e systematics of the economically important, endangered hardwoods in Guaiacum are unclear with regard to taxonomic ranks, and the relationships among taxa. This is partially due to a lack of diagnostic characters and minimal geographic sampling in previous studies. Nevertheless, systematic relationships are important to inform trade regulations and management practices for these species. This is especially true for Mexico, which is both the primary exporter and the center of diversity for Guaiacum. Systematic and biogeographic issues in Guaiacum were investigated by analyses of nuclear and chloroplast DNA markers from specimens sampled throughout the range. Phylogenetic and statistical parsimony analyses showed well-supported divergences within the group, including a deep divergence between G. officinale and other taxa with additional biogeographically correlated subdivisions. There is also an indication that accessions from Chiapas, Mexico are genetically intermediate between G. sanctum and G. coulteri, while minor segregates of Guaiacum (G. palmeri, G. guatemalense) were not well distinguished by either morphological or molecular characters. The genetic structuring among the major groups of Guaiacum shows evidence of isolation induced by fragmentation of the range, with the structure largely maintained with only occasional long distance gene flow between remote populations.
La sistemática de las especies maderables, económicamente importantes y en peligro de extinción en Guaiacum aún no aclara con respeto a la clasificación taxonómica y las relaciones entre los taxa. Esto se debe parcialmente a la falta de rasgos diagnósticos y a reducidas muestras geográficas en estudios previos. Sin embargo, los aspectos en la sistemática y las relaciones filogenéticas de las especies son importantes a considerar en la regulación comercial y en las prácticas de manejo de las especies. Esto es especialmente importante para México ya que es tanto el exportador más importante, como el centro de diversidad para Guaiacum. En este trabajo se investigaron aspectos sistemáticos y biogeográficos en Guaiacum analizando marcadores nucleares y de cloroplastos de ADN de especímenes muestreados de su área de distribución. Los análisis filogenéticos y de parsimonia estadística mostraron divergencia bien sustentada dentro del grupo, incluyendo una divergencia profunda entre G. officinale y los otros taxa con subdivisiones adicionales asociadas a la biogeográfía. También hay evidencia de accesiones de Chiapas, México, como genéticamente intermedias entre G. sanctum y G. coulteri, mientras que otras especies de Guaiacum (G. palmeri, G. guatemalense) no se distinguen bien sea por rasgos morfológicos o moleculares. La organización genética entre los mayores grupos de Guaiacum muestra evidencia de un aislamiento provocado por fragmentación en su distribución, con la estructura en gran parte mantenida por un flujo ocasional de genes a larga distancia entre poblaciones lejanas.
Guaiacum L. (Zygophyllaceae R. Br.) is a New World genus comprised of a group of 4 to 8 commonly recognized taxa of tropical and subtropical hardwoods. These trees and shrubs are distributed primarily within the arid and semi-arid regions of the Caribbean basin and Mexico, and represent economically important species characterized by extremely dense and resinous wood.
Guaiacum sanctum L. is the most abundant and economically important species, and has therefore been the species of greatest conservation focus. Studies of population structure, density, and regeneration in the chief exporting country of Mexico have been conducted (López-Toledo et al., 2010, 2011b), and population level genetic studies have been conducted for Caribbean, Mexican, Floridian (Dertien and Duvall, 2009), and Costa Rican populations (Fuchs and Hamrick, 2010a, b). Furthermore, Guaiacum sanctum could act as an umbrella species in terms of conservation, as protection of populations could also benefit other endangered flora and fauna (López-Toledo et al., 2011a). Guaiacum coulteri A. Gray has been studied less extensively, primarily because it has lower economic importance and does not occur in protected areas (López-Toledo et al., 2010). The population genetics of G. unijugum Brandegee have also been studied for conservation purposes as it is a relatively rare endemic of the Cape region of Baja California, Mexico (McCauley et al., 2010).
Despite various conservation and genetic studies, the number of species within the genus remains unresolved, with as few as 4 and as many as 8 species commonly recognized. (Grow and Schwartzman, 2001a, b; Axelrod, 2002). The absence of clear species delimitation within the genus is particularly problematic for trade regulation by the Convention on International Trade in Endangered Species or Wild Fauna and Flora (UNEP-WCMC, 2007), developing management practices of Guaiacum species by exporting countries, and assessing extinction risk for species or local populations (IUCN, 2007). In practice, taxonomic distinctions are frequently based on geographic origin rather than morphological, reproductive, or genetic criteria. Currently, the entire genus is listed on CITES Appendix II because of the inability to distinguish timber of the different species and a taxonomy that is complicated by numerous synonyms, unsettled taxonomic ranks, and invalidly published names. The listing effectively renders all species of Guaiacum endangered from a regulation standpoint (López-Toledo et al., 2010) regardless of what is known about the status of individual populations.
Previous phylogenetic studies of Guaiacum have only included a single species as an exemplar, and were therefore not informative at the species level (Sheahan and Chase, 2000; Lia et al., 2001). Other studies attempting to delimit species using morphological characters were not inclusive of all species, and failed to identify characters suitable for consistent unambiguous identification (Grow and Schwartzman, 2001b). Specifically, suites of morphological character combinations yield conflicting results in taxonomic identifications for specimens found in southern Mexico and Central America (Grow and Schwartzman, 2001b), an area coincidentally of great importance to the harvesting and trade of Guaiacum. A more robust analysis of evolutionary relationships within Guaiacum could provide useful insight for a future taxonomic revision, and a clearer understanding of these phylogenetic relationships could be directly applicable to conservation practices.
This study is an attempt to resolve evolutionary relationships, identify consistent genetic patterns, and discover novel genetic patterns among Guaiacum species by using a combination of analyses appropriate for several taxonomic levels. Maximum parsimony methods can resolve deeper nodes but leave shallower relationships unresolved where variation is lacking among closely related accessions. Statistical parsimony analyses, however, can resolve relationships down to single mutations separating closely related individuals in a manner similar to DNA barcoding (Schindel and Miller, 2005; Collins and Cruickshank, 2013). Any contrasting patterns from nuclear and chloroplast DNA markers can indicate potential hybridization events (Soltis and Kuzoff, 1995), and geographic correlations with genetic patterns may indicate populations of greater taxonomic or conservation interest.
In addition to broadly resolving evolutionary relationships, this study is designed to address the following particular phylogenetic and taxonomic issues associated with the genus:
1) Guaiacum guatemalense Planch. ex Vail and Rydberg while often identified as a distinct species in herbarium collections, is most commonly accepted as a synonym of G. sanctum (http://www.theplantlist.org). Individual specimens identified as G. guatemalense possess intermediate and shared morphological characteristics of G. sanctum and G. coulteri, suggesting a hybrid origin (Porter, 1972). This putative species is found in a region near the intersection/overlap of the ranges G. coulteri and G. sanctum; 2) Guaiacum angustifolium Engelm. and its synonym Porlieria angustifolia (Engelm.) A. Gray, may be more accurately described as a subspecies or variety of G. coulteri; 3) Guaiacum unijugum is a species endemic to the Cape region of the Baja peninsula. The species status is questionable, as its unique morphology may be an environmental response of G. coulteri to the extreme desert conditions of the area, and 4) Guaiacum coulteri var. palmeri, while sometimes identified as the species G. palmeri Vail, may not have consistent diagnostic morphological characteristics or a unique genetic signature to warrant a distinction from Guaiacum coulteri var. coulteri.
