The Magnificent Frigatebird (Fregata magnificens) is a monogamous, widespread, neotropical, pelagic seabird with low flight costs, high dispersal capacity, sex-biased dispersal, and female mate choice. Here, we adopt an integrative approach to evaluate the influence of behavior, male-biased philopatry, female choice, and geographic distance as non-physical barriers to dispersal acting on the genetic structure of magnificent frigatebirds in the 4 largest Mexican breeding colonies. We integrate ecological dispersal observations from tagging studies, analyses of maternally and biparentally inherited genetic markers, and group-structured population modeling. Mitochondrial DNA (matrilineal) and randomly amplified polymorphic DNA (biparental) data provided evidence of genetic differentiation between ocean basins, as well as among Eastern Pacific breeding colonies, and absence of a genetic pattern of isolation by distance. Our ecological, genetic, and modeling results are concordant with the expected effects of sex-biased philopatry and female mate choice as non-physical barriers to interbreeding, and suggest that geographic distance does not play a significant role in the genetic distinction of breeding colonies. The genetic differentiation found between Eastern Pacific and Caribbean, as well as that among Eastern Pacific breeding colonies is of consequence for the management plans and conservation measures of the Magnificent Frigatebird in the region.
La fragata común (Fregata magnificens) es un ave marina monógama, ampliamente distribuida, neotropical y pelágica, con bajos costos de vuelo, alta capacidad de dispersión, dispersión sesgada por sexo y selección de pareja por parte de la hembra. Aquí, adoptamos una aproximación integrativa para evaluar la influencia del comportamiento, la filopatria sesgada hacia los machos, la selección de pareja por parte de las hembras y la distancia geográfica como barreras no físicas para la dispersión, actuando en la estructura genética de las fragatas comunes de las 4 colonias reproductivas más grandes de México. Integramos observaciones ecológicas de dispersión, producto de estudios de marcaje, análisis de marcadores genéticos heredados maternal y biparentalmente, y modelos poblacionales de estructura de grupos. Los datos de ADN mitocondrial (matrilineales) y de ADN polimórfico amplificado al azar (biparentales) proporcionaron evidencia de la diferenciación genética entre cuencas oceánicas, y entre las colonias reproductivas del Pacífico oriental, así como de la ausencia de un patrón genético de aislamiento por distancia. Nuestros resultados ecológicos, genéticos y de modelaje son concordantes con los efectos esperados de filopatria sesgada por sexo y de selección por parte de las hembras como barreras no físicas para el entrecruzamiento, y sugieren que la distancia geográfica no juega un papel significativo en la distinción genética de las colonias reproductivas. La diferencia genética encontrada entre el Pacífico oriental y el Caribe, así como entre las colonias reproductivas del Pacífico, es relevante para los planes de manejo y las medidas de conservación de la fragata común en la región.
Physical, ecological and behavioral mechanisms are responsible for the presence of genetic differentiation among populations; hence their understanding is central for the identification of barriers that promote breeding isolation. In some highly dispersive seabird species, strong genetic differentiation among populations may occur in spite of their ability to breach obvious physical barriers, such as landmasses. This implies the action of non-physical barriers disrupting gene flow (Friesen, 2007; Friesen et al., 2007). In the absence of physical barriers to dispersal, geographic distance may prevent panmixia provided that individual movements are shorter than the whole distributional area of the species, thereby favoring an isolation by distance model (Wright, 1943). More specifically, since seabird breeding colonies are often discontinuously distributed, sex-biased movements of migrating individuals are restricted to adjacent colonies similar to a stepping stone model (Kimura, 1953), because establishment in familiar surroundings increases the chance of breeding success (Rockwell and Barrowclough, 1987). Furthermore, usually monogamous birds show male-biased philopatry and female-biased dispersal (Liberg and Von Schantz, 1985), and most seabird species exhibit different mating patterns and strategies, as opposed to the common random mating assumption in genetics (Sugg et al., 1996). Both sex-biased philopatry and mate selection have a prime influence on population structure and genetic variation (Chesser, 1991a; Chesser, 1991b).
Sex-biased mating patterns and breeding dispersal are social behaviors that promote genetic variation, within and between neighboring colonies, and both influence the average relatedness (i.e., coancestry) of colony members, usually among those of the philopatric sex (Sugg et al., 1996; Ross, 2001). Because in seabird species with high dispersal capability, such as frigatebirds, distance per se seems an unlikely mechanism for restricting gene flow, genetic structure has been related to behavioral factors promoting breeding isolation, such as sex-specific post-breeding movement patterns and mating preferences (Dearborn et al., 2003; Cohen and Dearborn, 2004; González-Jaramillo and Rocha-Olivares, 2011; Hailer et al., 2011). Assessing the influence of the breeding system and sex-biased dispersal on the genetic structure of populations can be achieved with the complementary use of genetic markers with different types of inheritance (e.g., biparental nuclear DNA and matrilineal mitochondrial DNA, Scribner et al., 2001).
