.17 However, additional tests were not performed, in order to know the genetic background of these strains and knowledge their possible acquisition of resistance genes. The aim of this work was research a correlation between pattern antibiotic resistances in bacterial strains isolated UCBU for transplant and the detection of genes coding for antibiotic resistance using Polymerase Chain Reaction (PCR) directed against β-lactamics genes.
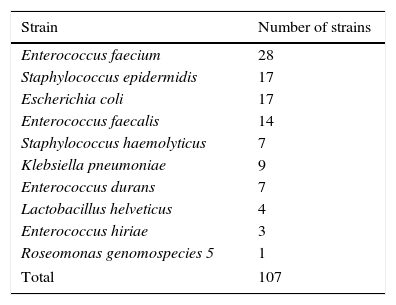
Material and methodsOrigin of the bacterial strainsThe bacterial strains used in this study are listed in Table 1. All strains were isolated between 2003 and 2013 from discharged UCBU of the NCBT in Mexico City.17 16S rRNA gene was amplified in all strains using the universal primers (27F and R1492) for bacteria and was sequenced. Additionally, ERIC–PCR assays to differentiate between unique and clones were done. All strains grew in Luria-Bertani (LB) broth (10g/L tryptone, 5g/L yeast extract, 5g/L NaCl), or LB agar (supplemented with 1.5% agar).
Strains used in this study.
| Strain | Number of strains |
|---|---|
| Enterococcus faecium | 28 |
| Staphylococcus epidermidis | 17 |
| Escherichia coli | 17 |
| Enterococcus faecalis | 14 |
| Staphylococcus haemolyticus | 7 |
| Klebsiella pneumoniae | 9 |
| Enterococcus durans | 7 |
| Lactobacillus helveticus | 4 |
| Enterococcus hiriae | 3 |
| Roseomonas genomospecies 5 | 1 |
| Total | 107 |
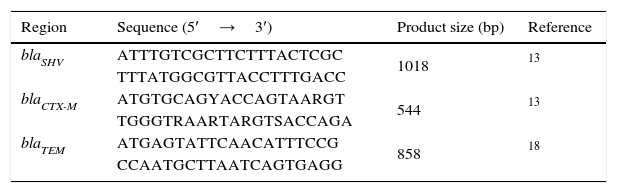
Genomic DNA was extracted using the QIAamp DNA Mini QIAcube Kit (QIAGEN, Germany). The reactions were performed in a Touchgene Gradient thermal cycler FTGRAD2D (TECHNE DUXFORT, Cambridge, UK) using MasterMix PCR 1×(Roche Diagnostics, Germany), 200pmol of each primer and 200ng of template DNA. Polimerase Chain Reactions of blaTEM, blaSHV, blaCTX-M genes were conducted in multiplex amplification using the conditions recommended by Jemima and Verghese and Tofteland et al. (Table 2).13,18 PCR products were run in 1×TBE buffer, pH 8.3 on horizontal electrophoresis in 1% agarose gels, visualized and photographed under UV illumination.