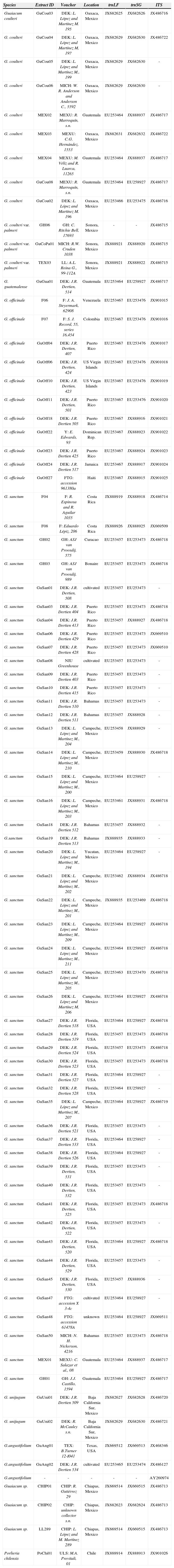
Materials and methodsTaxon samplingA total of 98 accessions of Guaiacum species were obtained from herbarium collections, living collections, or field-collected from populations in Mexico, Puerto Rico, and the Florida Keys (Table 1). Exemplars representing all currently recognized species were collected, as well accessions of putative species and varieties. Specimens were also collected from major geographic regions covering the extent of the generic distribution, as well as specific regions of potential biogeographic (i. e., Curasao) or other evolutionary interest. Multiple accessions of Guaiacum were collected from regions where species identification is difficult or inconsistent, particularly southern Mexico (states of Oaxaca, Chiapas, and Campeche) and Guatemala.
Collection information and GenBank accessions for all taxa used in this study. Specimens with shared accession numbers indicate identical DNA sequences for that marker, represented by a single GenBank submission
| Species | Extract ID | Voucher | Location | trnLF | trnSG | ITS |
|---|---|---|---|---|---|---|
| Guaiacum coulteri | GuCou03 | DEK: L. López and Martínez M. 195 | Oaxaca, Mexico | JX682625 | JX682626 | JX486716 |
| G. coulteri | GuCou04 | DEK: L. López and Martínez M, 197 | Oaxaca, Mexico | JX682629 | JX682630 | JX486722 |
| G. coulteri | GuCou05 | DEK: L. López and Martínez M., 199 | Oaxaca, Mexico | JX682629 | JX682630 | - |
| G. coulteri | GuCou06 | MICH: W. R. Anderson and Anderson C., 5592 | Oaxaca, Mexico | JX682629 | JX682630 | - |
| G. coulteri | MEX02 | MEXU: R. Marroquín, s.n. | Guatemala | EU253464 | JX888937 | JX486717 |
| G. coulteri | MEX03 | MEXU: C.G. Hernández, 1553 | Oaxaca, Mexico | JX682631 | JX682632 | JX486722 |
| G. coulteri | MEX04 | MEXU: M. Véliz and R. Luarca, 11265 | Guatemala | EU253464 | JX888937 | JX486717 |
| G. coulteri | GuCou08 | MEXU: R. Marroquín, s.n. | Guatemala | EU253464 | EU258927 | JX486717 |
| G. coulteri | GuCou02 | DEK: L. López and Martínez M. 196 | Oaxaca, Mexico | EU253466 | EU253475 | JX486716 |
| G. coulteri var. palmeri | GH06 | GH: C. Ritchie Bell, 17693 | Sonora, Mexico | - | - | JX486715 |
| G. coulteri var. palmeri | GuCoPa01 | MICH: R.W. Cruden 1038 | Sonora, Mexico | JX888921 | JX888920 | JX486715 |
| G. coulteri var. palmeri | TEX03 | LL: A.L. Reina G., 99-112A | Sonora, Mexico | JX888921 | JX888922 | JX486715 |
| G. guatemalense | GuGua01 | DEK: J.R. Dertien, 514 | Guatemala | EU253464 | EU258927 | JX486717 |
| G. officinale | F06 | F: J. A. Steyermark, 62908 | Venezuela | EU253467 | EU253476 | JX901015 |
| G. officinale | F07 | F: S. J. Record, 55, series 16,454 | Colombia | EU253467 | EU253476 | JX901016 |
| G. officinale | GuOff04 | DEK: J.R. Dertien, 407 | Puerto Rico | EU253467 | EU253476 | JX901017 |
| G. officinale | GuOff06 | DEK: J.R. Dertien, 424 | US Virgin Islands | EU253467 | EU253476 | JX901018 |
| G. officinale | GuOff10 | DEK: J.R. Dertien, 423 | US Virgin Islands | EU253467 | EU253476 | JX901019 |
| G. officinale | GuOff11 | DEK: J.R. Dertien, 501 | Puerto Rico | EU253467 | EU253476 | JX901020 |
| G. officinale | GuOff18 | DEK: J.R. Dertien 505 | Puerto Rico | EU253467 | JX888916 | JX901021 |
| G. officinale | GuOff22 | Y: E. Edwards, 93 | Dominican Rep. | EU253467 | JX888923 | JX901022 |
| G. officinale | GuOff23 | DEK: J.R. Dertien 425 | Puerto Rico | EU253467 | JX888924 | JX901023 |
| G. officinale | GuOff24 | DEK: J.R. Dertien 517 | Jamaica | EU253467 | JX888917 | JX901024 |
| G. officinale | GuOff27 | FTG: accession 961380a | Haiti | EU253467 | JX888915 | JX901025 |
| G. sanctum | F04 | F: R. Espinosa and R. Aguilar 1035 | Costa Rica | JX888919 | JX888918 | JX486714 |
| G. sanctum | F08 | F: Eduardo Lépiz, 206 | Costa Rica | JX888926 | JX888925 | JX669509 |
| G. sanctum | GH02 | GH: ASJ van Proosdij, 575 | Curacao | EU253457 | EU253473 | JX486718 |
| G. sanctum | GH03 | GH: ASJ van Proosdij, 989 | Bonaire | EU253457 | EU253473 | JX486718 |
| G. sanctum | GuSan01 | DEK: J.R. Dertien, 508 | cultivated | EU253457 | EU253473 | - |
| G. sanctum | GuSan03 | DEK: J.R. Dertien 404 | Puerto Rico | EU253457 | EU253473 | JX486718 |
| G. sanctum | GuSan04 | DEK: J.R. Dertien 413 | Puerto Rico | EU253457 | JX888927 | JX486718 |
| G. sanctum | GuSan06 | DEK: J.R. Dertien 429 | Puerto Rico | EU253457 | EU253473 | JX669510 |
| G. sanctum | GuSan07 | DEK: J.R. Dertien 428 | Puerto Rico | EU253457 | EU253473 | JX669510 |
| G. sanctum | GuSan08 | NIU Greenhouse | cultivated | EU253457 | EU253473 | - |
| G. sanctum | GuSan09 | DEK: J.R. Dertien 403 | Puerto Rico | EU253457 | EU253473 | - |
| G. sanctum | GuSan10 | DEK: J.R. Dertien 415 | Puerto Rico | EU253457 | EU253473 | - |
| G. sanctum | GuSan11 | DEK: J.R. Dertien 510 | Bahamas | EU253457 | EU253473 | - |
| G. sanctum | GuSan12 | DEK: J.R. Dertien 511 | Bahamas | EU253457 | JX888928 | - |
| G. sanctum | GuSan13 | DEK: L. López and Martínez M., 204 | Campeche, Mexico | EU253458 | JX888929 | - |
| G. sanctum | GuSan14 | DEK: L. López and Martínez M., 210 | Campeche, Mexico | EU253459 | JX888930 | JX486718 |
| G. sanctum | GuSan15 | DEK: L. López and Martínez M., 200 | Campeche, Mexico | EU253464 | EU258927 | - |
