Cytochrome P450 2C9 (CYP2C9) is an important enzyme in the metabolism of many drugs, including NSAIDs, antidepressants and anticoagulants. In vitro and in vivo studies have demonstrated that CYP2C9 polymorphisms modify the activity of the enzyme and subsequently the metabolism of different drugs.
ObjectiveTo characterize the diclofenac pharmacokinetics in healthy subjects with the wild type form of CYP2C9 genotype.
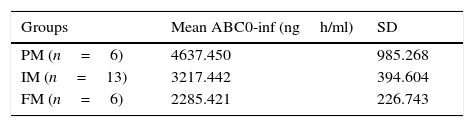
MethodsTwenty five healthy women were included in the study; a single dose of diclofenac (50mg) was administered orally. Pharmacokinetic analyses at 12 different times were performed. DNA was extracted from peripheral blood and analyzed through direct sequencing. The CYP2C9*1/*1 allele was found in all subjects. Using the AUC 0-inf parameter, we classified the 25 volunteers as poor, intermediate and extensive metabolizer (2285.421/3217.442/4637.450, respectively). We detected statistical significant differences between the groups, especially between poor metabolizers versus intermediate and extensive metabolizers.
ConclusionsThis data indicate that CYP2C9 is not the only enzyme responsible of the metabolism of diclofenac, so it is important to analyze other cytochromes and their variants potentially involved in the metabolism of this drug.
El citocromo P450 2C9 (CYP2C9) es una enzima importante en el metabolismo de muchos fármacos, incluyendo AINEs, antidepresivos y anticoagulantes. Estudios in vitro e in vivo han demostrado que los polimorfismos del CYP2C9 modifican la actividad de la enzima y posteriormente el metabolismo de diferentes fármacos.
ObjetivoCaracterizar la farmacocinética del diclofenaco en sujetos sanos con la forma silvestre del genotipo CYP2C9.
MétodosVeinticinco mujeres sanas fueron incluidas en el estudio; Se administró por vía oral una dosis única de diclofenaco (50mg). Se realizaron análisis farmacocinéticos a 12 tiempos diferentes. El ADN se extrajo de sangre periférica y se analizó mediante secuenciación directa. El alelo CYP2C9 * 1 / * 1 se encontró en todos los sujetos. Utilizando el parámetro AUC 0-inf, se clasificaron a los 25 voluntarios como metabolizadores pobres, intermedios y rápidos (2285.421 / 3217.442 / 4637.450, respectivamente). Se detectaron diferencias estadísticamente significativas entre los grupos, especialmente entre metabolizadores pobres frente a metabolizadores intermedios y rápidos.
ConclusionesEstos datos indican que CYP2C9 no es la única enzima responsable del metabolismo del diclofenaco, por lo que es importante analizar otros citocromos y sus variantes potencialmente implicadas en el metabolismo de este fármaco.
The interindividual variability in drug response is a major cause of adverse effects. In many cases, this variability is linked to polymorphisms of genes encoding enzymes responsible for the biotransformation of these drugs.1 Carriers of mutations in genes encoding drug metabolizing enzymes, when treated with standard doses of certain drugs, usually have higher plasma levels. They present lower clearance and an increase in the frequency and severity of adverse reactions secondary to the use of the drug.2 Cytochrome P450s superfamily (CYP) comprises enzymes involved in the metabolism of endogenous substances and biotransformation of drug.3
The CYP2C subfamily consists of 4 members clustered on chromosome 10q24 (Cen-CYP2C18-CYP2C19-CYP2C9-CYP2C8 – Tel) and constitute approximately 20% of all P450 enzymes in the liver. They also are expressed in other tissues such as kidney, intestine, brain, heart, lung and aorta. The CYP2C enzymes are well known to metabolize just over 20% of all drugs.4–9 To date, 33 different alleles have been described in CYP2C9. The most studied alleles CYP2C9 *2 (R144C) and CYP2C9 *3 (I359L) lead to a decrease in the enzymatic activity.6,9,10
Cytochrome P450 2C9 (CYP2C9) is an important enzyme involved in the biotransformation of many drugs, including non-steroidal anti-inflammatory drug (NSAIDs), antidepressants and anticoagulants.1 Diclofenac sodium is a NSAID, humans metabolize diclofenac in a great number of hydroxylated metabolites, the main metabolite in plasma and urine is 4′-hydroxy (OH) diclofenac whereas 3′-OH diclofenac and 5-OH diclofenac are minor metabolites.11 Apart from CYP2C9, other CYP2C enzymes such as CYP2C8 seem to be important in diclofenac metabolism.2 Studies in vitro and in vivo have demonstrated that CYP2C9 gene polymorphisms modify the enzymatic activity of CYP2C9 and alter the biotransformation of various drugs.7
The aim of this study was to characterize the pharmacokinetic of diclofenac in healthy women with the wild type form to exclude the exclusive participation of CYP2C9 in the metabolism of diclofenac.