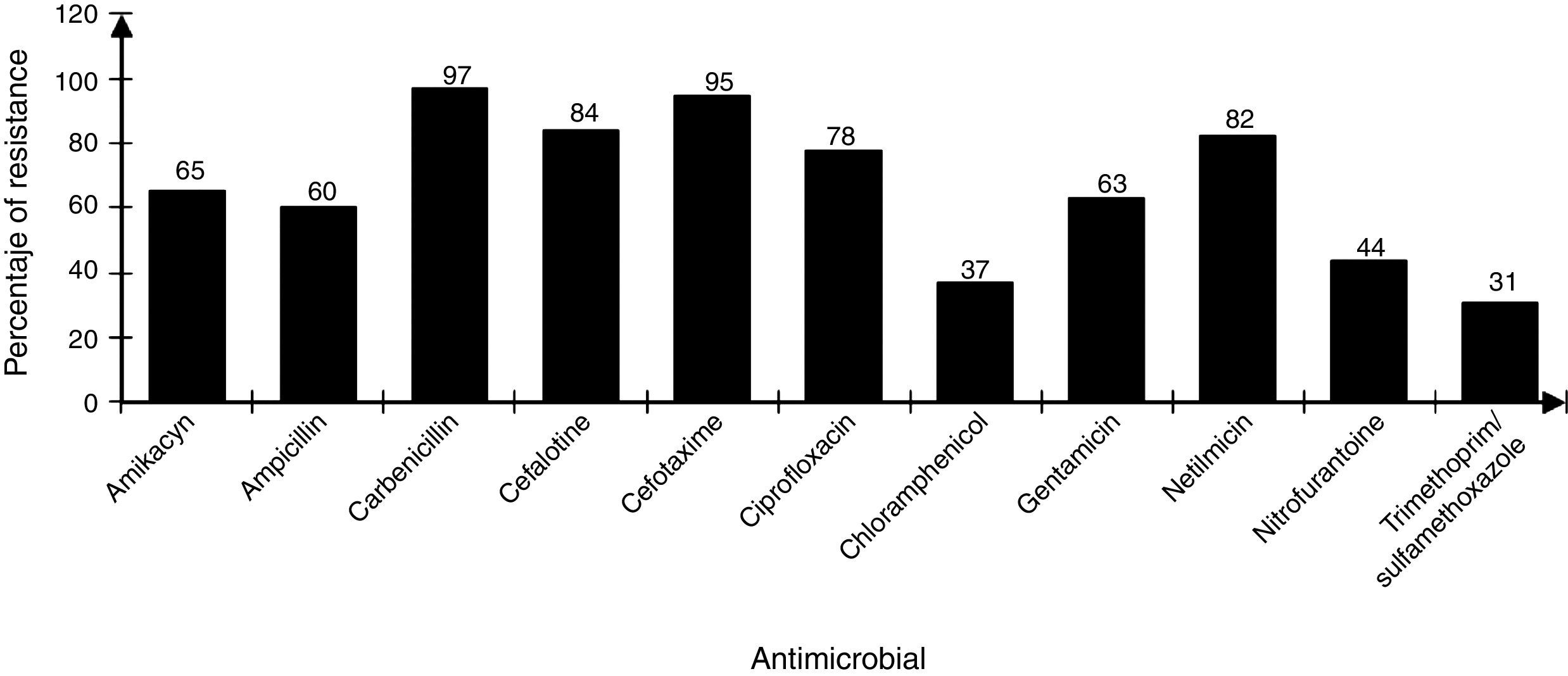
Sensibility/resistance assaysResistance/sensibility of all strains to different antibiotics were determined using the disk diffusion method on Mueller–Hilton agar plates according to the recommendations of the Clinical and Laboratory Standards Institute (CLSI).19 The antibiotics used were: amikacyn (30μg), ampicillin (30μg), carbenicillin (100μg), cefalotine (30μg), cefotaxime (30μg), ciprofloxacin (5μg), chloramphenicol (30μg), gentamicin (10μg), netilmicin (30μg), nitrofurantoine (300μg) and, trimethoprim/sulfamethoxazole (25μg). Antibiotics used in this test are those that are commonly prescribed prophylactic against Gram positive and Gram-negative bacteria. Pseudomonas aeruginosa ATCC 27853, E. coli ATCC 25922 and S. aureus ATCC 25923 strains were used as controls. Results were interpreted as susceptible or resistant by measuring the diameter of inhibition zone according to the criteria stipulated by the CLSI.26
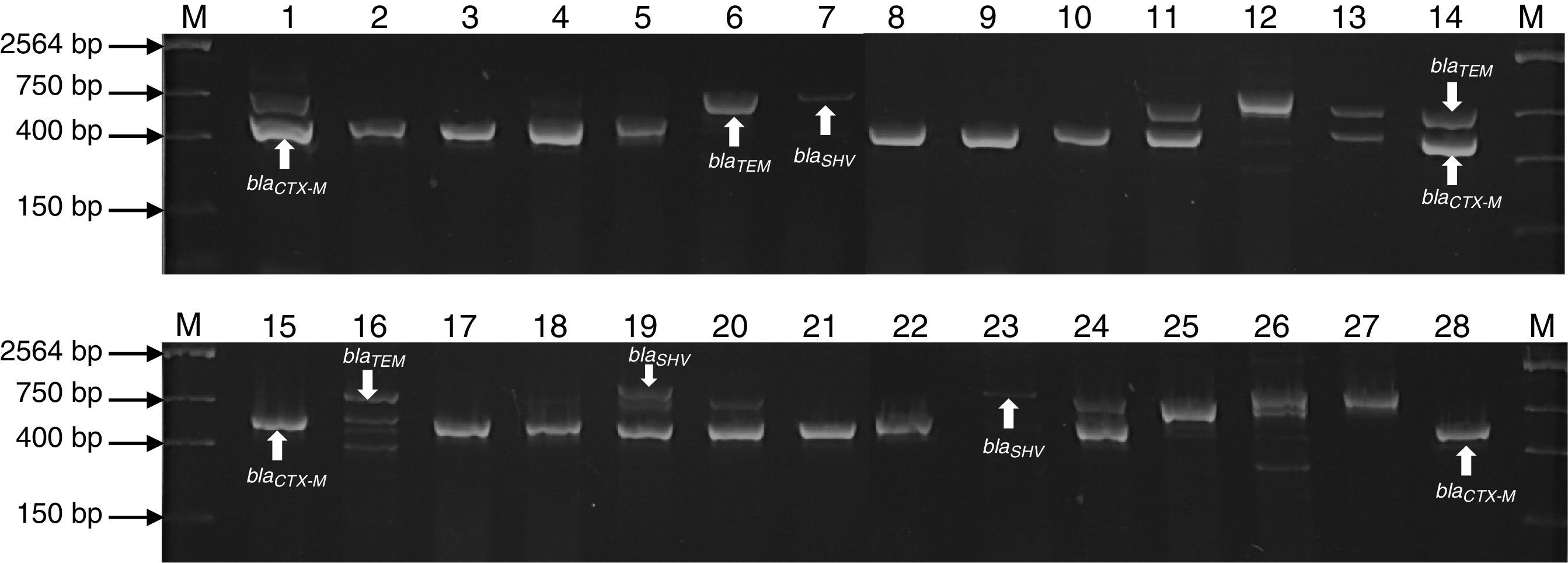
ResultsMultiplex amplification of genes blaTEM, blaSHV, blaCTX-MOne hundred and seven bacterial strains isolated of Umbilical Cord Blood Unit for hematopoietic transplant were identified by analysis of sequence of 16S rRNA and subjected to multiplex amplification by PCR in order to detect encoding genes β-lactamases (blaTEM, blaSHV, blaCTX-M). Twenty-eight (26.1%) strains were positive to blaSHV gene, 40 (37.3%) to blaCTX-M gene and 48 (54.2%) blaTEM gene. Twelve strains were positive (11.2%) to blaSHV/TEM, 3 (2.8%) to blaSHV/CTX and 10 (9.34%) to blaCTX/TEM. A summary of genotype detected in all strains is shown in Table 3. The genes were predominantly detected in Enterobacteriaceae (E. coli and K. pneumoniae), Staphylococcaceae (S. epidermidis and S. haemolyticus) and Enterococcaceae (E. faecium and E. faecalis) family members (Fig. 1). The ATTC strains (P. aeruginosa) ATCC 27853, E. coli ATCC 25922 and S. aureus ATCC 25923 were used as negative controls of amplification (data not shown).