| G. sanctum | GuSan16 | DEK: L. López and Martínez M., 203 | Campeche, Mexico | EU253461 | JX888931 | JX486718 |
| G. sanctum | GuSan18 | DEK: J.R. Dertien 512 | Bahamas | EU253457 | JX888932 | - |
| G.sanctum | GuSan19 | DEK: J.R. Dertien 513 | Bahamas | JX888935 | JX888933 | - |
| G. sanctum | GuSan20 | DEK: L. López and Martínez M., 194 | Yucatan, Mexico | EU253464 | EU258927 | - |
| G. sanctum | GuSan21 | DEK: L. López and Martínez M., 202 | Campeche, Mexico | EU253462 | JX888934 | JX486718 |
| G. sanctum | GuSan22 | DEK: L. López and Martínez M., 201 | Campeche, Mexico | JX888935 | EU253469 | JX486718 |
| G. sanctum | GuSan23 | DEK: L. López and Martínez M., 209 | Campeche, Mexico | EU253464 | EU258927 | JX486718 |
| G. sanctum | GuSan24 | DEK: L. López and Martínez M., 211 | Campeche, Mexico | EU253464 | EU258927 | JX486718 |
| G. sanctum | GuSan25 | DEK: L. López and Martínez M., 205 | Campeche, Mexico | EU253463 | EU253470 | JX486718 |
| G. sanctum | GuSan26 | DEK: L. López and Martínez M, 206 | Campeche, Mexico | EU253464 | EU258927 | JX486718 |
| G. sanctum | GuSan27 | DEK: J.R. Dertien 518 | Florida, USA | EU253464 | EU258927 | JX486718 |
| G. sanctum | GuSan28 | DEK: J.R. Dertien 519 | Florida, USA | EU253457 | EU253473 | JX486718 |
| G. sanctum | GuSan29 | DEK: J.R. Dertien 524 | Florida, USA | EU253457 | EU253473 | JX486718 |
| G. sanctum | GuSan30 | DEK: J.R. Dertien 523 | Florida, USA | EU253457 | EU253473 | JX486718 |
| G. sanctum | GuSan31 | DEK: J.R. Dertien 527 | Florida, USA | EU253464 | EU258927 | - |
| G. sanctum | GuSan32 | DEK: J.R. Dertien 528 | Florida, USA | EU253464 | EU258927 | - |
| G. sanctum | GuSan35 | DEK: L. López and Martínez M., 207 | Campeche, Mexico | EU253464 | EU258927 | JX486719 |
| G. sanctum | GuSan36 | DEK: J.R. Dertien 521 | Florida, USA | EU253457 | EU253473 | - |
| G. sanctum | GuSan37 | DEK: J.R. Dertien 533 | Florida, USA | EU253464 | EU258927 | - |
| G. sanctum | GuSan38 | DEK: J.R. Dertien 526 | Florida, USA | EU253464 | EU258927 | - |
| G. sanctum | GuSan39 | DEK: J.R. Dertien, 531 | Florida, USA | EU253457 | EU253473 | - |
| G. sanctum | GuSan40 | DEK: J.R. Dertien, 532 | Florida, USA | EU253457 | EU253473 | - |
| G. sanctum | GuSan41 | DEK: J.R. Dertien, 525 | Florida, USA | EU253457 | EU253473 | JX486718 |
| G. sanctum | GuSan42 | DEK: J.R. Dertien, 522 | Florida, USA | EU253457 | EU253473 | - |
| G. sanctum | GuSan43 | DEK: J.R. Dertien, 520 | Florida, USA | EU253464 | EU258927 | JX486718 |
| G. sanctum | GuSan44 | DEK: J.R. Dertien, 529 | Florida, USA | EU253457 | EU253473 | - |
| G. sanctum | GuSan45 | DEK: J.R. Dertien, 530 | Florida, USA | EU253457 | JX888936 | - |
| G. sanctum | GuSan47 | FTG: accession X 3-4c | cultivated | EU253464 | EU258927 | - |
| G. sanctum | GuSan48 | FTG: accession 61478A | unknown | EU253464 | EU258927 | JX669511 |
| G. sanctum | GuSan50 | MICH: N. H. Nickerson, 4216 | Bahamas | EU253457 | EU253473 | JX486718 |
| G. sanctum | MEX01 | MEXU: C. Salazar et al., 08 | Guatemala | EU253464 | JX888937 | JX486717 |
| G. sanctum | GH01 | GH: J.J. Castillo, 1594 | Guatemala | EU253464 | EU258927 | JX486717 |
| G. unijugum | GuUni01 | DEK: J.R. Dertien 509 | Baja California Sur, Mexico | JX682627 | JX682628 | JX486720 |
| G. unijugum | GuUni02 | DEK: R. McCauley s.n. | Baja California Sur, Mexico | JX682629 | JX682630 | JX486721 |
| G.angustifolium | GuAng01 | TEX: B.Turner 12-I041 | Texas, USA | JX669512 | JX669513 | JX468346 |
| G.angustifolium | GuAng02 | DEK: J.R. Dertien 534 | cultivated | EU253465 | EU253474 | JX486127 |
| G.angustifolium | - | - | - | - | - | AY260974 |
| Guaiacum sp. | CHIP01 | CHIP: R. Gutiérrez 29 | Chiapas, Mexico | JX669514 | JX669515 | JX486713 |
| Guaiacum sp. | CHIP02 | CHIP: unknown collector s.n. | Chiapas, Mexico | JX682623 | JX682624 | JX486713 |
| Guaiacum sp. | LL289 | CHIP: L. López and M. Martínez 289 | Chiapas, Mexico | JX669514 | JX669515 | JX486713 |
| Porlieria chilensis | PoChi01 | ULS: M.A. Previtali, 01 | Chile | JX888914 | JX888913 | JX901026 |
Porlieria chilensis was chosen as an outgroup based on previous phylogenetic studies that included Guaiacum and related genera (Sheahan and Chase, 2000; Lia et al., 2001).
DNA Extraction, PCR, and purification and sequencingDNA was extracted from leaflets stored in silica gel or from herbarium specimens using either a modified CTAB method (Doyle and Doyle, 1987) or the DNeasy Plant Mini Kit (Qiagen, Valencia, California). The manufacturer’s protocol was followed for the DNeasy Plant Mini Kit; however modifications were applied to the extraction protocol to increase DNA yield from degraded herbarium material. Modifications included an extended elution time of 10-20 minutes (Draábková et al., 2002) and in some cases, an overnight alcohol precipitation prior to RNAse digestion. A PEG/NaCl (polyethylene glycol/salt) precipitation following RNAse digestion was used to remove potentially PCR inhibiting soluble polysaccharides and secondary metabolites (Li et al., 1994).
Genetic markers were amplified via polymerase-chain reaction (PCR) using the FailSafe PCR system (Epicentre Technologies, Madison, Wisconsin) with buffer premixes “I” or “H”.