The Magnificent Frigatebird (Fregata magnificens, hereafter frigatebird) is a high concern (Kushlan et al., 2002), sequentially monogamous (Diamond, 1972; Diamond, 1973), long-lived (>40 years for Fregatidae Family; Madsen et. al., 2007a), colonial, pelagic seabird, that breeds in tropical and subtropical Eastern Pacific and American Atlantic Oceans. In the Eastern Pacific, frigatebirds nest from Ecuador, including Galapagos Islands, to Baja California and in the American Atlantic from Brazil, including Cape Verde Islands, to the northern Gulf of Mexico (Harrington et al., 1972; Harrison, 1983). It is the only species of the family Fregatidae that breeds in the Caribbean (Nelson, 1975). In Mexico, the largest breeding frigatebird colonies are in Santa Margarita (24°25' N, 111°07' W), Tunitas (25°00' N, 108°10' W), and Isabel (21°51' N, 105°54' W) islands in the Eastern Pacific Ocean, as well as in Contoy island (21°27' N, 86°46' W) in the Caribbean Sea (Fig. 1). Because frigatebirds have high dispersal capability (Weimerskirch et al., 2006) and low flight costs (Harrington et al., 1972; Pennycuick, 1983; Weimerskirch et al., 2003), they can fly from one ocean to another, breaching physical barriers such as the Panama Isthmus (Hailer et al., 2011). Nevertheless, this species exhibits male-biased breeding philopatry (González and de la Cueva, 2007), male site fidelity (Osorno, 1996), female mate choice (Madsen et al., 2004; Madsen et al., 2007a; Madsen et al., 2007b), as well as female-biased parental care investment for their single altricial chick (up to 17 months vs. 3 months for males, Osorno, 1999; Osorno and Székely, 2004). The resulting breeding cycle, biennial for successful females and annual for males (Diamond, 1975; Osorno, 1996), and its breeding characteristics and behavior are of consequence for population genetics (Sugg et al., 1996; Ross, 2001).
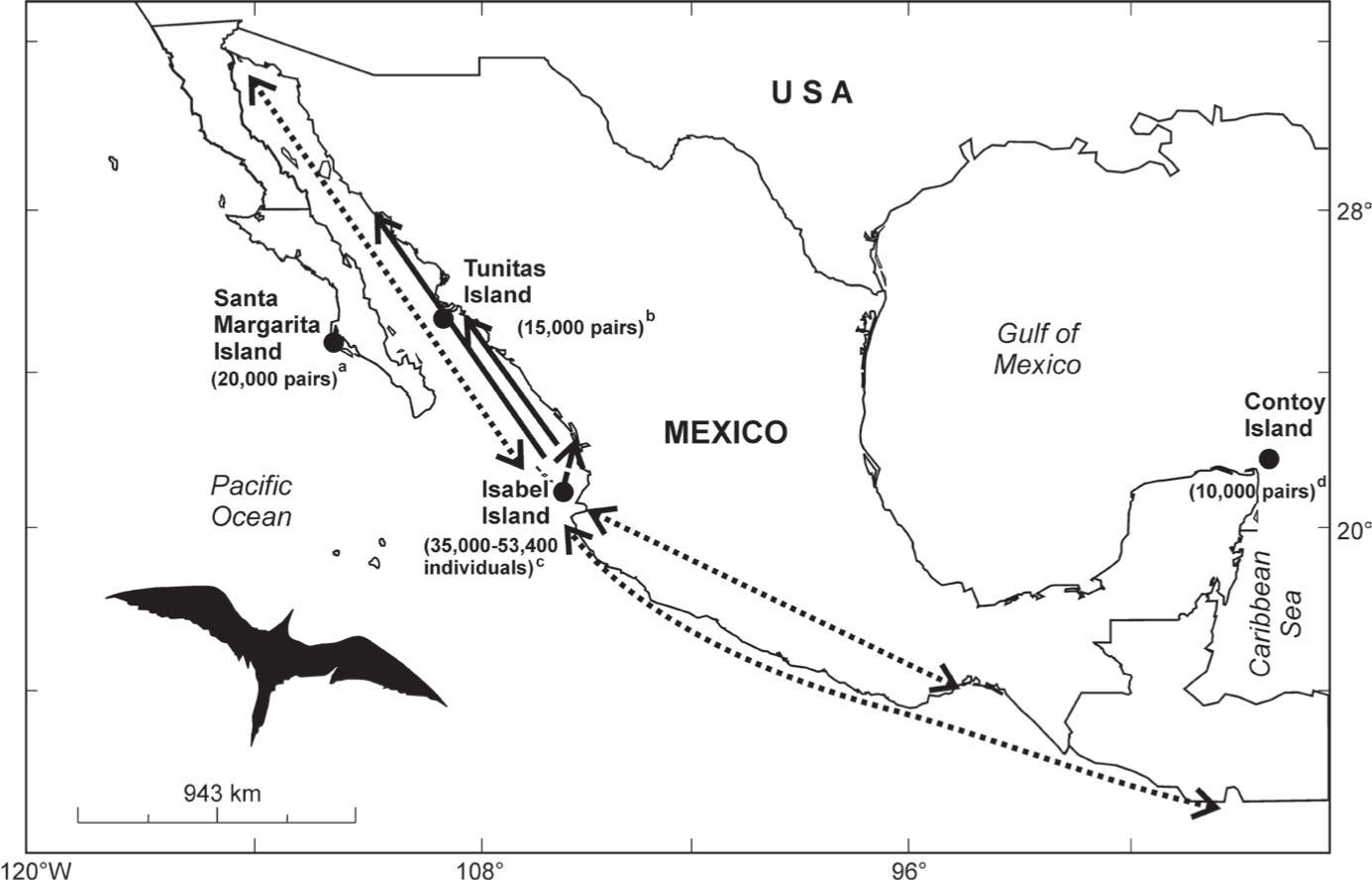
Breeding colonies and census numbers of frigatebirds Fregata magnificens in Eastern Pacific and Caribbean Sea, México, analyzed in this study. aMoreno and Carmona (1988), bCarmona and Daneman (1994),cMadsen (2005). Trajectories of banded or satellite tracked organisms are indicated by arrows (solid line: females, dotted lines: males). Photograph by R.S. Quintero-Félix.