Genotype detected of antibiotic resistance in bacteria strains isolated from Umbilical Cord Units for transplant.
| Tested strains | Detected genotype | ||||||
|---|---|---|---|---|---|---|---|
| bla variants detected | bla combinations detected | ||||||
| blaSHV | blaCTX-M | blaTEM | blaSHV/TEM | blaSHV/CTX | blaCTX/TEM | ||
| Enterococcus faecium | 28 | 5 (17.8) | 9 (32.1) | 10 (35.7) | 4 (14.2) | 0 (0) | 2 (7.1) |
| Staphylococcus epidermidis | 17 | 5 (29.4) | 4 (23.5) | 6 (35.2) | 1 (5.8) | 0 (0) | 2 (11.7) |
| Escherichia coli | 17 | 6 (35.2) | 10 (58.8) | 11 (64.7) | 1 (5.8) | 2 (11.7) | 1 (5.8) |
| Enterococcus faecalis | 14 | 2 (14.2) | 5 (35.7) | 6 (42.8) | 2 (14.2) | 0 (0) | 1 (7.1) |
| Staphylococcus haemolyticus | 7 | 2 (28.5) | 4 (57.1) | 3 (42.8) | 1 (14.2) | 0 (0) | 2 (28.5) |
| Klebsiella pneumoniae | 9 | 4 (44.4) | 4 (44.4) | 6 (66.6) | 2 (22.2) | 1 (11.1) | 1 (11.1) |
| Enterococcus durans | 7 | 2 (28.5) | 2 (28.5) | 3 (42.8) | 0 (0) | 0 (0) | 0 (0) |
| Lactobacillus helveticus | 4 | 0 (0) | 0 (0) | 1 (25) | 0 (0) | 0 (0) | 0 (0) |
| Enterococcus hiriae | 3 | 1 (33.3) | 2 (66.6) | 1 (33.3) | 0 (0) | 0 (0) | 1 (33.3) |
| Roseomonas gensp 5 | 1 | 1 (100) | 0 (0) | 1 (100) | 1 (100) | 0 (0) | 0 (0) |
| Total | 107 | 28 (26.1) | 40 (37.3) | 48 (44.8) | 12 (11.2) | 3 (2.8) | 10 (9.34) |
R, resistant; S, sensible.
The numbers indicated in parentheses is the percentage (%) of bla-gene detection.
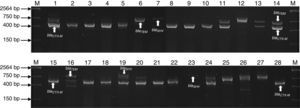
Agarose gel electrophoresis representative of the PCR products of encoding genes β-lactamases (blaTEM, blaSHV and blaCTX-M) of bacterial strains isolated of UBCU for transplant. Lanes: M: Molecular size marker (One lambda) Thermo Fisher Scientific, Brand, 1–28: PCR products of blaTEM (858bp), blaSHV (1018bp), blaCTX-M (544bp) in some strains.
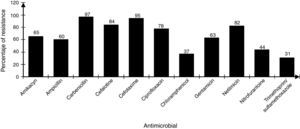
The strains showed differences in susceptibility and resistance to the antimicrobials tested. Sulfonamides, folate antagonists, nitrofurans, and first generation of cephalosporins had the best antimicrobial activity. The broad-spectrum penicillins, third generation of cephalosporins, aminoglycosides, and fluoroquinolones showed lower activity (Fig. 2).
Statistical analysisFor statistical analysis, the following was considered: null hypothesis (Ho)=independent variable (presence of bla genes in different strains), alternative hypothesis (H1): dependent variable (bla genes related to the strain or type of bla gene). Considering p>0.05, the null hypothesis is accepted and the alternative hypothesis is rejected. p values of 0.242 (blaSHV), 0.242 (blaCTX-M) and 0.254 (blaTEM) were calculated. As in all cases the result was greater than 0.05, therefore be concluded that the results are significantly different at 95% confianze, indicating that there is no relationship between bacterial genera and the presence of the bla genes.