The trnL-F chloroplast marker was amplified using primer pairs “C” and “F” and by following the PCR protocol described in Taberlet et al. (1991). The trnS-G marker, containing both the trnS-trnG intergenic spacer and the trnG intron, was amplified using primers “trnS” and “3’tmG” and following the PCR protocol described in Shaw et al. (2005). Two additional internal primers, “trnG2G” and “trnG2S” (Shaw et al., 2005) were occasionally used in sequencing reactions in cases where short reads were obtained from external primers or where large poly-T regions created downstream sequencing artifacts. The complete nuclear ITS region was amplified (including ITS1, 5.8s, and ITS2) using the primer pair ITS4 and ITSL (White et al., 1990). Additional accessions were sampled for the ITS2 region alone using the primer pair P3K (Kim and Jansen, 1994) and P4 (White et al., 1990; Simpson et al., 2004).
PCR products were prepared for sequencing using either the Wizard SV PCR clean-up system (Promega Corp., Madison, Wisconsin) or the Microcon YM100 system (Millipore Corp., Billerica, Massachusetts). Automated capillary sequencing was performed with ABI3730xl DNA analyzers (Macrogen, Seoul, South Korea) or Beckman Coulter CEQ-8000 DNA analyzers at the Northern Illinois University Core Sequencing Facility (NIU, DeKalb, Illinois).
Sequence alignment and analysesSequences were machine-aligned using the “Geneious” align function within the Geneious Pro V. 4.5 software package (Drummond et al., 2008). Parameters were set for a global alignment with free end gaps of 70% similarity (IUB 5.0/- 4.5) with a gap opening penalty setting of 12 and gap extension penalty setting of 3. Minor manual adjustments were made to alignments to match patterns of tandem repeats within regions of insertion/deletion mutations (indels) within the sequence matrix. Variable positions were confirmed against original chromatograms using Geneious Pro V. 4.5 (Drummond et al., 2008) to confirm the nucleotide identity and signal strength at the variable position. Positions of variable nucleotides that could not be confidently confirmed due to weak or conflicting signal in chromatograms were treated as sequencing artifacts and excluded from analyses. Regions of high variability and ambiguous alignment were removed prior to analysis, as were regions of poly-nucleotide runs in which proper homology could not be assessed. Additionally, portions of the trnS-G marker were excluded as a result of poor quality or little overlap between forward and reverse primers in some accessions. Autapomorphic indel mutations in the outgroup taxon Porlieria chilensis, were also removed, including a 153 base pair (bp) insertion in the trnS-G region that was absent in all of the ingroup taxa. Indel mutations were coded as multi-state (0, 1, 2, 3) (González et al., 2006) and added to the sequence data matrix prior to phylogenetic analysis.
Phylogenetic analysisPhylogenetic relationships were inferred using PAUP* (Swofford, 2002). The Maximum Parsimony (MP) method was implemented to make use of parsimony informative indels. Heuristic searches with 100 random addition sequence replicates and tree-bisection reconnection branch swapping were performed to find the set of most parsimonious trees. A MP bootstrap analysis (Felsenstein, 1985) was performed with 1 000 bootstrap pseudoreplicates. Accessions with identical cpDNA sequences were removed from the data matrices prior to analysis, leaving exemplars of each unique haplotype. Phylogenetic analyses were conducted using a data matrix of combined cpDNA and nrDNA sequences (trnL-F, trnS- G, ITS1, 5.8s, ITS2) and separate data matrices of cpDNA (combined trnL-F, trnS-G) and nuclear (ITS1, 5.8s, ITS2) data.
Incongruence length difference testsWilcoxon signed- ranks tests (Templeton, 1983) and Kishino-Hasegawa (KH) tests (Kishino and Hasegawa, 1989) were conducted using PAUP* (Swofford, 2002) to test incongruence between chloroplast and nuclear trees.
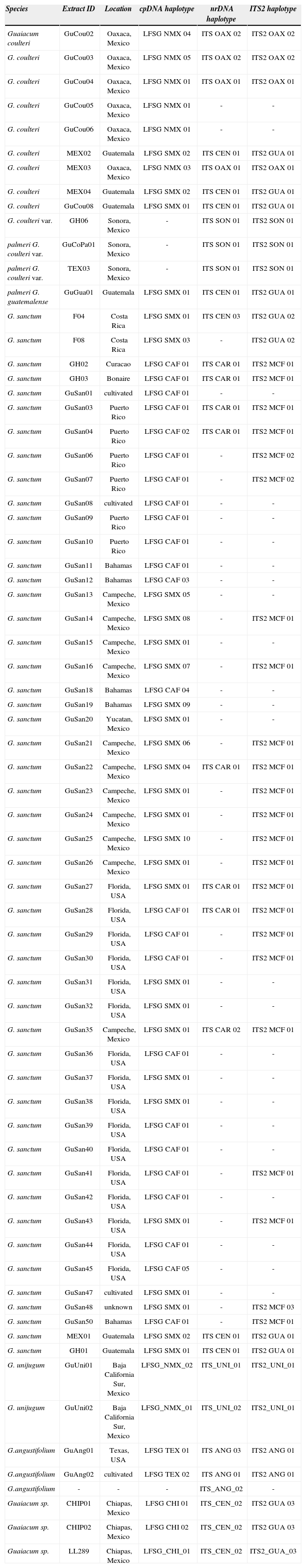
Statistical Parsimony AnalysisPhylogenetic networks of interconnected haplotypes were inferred through the statistical parsimony method of estimating gene genealogies, as implemented by the TCS v 1.21 software package (Clement et al., 2000). Unlike the maximum parsimony method of phylogenetic analyses, these phylogenetic reconstructions make use of multiple accessions with identical haplotypes (Table 2). Furthermore, the individual mutational steps separating unique haplotypes are mapped in the network. As such, biogeographic and similar data can be overlaid on these networks, making them useful for studies in which biogeographical elements, hybridization, and speciation are prominent components (Shaw and Small, 2005; Ran et al., 2006). Autapomorphies are similarly mapped, providing further insight into levels of genetic diversity.