Methods and subjectsTo prevent from gender bias, only women were included in the study. Twenty five unrelated healthy Mexican volunteers [mean age of 29±5 years and body mass index within normal parameters] were evaluated. Medical history was done in all subjects. Volunteers with history of adverse drug effects and those with any drug intake were excluded. The 25 women volunteers were nonsmokers and abstained from caffeine- or alcohol-containing beverages as well as foodstuffs from grapefruit during the course of the study. The pre-study health check consisted of physical examination, laboratory tests, including blood cell counts and hepatic function tests, urine analysis and an electrocardiogram. The subjects were informed about the aim of the study and they agreed to participate. The study was approved by the Ethics Committee of the General Hospital of Mexico.
To analyze the CYP2C9 gene, DNA was obtained from peripheral leukocytes under standard techniques. To perform PCR, oligonucleotides were designed considering the database NBCI of the CYP2C9 gene. The protocol was approved by the Ethics Committee of the General Hospital of Mexico.
Analysis concentrations of plasma diclofenacAfter a fasting period of 12h, the 25 volunteers were given a single oral dose of 50mg diclofenac sodium (Voltaren, Novartis, Switzerland). Blood samples were taken at 0, 0.33, 0.66, 1, 1.33, 1.66, 2, 2.5, 3, 4, 6 and 9h. Serum levels of diclofenac were assayed in the samples using a specific liquid chromatography, ultra high resolution coupled to tandem mass spectrometry. Mobile phase was: acetonitrile:water (62:38 v/v) Flow rate: 0.5ml/min, Column temperature: 35°C, autosampler temperature: 13°C, injection volume: 5μL, Column: Acquity UPLC C18. The method of acceptance criteria for the validation parameters is established in NOM – 177 – SSA1 199825, Mexico.
StatisticsDescriptive statistical analysis was performed on the sample of the 25 volunteers for the variables of sex, age, weight, height, BMI, systolic and diastolic blood pressure and heart rate. Later analysis with ABC variable 0–∞ was performed by ANOVA.
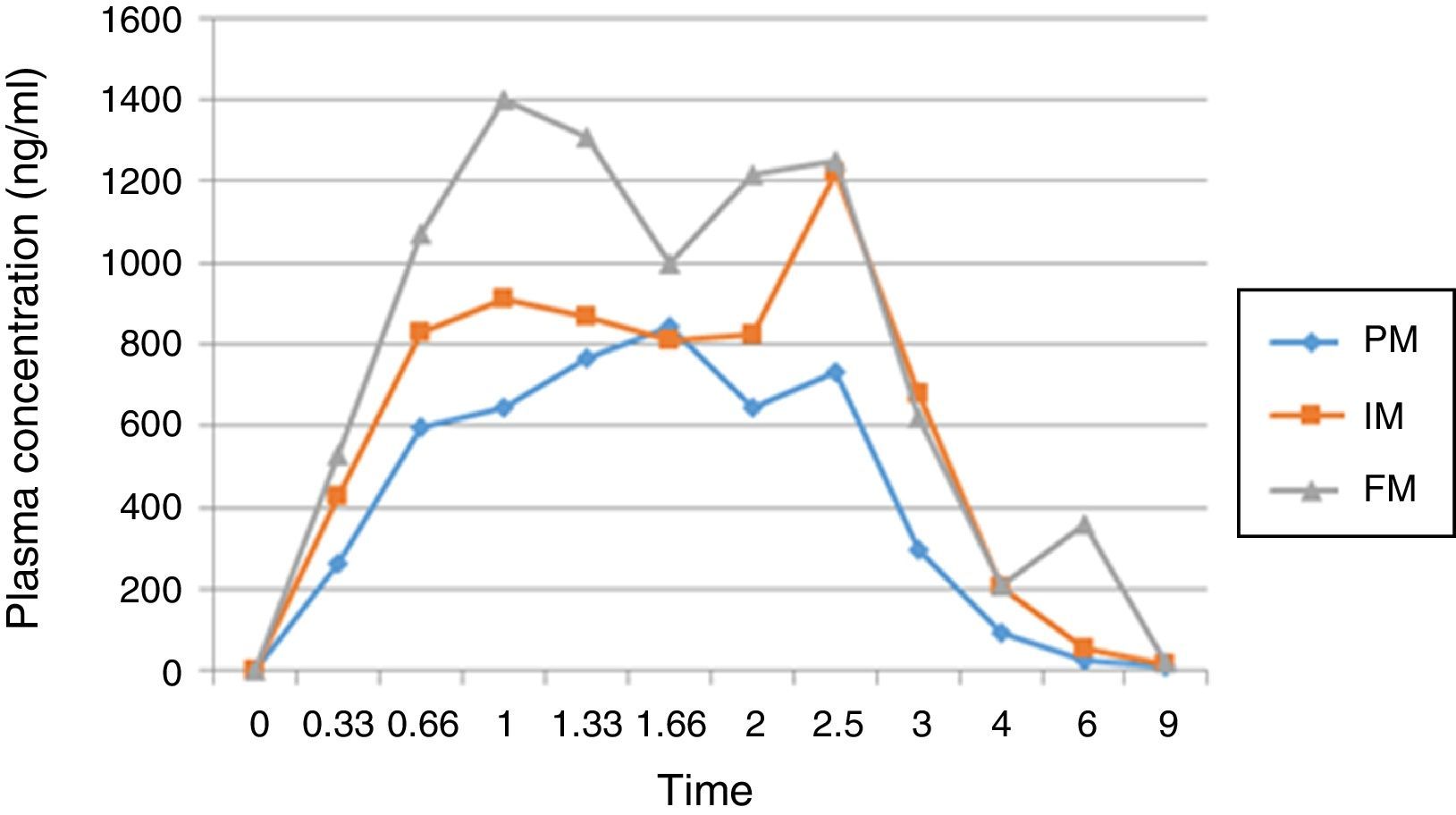
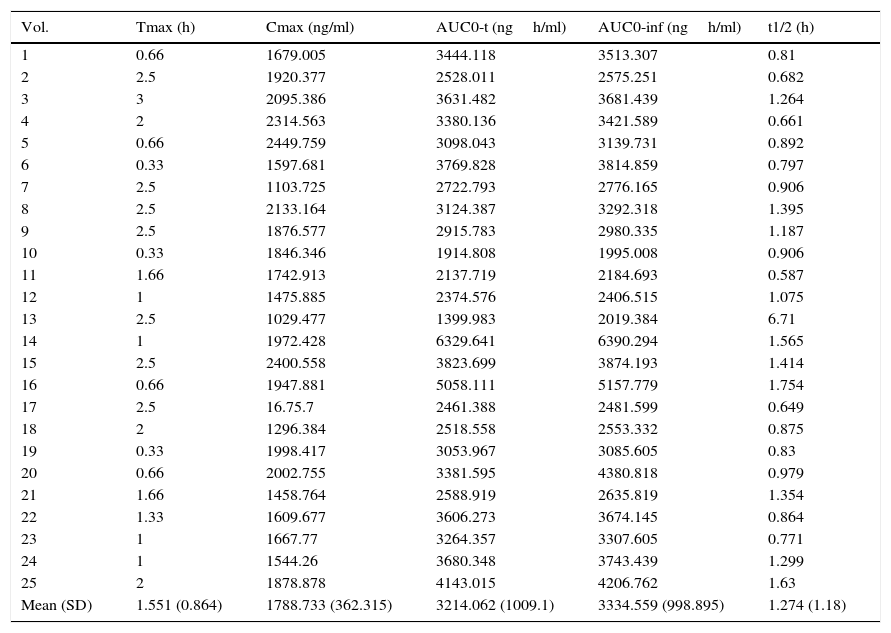
Results and discussionThe demographic data of 25 volunteers are shown in Table 1. Pharmacokinetic values are shown in Table 2 together with the mean and standard deviation. Using the values of ABC in infinite time, it was possible to make a classification between slow (ML), intermediate (MI) and fast (MR) metabolizers (Table 3).
Pharmacokinetic parameters.