DiscussionThe contamination of umbilical cord blood for transplant in the majority of cases occurs at the time of collection and processing or thawing of hematopoietic stem cells prior to transplantation.17 In general, the increased incidence of contamination in placental blood occurs in proportion with the increase in handling steps of the final UCBU, such as removing (purging) the excess red blood cells by not automated means and cryopreservation procedures, between others.20,26 The bacterial strains analyzed in this work are mainly family members Enterobacteriaceae, Enterococcaceae and Staphylococcaceae; therefore the source was: digestive tract and skin flora respectively. This allows the identification of critical points, from the time of collection of the UCB in the operating room to prevent bacterial contamination. Valuable information about the strains identified in UCBU for transplant, is related to resistance to antibiotics since from this depends the patient safety. As mentioned, before patients exposed to transplant protocols with HSC are subject to a preconditioning program (immunosuppression) to prevent graft rejection. Therefore, the patient is exposed to opportunistic infections caused by bacteria that are generally innocuous, potentially pathogenic or virulent. Knowledge about this resistance and its genotype, provides an opportunity to act efficiently in an emergency providing effective antimicrobial treatment. The identification of resistance phenotype by disk diffusion method and genotype by multiplex amplification by PCR in all strains, showed the presence of genes in some strains that are phylogenetically distant from the family Enterobacteriaceae (Enterococcaceae, Staphylococcaceae, Acetobacteraceae and Lactobacteriaceae). The presence of phylogenetically distant genes in bacteria has been previously reported; we speculate that the presence of these genes reported in the Enterobacteriaceae family is due to events of horizontal transfer of genetic material between bacteria by specific mechanisms (conjugation, transduction and transformation). These phenomenons are not new in their nature and less on bacteria enteric origin, where it is known that DNA transfer is incredibly high and clearly explains the presence of DNA transfer events. Previous studies demonstrated the transfer of bacterial DNA in the human digestive tract, therefore we suppose that this phenomenon is the reason of the presence of this type of genes in the isolates. Resistance to β-lactamics antibiotics is mainly attributable to the production of β-lactamases enzymes. The β-lactamases spread spectrum enzimes (ESBL) represent today a great risk due to a broad spectrum to antibiotic inactivation usually available for the treatment of infections. These strains producing β-lactamases exhibit hydrolytic activity against penicillin and cephalosporins. Since its initial description more than 300 different ESBL have been identified, and most of them belong to the TEM, SHV and CTX-M families (Paterson et al., 2005).21 The analysis of the presence of blaTEM, blaSHV, blaCTX-M genes in 107 bacterial strains isolated from UCBU indicated the presence of blaTEM gene (44.8%) followed by blaCTX-M (37.3%) and finally blaSHV (26.1%). These results are comparable to those obtained in tests of sensitivity/resistance and show that cefotaxime is the least effective antibiotic against these strains (95%) (Fig. 2); this is a third generation cephalosporin with an oximino group, being the most susceptible to inactivation by the β-lactamases.22 In the case of β-lactamics, ampicillin has a low level of activity, 60% of strains were resistant to this compound; bla gene has been reported as being responsible for ampicillin resistance in E. coli and other enterobacterias.17 Moreover aminoglycosides have limited activity against most ESBL producing strains being gentamicin the least active within this family (63%).23 In the case of amikacin, another aminoglycoside, antibiotic presented greater activity and only 65% of the strains were resistant. Similar results have been reported to amikacin before.24 The emergence of such resistance is mainly due to over-consumption of antibiotics, as well as the natural existence of genetic material and their acquisition by horizontal transfer among bacteria, this is the case of the presence of encoders as TEM, SHV and CTX-M β-lactamases.25 The presence of genes blaTEM, blaSHV, and blaCTX-M was demonstrated in all strains tested. It was determinated that third-generation cephalosporins and β-lactamics such as cefotaxime and ampicillin, respectively, showed low effective activity against the strains tested. Instead, chloramphenicol and amikacin proved less susceptible to inactivation by β-lactamases. The reduction in the spread of resistance genes mediated requires the identification of the genes involved with the purpose of controlling the movement of this mechanism of resistance.
Ethical disclosuresProtection of human and animal subjectsThe authors declare that no experiments were performed on humans or animals for this study.
Confidentiality of dataThe authors declare that no patient data appear in this article.
Right to privacy and informed consentThe authors declare that no patient data appear in this article.
Conflict of interestThe authors declare that they have no conflict of interests.