Haplotype designations for all specimens used in this study
| Species | Extract ID | Location | cpDNA haplotype | nrDNA haplotype | ITS2 haplotype |
|---|---|---|---|---|---|
| Guaiacum coulteri | GuCou02 | Oaxaca, Mexico | LFSG NMX 04 | ITS OAX 02 | ITS2 OAX 02 |
| G. coulteri | GuCou03 | Oaxaca, Mexico | LFSG NMX 05 | ITS OAX 02 | ITS2 OAX 02 |
| G. coulteri | GuCou04 | Oaxaca, Mexico | LFSG NMX 01 | ITS OAX 01 | ITS2 OAX 01 |
| G. coulteri | GuCou05 | Oaxaca, Mexico | LFSG NMX 01 | - | - |
| G. coulteri | GuCou06 | Oaxaca, Mexico | LFSG NMX 01 | - | - |
| G. coulteri | MEX02 | Guatemala | LFSG SMX 02 | ITS CEN 01 | ITS2 GUA 01 |
| G. coulteri | MEX03 | Oaxaca, Mexico | LFSG NMX 03 | ITS OAX 01 | ITS2 OAX 01 |
| G. coulteri | MEX04 | Guatemala | LFSG SMX 02 | ITS CEN 01 | ITS2 GUA 01 |
| G. coulteri | GuCou08 | Guatemala | LFSG SMX 01 | ITS CEN 01 | ITS2 GUA 01 |
| G. coulteri var. | GH06 | Sonora, Mexico | - | ITS SON 01 | ITS2 SON 01 |
| palmeri G. coulteri var. | GuCoPa01 | Sonora, Mexico | - | ITS SON 01 | ITS2 SON 01 |
| palmeri G. coulteri var. | TEX03 | Sonora, Mexico | - | ITS SON 01 | ITS2 SON 01 |
| palmeri G. guatemalense | GuGua01 | Guatemala | LFSG SMX 01 | ITS CEN 01 | ITS2 GUA 01 |
| G. sanctum | F04 | Costa Rica | LFSG SMX 01 | ITS CEN 03 | ITS2 GUA 02 |
| G. sanctum | F08 | Costa Rica | LFSG SMX 03 | - | ITS2 GUA 02 |
| G. sanctum | GH02 | Curacao | LFSG CAF 01 | ITS CAR 01 | ITS2 MCF 01 |
| G. sanctum | GH03 | Bonaire | LFSG CAF 01 | ITS CAR 01 | ITS2 MCF 01 |
| G. sanctum | GuSan01 | cultivated | LFSG CAF 01 | - | - |
| G. sanctum | GuSan03 | Puerto Rico | LFSG CAF 01 | ITS CAR 01 | ITS2 MCF 01 |
| G. sanctum | GuSan04 | Puerto Rico | LFSG CAF 02 | ITS CAR 01 | ITS2 MCF 01 |
| G. sanctum | GuSan06 | Puerto Rico | LFSG CAF 01 | - | ITS2 MCF 02 |
| G. sanctum | GuSan07 | Puerto Rico | LFSG CAF 01 | - | ITS2 MCF 02 |
| G. sanctum | GuSan08 | cultivated | LFSG CAF 01 | - | - |
| G. sanctum | GuSan09 | Puerto Rico | LFSG CAF 01 | - | - |
| G. sanctum | GuSan10 | Puerto Rico | LFSG CAF 01 | - | - |
| G. sanctum | GuSan11 | Bahamas | LFSG CAF 01 | - | - |
| G. sanctum | GuSan12 | Bahamas | LFSG CAF 03 | - | - |
| G. sanctum | GuSan13 | Campeche, Mexico | LFSG SMX 05 | - | - |
| G. sanctum | GuSan14 | Campeche, Mexico | LFSG SMX 08 | - | ITS2 MCF 01 |
| G. sanctum | GuSan15 | Campeche, Mexico | LFSG SMX 01 | - | - |
| G. sanctum | GuSan16 | Campeche, Mexico | LFSG SMX 07 | - | ITS2 MCF 01 |
| G. sanctum | GuSan18 | Bahamas | LFSG CAF 04 | - | - |
| G. sanctum | GuSan19 | Bahamas | LFSG SMX 09 | - | - |
| G. sanctum | GuSan20 | Yucatan, Mexico | LFSG SMX 01 | - | - |
| G. sanctum | GuSan21 | Campeche, Mexico | LFSG SMX 06 | - | ITS2 MCF 01 |
| G. sanctum | GuSan22 | Campeche, Mexico | LFSG SMX 04 | ITS CAR 01 | ITS2 MCF 01 |
| G. sanctum | GuSan23 | Campeche, Mexico | LFSG SMX 01 | - | ITS2 MCF 01 |
| G. sanctum | GuSan24 | Campeche, Mexico | LFSG SMX 01 | - | ITS2 MCF 01 |
| G. sanctum | GuSan25 | Campeche, Mexico | LFSG SMX 10 | - | ITS2 MCF 01 |
| G. sanctum | GuSan26 | Campeche, Mexico | LFSG SMX 01 | - | ITS2 MCF 01 |
| G. sanctum | GuSan27 | Florida, USA | LFSG SMX 01 | ITS CAR 01 | ITS2 MCF 01 |
| G. sanctum | GuSan28 | Florida, USA | LFSG CAF 01 | ITS CAR 01 | ITS2 MCF 01 |
| G. sanctum | GuSan29 | Florida, USA | LFSG CAF 01 | - | ITS2 MCF 01 |
| G. sanctum | GuSan30 | Florida, USA | LFSG CAF 01 | - | ITS2 MCF 01 |
| G. sanctum | GuSan31 | Florida, USA | LFSG SMX 01 | - | - |
| G. sanctum | GuSan32 | Florida, USA | LFSG SMX 01 | - | - |
| G. sanctum | GuSan35 | Campeche, Mexico | LFSG SMX 01 | ITS CAR 02 | ITS2 MCF 01 |
| G. sanctum | GuSan36 | Florida, USA | LFSG CAF 01 | - | - |
| G. sanctum | GuSan37 | Florida, USA | LFSG SMX 01 | - | - |
| G. sanctum | GuSan38 | Florida, USA | LFSG SMX 01 | - | - |
| G. sanctum | GuSan39 | Florida, USA | LFSG CAF 01 | - | - |
| G. sanctum | GuSan40 | Florida, USA | LFSG CAF 01 | - | - |
| G. sanctum | GuSan41 | Florida, USA | LFSG CAF 01 | - | ITS2 MCF 01 |
| G. sanctum | GuSan42 | Florida, USA | LFSG CAF 01 | - | - |
| G. sanctum | GuSan43 | Florida, USA | LFSG SMX 01 | - | ITS2 MCF 01 |
| G. sanctum | GuSan44 | Florida, USA | LFSG CAF 01 | - | - |
| G. sanctum | GuSan45 | Florida, USA | LFSG CAF 05 | - | - |
| G. sanctum | GuSan47 | cultivated | LFSG SMX 01 | - | - |
| G. sanctum | GuSan48 | unknown | LFSG SMX 01 | - | ITS2 MCF 03 |
| G. sanctum | GuSan50 | Bahamas | LFSG CAF 01 | - | ITS2 MCF 01 |
| G. sanctum | MEX01 | Guatemala | LFSG SMX 02 | ITS CEN 01 | ITS2 GUA 01 |
| G. sanctum | GH01 | Guatemala | LFSG SMX 01 | ITS CEN 01 | ITS2 GUA 01 |
| G. unijugum | GuUni01 | Baja California Sur, Mexico | LFSG_NMX_02 | ITS_UNI_01 | ITS2_UNI_01 |
| G. unijugum | GuUni02 | Baja California Sur, Mexico | LFSG_NMX_01 | ITS_UNI_02 | ITS2_UNI_01 |
| G.angustifolium | GuAng01 | Texas, USA | LFSG TEX 01 | ITS ANG 03 | ITS2 ANG 01 |
| G.angustifolium | GuAng02 | cultivated | LFSG TEX 02 | ITS ANG 01 | ITS2 ANG 01 |
| G.angustifolium | - | - | - | ITS_ANG_02 | - |
| Guaiacum sp. | CHIP01 | Chiapas, Mexico | LFSG CHI 01 | ITS_CEN_02 | ITS2 GUA 03 |
| Guaiacum sp. | CHIP02 | Chiapas, Mexico | LFSG CHI 02 | ITS_CEN_02 | ITS2 GUA 03 |
| Guaiacum sp. | LL289 | Chiapas, Mexico | LFSG_CHI_01 | ITS_CEN_02 | ITS2_GUA_03 |
Separate aligned sequence data matrices were generated for nrDNA (ITS1, 5.8s, ITS2), the ITS2 marker, and cpDNA (trnL-F + trnS-G). Sites of ambiguous base calls or unconfirmed mutations were removed prior to analysis. Statistical parsimony networks were constructed using the 95% connection limit criterion, and reticulating patterns in networks were resolved following guidelines outlined by Crandall (1994).