| Vol. | Tmax (h) | Cmax (ng/ml) | AUC0-t (ngh/ml) | AUC0-inf (ngh/ml) | t1/2 (h) |
|---|---|---|---|---|---|
| 1 | 0.66 | 1679.005 | 3444.118 | 3513.307 | 0.81 |
| 2 | 2.5 | 1920.377 | 2528.011 | 2575.251 | 0.682 |
| 3 | 3 | 2095.386 | 3631.482 | 3681.439 | 1.264 |
| 4 | 2 | 2314.563 | 3380.136 | 3421.589 | 0.661 |
| 5 | 0.66 | 2449.759 | 3098.043 | 3139.731 | 0.892 |
| 6 | 0.33 | 1597.681 | 3769.828 | 3814.859 | 0.797 |
| 7 | 2.5 | 1103.725 | 2722.793 | 2776.165 | 0.906 |
| 8 | 2.5 | 2133.164 | 3124.387 | 3292.318 | 1.395 |
| 9 | 2.5 | 1876.577 | 2915.783 | 2980.335 | 1.187 |
| 10 | 0.33 | 1846.346 | 1914.808 | 1995.008 | 0.906 |
| 11 | 1.66 | 1742.913 | 2137.719 | 2184.693 | 0.587 |
| 12 | 1 | 1475.885 | 2374.576 | 2406.515 | 1.075 |
| 13 | 2.5 | 1029.477 | 1399.983 | 2019.384 | 6.71 |
| 14 | 1 | 1972.428 | 6329.641 | 6390.294 | 1.565 |
| 15 | 2.5 | 2400.558 | 3823.699 | 3874.193 | 1.414 |
| 16 | 0.66 | 1947.881 | 5058.111 | 5157.779 | 1.754 |
| 17 | 2.5 | 16.75.7 | 2461.388 | 2481.599 | 0.649 |
| 18 | 2 | 1296.384 | 2518.558 | 2553.332 | 0.875 |
| 19 | 0.33 | 1998.417 | 3053.967 | 3085.605 | 0.83 |
| 20 | 0.66 | 2002.755 | 3381.595 | 4380.818 | 0.979 |
| 21 | 1.66 | 1458.764 | 2588.919 | 2635.819 | 1.354 |
| 22 | 1.33 | 1609.677 | 3606.273 | 3674.145 | 0.864 |
| 23 | 1 | 1667.77 | 3264.357 | 3307.605 | 0.771 |
| 24 | 1 | 1544.26 | 3680.348 | 3743.439 | 1.299 |
| 25 | 2 | 1878.878 | 4143.015 | 4206.762 | 1.63 |
| Mean (SD) | 1.551 (0.864) | 1788.733 (362.315) | 3214.062 (1009.1) | 3334.559 (998.895) | 1.274 (1.18) |
AUC: area under the curve.
AUC values in infinite time showed statistically significant differences between groups (p<0.05). Post hoc analysis (Scheffé) was conducted to determine differences between groups; the group of poor metabolizing showed significant differences against the fast and intermediate metabolizers (p<0.05) (Fig. 1).
The present study aimed to evaluate the effect of the CYP2C9 wild type on the pharmacokinetics of diclofenac in healthy Mexican volunteers. Twenty five subjects with genotype * 1/* 1 (wild type) were included in the study, all were females because it has been reported that some NSAIDs biotransformation by CYP2C9 is affected depending on gender.12 Within the studied parameters, we established the area under the curve from zero time to infinity (AUC0-inf) as the most important one, it was done because it permits us to have a better estimate of the transformation of the drug. Based on this result, quartile separation allowed us to classify the volunteers in 6 PM, 13 IM and 6 FM. We found statistically significant differences (p<0.05) among the 3 groups and when we compared the PM group against IM and FM together we detected (p<0.05).
Studies about the biotransformation of diclofenac and CYP2C9 variants divide subjects into groups depending on their genotype and compare the pharmacokinetic parameters. An example would be the study of Yasar et al. in 2001, which administered a dose of 50mg oral diclofenac to 20 healthy subjects and compared the values of Cmax, AUC, time and clearance of the following groups: * 1/* 2, * 1/* 3, * 2/* 2, * 2/* 3 and * 3/* 3 against * 1/* 1 group; the authors did not find statistical differences.13
Our findings agree with authors like Zi et al., who refers that the involvement of CYP2C9 in the biotransformation of diclofenac cannot be exclusive and there may be other enzymes involved as CYP3A4, CYP2C19, CYP2C8 and CYP2C18.6 It is also important to consider the variations in the POR gene encoding an enzyme involved in the proper functioning of CYP2C919.
In conclusion, the variability in the metabolism of diclofenac in the * 1/* 1 genotype of CYP2C9 indicates that CYP2C9 is not the only enzyme responsible for the metabolism of diclofenac. Different responses in the same genotype mean that other cytochromes could participate in the metabolism of this drug. It is important to characterize other cytochromes possibly related to diclofenac such as CYP2C8, CYP2C18, CYP2C19 and CYP2B614 or even POR gene polymorphism.
Ethical disclosuresProtection of human and animal subjectsThe authors declare that no experiments were performed on humans or animals for this study.
Confidentiality of dataThe authors declare that they have followed the protocols of their work center on the publication of patient data.
Right to privacy and informed consentThe authors declare that no patient data appear in this article.
Conflict of interestThe authors have no conflict of interest to declare.