ResultsThe aligned data matrix for the combined analysis of cpDNA and nrDNA(trnL-F + trnS-G + ITS1, 5.8s, ITS2) consisted of 3 055 bp for 41 ingroup taxa and 1 outgroup taxon. Maximum parsimony analysis of the combined sequence data included a total of 2 324 characters, with 119 being parsimony informative. The result was 24 equally parsimonious trees with a tree length of 351 steps, with a consistency index (CI)= 0.8433 and a retention index (RI)= 0.9417. The strict consensus tree included 18 nodes resolved with a bootstrap support value exceeding 70%, and ingroup taxa divided into 4 strongly supported clades (Fig. 1, clades A-D).
The basal clade is a monophyletic group inclusive of all accessions identified as Guaiacum officinale (100% bootstrap support) that is sister to all other Guaiacum species. These data also indicate significant intraspecific genetic structuring within this clade with 2 strongly supported subclades (100% bootstrap support each) separating accessions from Jamaica (GuOff24), Haiti (GuOff27), and the Dominican Republic (GuOff22) from accessions collected in Colombia, Venezuela, Puerto Rico, and the Virgin Islands. A second well-supported clade (98% bootstrap support) includes accessions confidently identified as G. sanctum, as well as accessions collected from Guatemala dubiously identified as G. sanctum, G. coulteri, or G. guatemalense (Fig. 1, clade B). A third clade with 99% bootstrap support includes accessions identified as G. angustifolium, G. unijugum, G. coulteri var. palmeri, and G. coulteri (var. coulteri) of non-Guatemalan origin (Fig. 1, clade C). This clade has a sister relationship (100% bootstrap support) to all accessions identified as G. sanctum based on morphology. The fourth clade (Fig. 1, clade D) contains 3 accessions (CHIP01, CHIP02, LL289) collected from Chiapas, Mexico lacking clear morphological. character states that readily distinguish them as either G. coulteri, G. sanctum, or G. guatemalense. This clade shows only moderate support (73% bootstrap) for a sister relationship to the G. sanctum clade.
Sequence data were divided into separate groupings of cpDNA (trnL-F + trnS-G) and nrDNA (ITS1, 5.8s, ITS2). Maximum parsimony analysis of cpDNA included 1 782 bp, with 48 parsimony informative characters (~2.7%). Three equally parsimonious trees with a length of 135 steps (CI= 0.9185, RI= 0.8935) were produced (Fig. 2).
The strict consensus tree includes 8 nodes resolved with a bootstrap support value exceeding 70%. Analysis of nrDNA included 542 characters, with a total of 71 characters being parsimony informative. (~13%). Maximum Parsimony produced 743 equally parsimonious trees with tree lengths of 208 steps; CI= 0.8269, RI= 0.9332. (Fig. 2) The strict consensus tree includes 13 nodes resolved with a bootstrap support value exceeding 70%. The nrDNA contains more parsimony informative characters than the cpDNA data (71 vs. 48, respectively). Furthermore, the nrDNA contains 8 parsimony informative indel mutations, compared to the 5 indels scored in the cpDNA data set. The indels in the nrDNA are small indels of 1 to 2 bp, whereas the cpDNA indels range from 1 to 38 bp.
Topologies for strict consensus trees generated from cpDNA and nrDNA are largely similar; however, both KH and Templeton tests of incongruence conducted between cpDNA and nrDNA indicate strongly incongruent tree topologies between the 2 data subsets (p< 0.001).
A sister relationship between G. officinale and other Guaiacum species is strongly supported in both trees (Fig. 2).
The cpDNA tree unites G. angustifolium, G. unijugum, and non-Guatemalan G. coulteri with strong support (B’, 100% bootstrap support). A second strongly-supported clade unites accessions of G. sanctum, G. coulteri of Guatemalan origin, and G. guatemalense (C’, 96% bootstrap support) with moderate support (79% bootstrap) uniting accessions from Guatemala, regardless of identity based on morphology. A third clade unites accessions collected from Chiapas Mexico (D’, 99% bootstrap support) and is positioned as sister to clade containing G. angustifolium, non-Guatemalan G. coulteri, and G. unijugum and the clade containing G. sanctum and Guatemalan accessions.
Incongruence in topology is notable for the accessions from Chiapas (D, D’). These accessions are united with accessions of G. sanctum in the nrDNA tree, although support for this clade (C) is weak (53% bootstrap support). The cpDNA reconstruction places these accessions in a sister relationship to other (non-officinale) Guaiacum species.
The nrDNA data failed to resolve a single clade containing all of the accessions the G. sanctum. These data also failed to support a clade containing the accessions of non-Guatemalan G. coulteri, G. unijugum, and G. angustifolium.
Conversely, the cpDNA phylogenetic reconstruction shows strong support for a clade uniting G. sanctum (96% bootstrap support) and another clade in which accessions of G. unijugum, and G. angustifolium were united with non-Guatemalan accessions of G. coulteri (100% bootstrap support). This reconstruction fails to resolve sister relationships among the 3 well-supported clades containing non-officinale accessions.
Phylogenetic network estimation using statistical parsimonyThe phylogenetic network constructed from combined nrDNA data (Fig. 3) connects 30 taxa at a 95% confidence interval from a data matrix consisting of 517 bp. Guaiacum sanctum accessions represent a majority of haplotypes that are separated from Central American and Chiapas accessions by 3 mutational steps. Accessions of G. coulteri from Oaxaca, Mexico are divided into 2 haplotypes separated by 6 mutational steps. Accessions identified as G. coulteri var. palmeri are derived from one of these G. coulteri haplotypes with a single additional mutational step. Accessions from Baja California Sur identified as G. unijugum have the closest connection in the network to G. coulteri, with 8 mutational steps separating the nearest accession of G. unijugum from either sampled haplotype of G. coulteri.
Haplotype network of Guaiacum from nrDNA (ITS1, 5.8s, ITS2) for all taxa excluding G. officinale. Large circles represent sampled haplotypes, with numbers and relative size indicating frequency of observation. Dots represent unobserved haplotypes with inferred mutational steps. Lines connecting haplotypes represent mutational steps.
The phylogenetic network constructed from the ITS2 marker data (Fig. 4) connects 45 taxa at the 95% confidence interval. The ITS2 network is constructed with fewer mutational steps separating sampled haplotypes; however the overall network patterns are visually congruent with the combined nrDNA network.
Haplotype network of Guaiacum from ITS2 region of nrDNA for all taxa excluding G. officinale. Large circles represent sampled haplotypes, with numbers and relative size indicating frequency of observation. Dots represent unobserved haplotypes with inferred mutational steps. Lines connecting haplotypes represent mutational steps.
The phylogenetic network constructed from cpDNA (Fig. 5) connects 66 taxa at a 95% confidence interval. This network segregates G. sanctum into 2 major clades, consistent with the patterns outlined in Dertien and Duvall (2009). Accessions identified as G. coulteri, and G. unijugum form a complex containing shared haplotypes, and G. angustifolium is connected to this complex by a single mutational step. Accessions from Chiapas, Mexico, show intermediacy, being connected to the nearest sampled haplotypes of the G. sanctum and G. coulteri complexes by 10 and 14 mutational steps, respectively.
Haplotype network of Guaiacum from cpDNA for all taxa excluding G. officinale. Large circles represent sampled haplotypes, with numbers and relative size indicating frequency of observation. Dots represent unobserved haplotypes with inferred mutational steps. Solid lines connecting haplotypes represent single nucleotide polymorphisms and dashed lines represent indel mutations.
Two equally parsimonious networks are generated from cpDNA, representing a differential placement of Chiapas accessions in the network. Although equally parsimonious, one network depicts the reversal of an ordered state indel mutation. Given the mutational mechanism of slipped- strand mispairing generating the derived states of either an inserted or deleted copy of a tandem repeat found in an ancestral condition, this network can be ruled out because it requires an insertion from a condition in which the tandem repeat has already been removed through deletion. The alternate network (Fig. 5) does not require such a reversal, but rather a parallel insertion from the ancestral condition, and is therefore considered the more probable network. Specific details about the ancestral and derived states of this mutation are explained in Dertien and Duvall (2009).
DiscussionGuaiacum officinaleThe phylogenetic relationship of G. officinale as sister to the remaining taxa is unambiguous. This result based on the genetic evidence is expected given the high levels of morphological divergence between G. officinale and remaining Guaiacum taxa. Nevertheless, misidentification between G. officinale and G. sanctum occasionally occurs among specimens collected in areas of the Caribbean where both species occur sympatrically. A screening of 1 694 specimens from 11 herbaria showed misidentification to occur at a rate of approximately 10%. Misidentified specimens between these species did not demonstrate ambiguous or intermediate character states, and it is therefore likely that such misidentification is attributable to error.
Genetic divergence and intraspecific structuring for G. officinale was evident in all trees (Figs. 1, 2). These data show strong support for a genetic divergence between accessions from Jamaica/Hispaniola (GuOff22, GuOff24, GuOff27) from those collected in the rest of the Caribbean. The monophyly of the sister group to the Jamaica/Hispaniola clade was resolved more strongly with the nrDNA data, which could be explained by a stronger phylogenetic signal from the more variable nrDNA, or indicative of chloroplast capture from a past hybridization event (Soltis and Kuzoff, 1995) confounding the phylogenetic signal. It is anticipated that additional sampling of G. officinale from Caribbean populations could reveal a complex evolutionary pattern coinciding with the region’s rich geologic history.
Guaiacum sanctum, G. guatemalense, G. coulteriGuaiacum along the Pacific coast of Mexico are generally identified as G. coulteri based on having linear to linear oblong leaflets whereas Guaiacum growing in Yucatán and Campeche are distinguished as G. sanctum based on their oblong to obovate leaflets. However, these morphological character states converge in parts of southern Mexico and Central America (Grow and Schwartzman, 2001b) making identification difficult. It has been hypothesized that populations of G. coulteri and G. sanctum could be occurring sympatrically or possibly hybridizing in these regions, where species distributions presumably overlap (Grow and Schwartzman, 2001b) Furthermore, Guaiacum found in Guatemala and parts of Central America have been treated as G. guatemalense, a novel species that possesses some intermediate characteristics of G. sanctum and G. coulteri. However, this distinction is often indiscernible or otherwise ill-defined because of the absence of discrete character states (e. g., leaf and petal shape, and hairiness of abaxial leaf surfaces).
The difficulty in distinguishing and defining species in these regions is not only interesting in the context of systematic botany and taxonomy, but also has potential impact on resource management and regulation of trade of Guaiacum, as Mexico is the largest global exporter (CITES, 2002).
The molecular data of this study show similar challenges as morphological data in efforts to clearly distinguish between G. coulteri and G. sanctum. Most accessions are readily distinguished by mutation patterns in both cpDNA and nrDNA. However, some accessions from southern Mexico show intermediate patterns in cpDNA. Specifically, 3 accessions collected from Chiapas, Mexico represent a genetic intermediate between G. sanctum and G. coulteri in the statistical parsimony analyses of cpDNA. The cpDNA network places these accessions in a position that is nearly equidistant from the nearest accession of G. sanctum (15 mutational steps) or of G. coulteri (14 mutational steps). Furthermore, statistical parsimony networks generated from the cpDNA data matrix with these accessions removed failed to place remaining accessions in a single network at the 95% confidence limit (networks not shown). Networks generated from the nrDNA dataset demonstrated a different pattern in which the Chiapas accessions are closely connected to the subgroup of Guaiacum collected from Guatemala and Costa Rica. The intermediate cpDNA pattern and different pattern in nrDNA is the most likely explanation for the difference in bootstrap support and topology in the maximum parsimony analyses. However, removal of these accessions failed to induce congruence between the 2 trees, most likely due to further differences in topology involving the G. coulteri, G. angustifolium, G. unijugum accessions.
These data show no evidence of contemporary hybridization between G. sanctum and G. coulteri. However, the intermediate position of the Chiapas accessions in the cpDNA network can be explained by a past event of hybridization and introgression of ancestral populations that subsequently diverged further into what are now distinct populations of G. coulteri and G. sanctum. Specifically, we hypothesize that the Chiapas region of Mexico could have served as an ancient refuge, thereby becoming a center of diversity during range expansion of what are now populations of G. sanctum in the Yucatan peninsula, Caribbean and Central America, and G. coulteri along the western coast of Mexico. Under this scenario, Chiapas populations later hybridized with Central American populations, retaining the intermediate cpDNA molecular signature while gaining an nrDNA pattern that is most closely related to the Central American populations. Interestingly, this scenario aligns with the suggestion by Porter (1972) that G. guatemalense is the result of hybridization and introgression between G. sanctum and G. coulteri. However, the data from this study show that such a scenario can only be applied to the Chiapas population(s), rather than to all accessions dubiously identified as G. guatemalense.
The species status of Guaiacum guatemalense is supported by these data inasmuch as accessions from Guatemala are monophyletic in the phylogenetic trees and gene networks for nrDNA. However, accessions from Costa Rica and Chiapas appear to be derived from Guatemalan nrDNA haplotypes (Figs. 3, 4), and there is strong support uniting most accessions collected from the same geographic region. Therefore, it is suggested that these phylogenetic patterns are best interpreted as significant intraspecific structuring within G. sanctum rather than recognizing G. guatemalense as a separate species, especially in the absence of distinguishing morphological characters. Additional sequence data would most likely raise bootstrap support values for subclades within G. sanctum, and raising G. guatemalense to a full species based on a molecular signature would likely warrant the splitting of other regional variants (e. g., Aruba and Curasao, Costa Rica, Chiapas) to species status. Conversely, sampling of additional populations (e. g., Honduras, Nicaragua) may reveal additional intermediate haplotypes that could reduce support for these subclades. This interpretation is congruent with the hypothesis that gene flow is limited and populations remain relatively isolated from one another due to natural and anthropologic habitat fragmentation. The haplotype networks and tree topologies for cpDNA and nrDNA data indicate that Guaiacum from Guatemala is not likely G. coulteri, and species identifications should be treated with care.
Guaiacum unijugum, G. coulteri and G. angustifoliumMaximum parsimony and statistical parsimony methods of phylogenetic reconstruction fail to clearly resolve sister relationships among accessions of G. unijugum, G. coulteri and G. angustifolium. This result is somewhat unexpected given that these taxa are readily distinguished based on morphology. Floral characters such as carpel number and basal filament appendages are generally distinct and unambiguous for species identification and sterile specimens can be readily distinguished based on leaf morphology, with leaflet number and width to length ratio being particularly useful characters.
The lack of resolution in the maximum parsimony analyses and patterns in the statistical parsimony networks are indicative of either recent or incomplete speciation, as well as possible hybridization among taxa. One reason the molecular data fail to resolve these relationships is the lack of cpDNA divergence and presence of shared haplotypes among taxa in this group, which can be explained by the slower mutational rate of the chloroplast genome (Wolf et al., 1987). Accessions of G. coulteri, G. coulteri var. coulteri, and G. unijugum share cpDNA haplotypes, and G. angustifolium is derived from this complex by a single mutation. Conversely, the nrDNA haplotypes do not show G. angustifolium as derived from the G. coulteri complex. Accessions of G. coulteri have multiple nrDNA haplotypes, with G. unijugum and G. coulteri var. palmeri being derived from different G. coulteri ancestral haplotypes.
This species complex may have arisen from a polyploidization event. Evidence of polyploidy in both G. coulteri and G. angustifolium (McCauley et al., 2008) has been identified; however karyotyping has not been conducted to resolve the full extent of these events. As such, the paraphyly in G. coulteri may be the result of differential amplification and sequencing of paralogous ITS loci.
Guaiacum angustifolium appears to have evolved from a shared ancestor with the G. coulteri complex, as evidenced by the nrDNA network. However, a subsequent hybridization in which G. coulteri cpDNA was introgressed back into the population may have occurred, which could explain the close, derived condition seen in the cpDNA network. Nevertheless, this taxon displays significant differences in morphology from other members in this clade, and the designation as a species is likely valid despite low phylogenetic resolution.
Taxonomically, we recommend that G. unijugum retain species status based on morphological, geographic, and molecular criteria. Morphologically, G. unijugum possess a suite of characters, both floral and vegetative that make it readily distinguishable from other taxa. The distribution of this taxon is also highly limited to a small region of the Baja peninsula that does not overlap with other taxa (Porter, 1963). Finally, nrDNA data show that G. unijugum is as divergent from G. coulteri as G. coulteri is from G. sanctum and G. angustifolium (8 mutational steps, Fig. 3). This is consistent with conclusions of McCauley et al. (2010) that G. unijugum and G. coulteri have evolved independently from a common ancestor, with microsatellite data also suggesting that remnant populations of G. unijugum are maintained primarily by selfing with little evidence of outcrossing or hybridization with outside populations.
Accessions identified as G. coulteri var. palmeri share a nrDNA haplotype that is derived from 1 of the 2 haplotypes found in other G. coulteri accessions, which is indicative of isolation and a separate evolutionary trajectory from other G. coulteri populations. However, this haplotype is not significantly divergent from other G. coulteri (var. coulteri) in cpDNA and nrDNA phylogenetic networks where it is separated by a single mutational step. Furthermore, the 2 nrDNA haplotypes found in G. coulteri var. coulteri accessions are more divergent from one another than the G. coulteri var. palmeri haplotype (Fig. 3), indicating that G. coulteri may have intraspecific genetic structuring similar to that seen in G. sanctum.
Accessions of G. coulteri var. palmeri are distinguished by a single morphological character, i. e., the presence of a pubescent ovary. This character is not discrete, and intermediate or partially pubescent ovaries were observed in several accessions. The pubescent ovary is also shared with G. unijugum and G. angustifolium, and it has been suggested that the pubescent character state is an environmental response shared by specimens in the northern distributional range of Guaiacum. An herbarium specimen identified as G. coulteri var. palmeri retained the pubescent ovary when planted outside of its natural range as an ornamental specimen (R. Grether 2513, 14-Aug-1989, MEXU), thereby providing some evidence of a genetic condition and not a strict environmental response. Ovary pubescence is retained in the developing and immature fruits, but the character becomes less discernible as the fruit develops and ripens. Sterile specimens of G. coulteri var. palmeri are indistinguishable from G. coulteri var. coulteri.
The lack of clear morphological differences and minimal genetic divergence lead to the conclusion that the distinction of varieties is likely unwarranted in G. coulteri.
Taxonomic conclusionsThe phylogenetic relationships resolved in this study do not suggest major taxonomic changes such as splitting of species are necessary in Guaiacum, although many nomenclatural issues remain to be addressed. While accessions from Guatemala form a moderately supported clade, these data do not show strong support of G. guatemalense as a distinct species based on genetic divergence. The majority of publications involving Guaiacum already treat G. guatemalense as a synonym of G. sanctum and from an application standpoint the absence of diagnostic morphological characters would make species identification difficult.
Varietal distinctions in G. coulteri are not supported by monophyly or unique haplotypes. The formal dissolution of these taxa would not have a large impact as the varietal distinction of var. palmeri is rarely referenced in the literature. Taxonomic distinction of G. coulteri var. coulteri was not observed in any of the 1 800+ herbarium specimens reviewed for this study, and distinction of G. coulteri var. coulteri from G. coulteri var. palmeri was not included among the 31 Guaiacum taxa listed in the International Plant Names Index (2009), nor listed in Tropicos, the leading database for tropical plant taxa (2009). The distinction between varieties does not follow the guidelines established by the International Code of Botanical Nomenclature (2009), and it is therefore assumed that a formal taxonomic revision is not necessary or practical for taxa that are so rarely recognized. The intermediate genetic fingerprint of the accessions from Chiapas provide evidence that past hybridization may have occurred, and contemporary hybridizations could be possible. These accessions also show that the taxonomic separation of G. sanctum and G. coulteri is not distinct at the molecular level. While it can be argued that speciation has not fully occurred between these taxa, to combine them as one species would not be prudent, as there is significant genetic divergence between the 2, and the range of overlap is relatively small.
Conservation applicationThe most significant finding of this study that is directly applicable to conservation is the biogeographic genetic structuring found within the various taxa. The Guaiacum taxa that were sampled in multiple locations display intraspecific genetic structuring, with G. officinale, G. sanctum, and G. coulteri all possessing unshared haplotypes among sampled populations. Given the resolution of the molecular methods utilized in this study, it can be concluded that this structuring is most likely a result of historical isolation and fragmentation, which is maintained by minimal long distance gene flow among distant populations. So while negative genetic effects caused by anthropogenic fragmentation are a conservation concern, this study indicates that much of the genetic variability among populations is more likely indicative of pre-fragmentation genetic patterns. This is likely attributable to the longevity of these trees, effective dispersal of seeds and efficient pollen transfer in what are generally naturally fragmented populations (Dertien and Duvall, 2009; Fuchs and Hamrick 2010a). This historical genetic structure is also consistent with the conclusions drawn from population level allozyme and microsatellite studies in G. sanctum (Fuchs and Hamrick, 2010a) and G. unijugum (MacCauley et al., 2010).
The historical genetic patterns demonstrate that individual populations are likely following different evolutionary trajectories than distant neighbors, with the implication that populations could be highly adapted to local environmental conditions or microclimates (Lopez-Toledo, 2011b). This could also explain the intermediate phylogenetic position between G. sanctum and G. coulteri of the Chiapas accessions. Genetic diversity levels of populations are not likely to be maintained through long distance gene flow, and even selective logging could have a high impact on a population that is genetically distinct because of its biogeographical history.
The current phylogenetic signature created from the dynamic biogeographical and evolutionary history of Guaiacum makes species delimitation difficult in a context useful for regulating harvesting and trade. As such, it may be more pragmatic to protect and regulate trade on the generic taxonomic level rather than the species level to provide proper levels of protection for remaining Guaiacum populations. However, the consistent correlation between distinct haplotypes and geography show promise that DNA barcoding methods could be developed as a tool for specimen identification and determining provenance.
AcknowledgementsThis work was supported in part by the Plant Molecular Biology Center and the Department of Biological Sciences at Northern Illinois University. The authors would like to thank Frank Axelrod (University of Puerto Rico at Río Piedras), Leonel López-Toledo (San Diego Zoo’s Institute for Conservation Research), and Ross A. McCauley (Fort Lewis College) for their insight and assistance in collecting specimens. Mr. Cosme Becerra (Northern Illinois University) graciously provided Spanish language translation.