Previous genetic surveys of frigatebird populations have underlined the importance of behavioral mechanisms as responsible for the observed patterns of differentiation. Using analyses of mitochondrial DNA (mtDNA) and nuclear loci, Hailer et al. (2011) unraveled the long-term isolation and endemism of the Galapagos frigatebird breeding colony relative to other colonies in the Pacific coast of Panamá, Colombia and in the Caribbean; finding no genetic heterogeneity among the latter. In that study, only 5 historical museum specimens (4 collected in 1895 and 1 in 1961) from the Pacific coast of Mexico were genetically analyzed (only for mtDNA). On the other hand, González-Jaramillo and Rocha-Olivares (2011) focused their mtDNA sequence analyses on contemporaneous organisms from the major frigatebird Mexican breeding colonies, 3 in the Pacific and 1 in the Caribbean, finding genetic differentiation between Pacific and Caribbean Mexican breeding colonies, but none among the Pacific colonies. Contrasting as they are, these 2 studies are not directly contradictory as both suggest behavior as a prime cause of the detected genetic differentiation between colonies (Galapagos vs. Pacific coast of Panama, and Pacific vs. Caribbean, respectively). Furthermore, they did not involve the same breeding colonies, as there was no geographical overlap in genetic analyses. Nevertheless, the question remains as to whether the patterns found in the mtDNA sequences relating to the differentiation of Mexican Caribbean and Pacific colonies, and the lack of differentiation among the latter, also extend to biparentally inherited nuclear markers or are constrained to the matrilineal mitochondrial genome.
Randomly Amplified Polymorphic DNAs (RAPDs) are bi-parentally inherited anonymous nuclear markers that have been successfully used in bird population genetics studies (e. g., Geese, Horn et al., 1996; Storm Petrels, Paterson and Snyder, 1999; Vireos, Zwartjes, 1999; Iberian Imperial eagle, Padilla et al., 2000). They allow the analysis of a large number of loci using short (10 base-pair long) synthetic primers of random sequences to PCR-amplify and can randomly sample the genome more completely than other nuclear markers based on specific coding regions (Hadrys et al., 1992; Lynch and Milligan, 1994; Parker et al., 1998). On the other hand, the use of RAPDs has been criticized on the basis of its sensibility to a variety of PCR artifacts that can limit their reproducibility across amplification experiments (Hadrys et al., 1992; Parker et al., 1998). Nevertheless, experimental steps can be taken to overcome this problem and increase the reliability and accuracy of genotypes, such as standardizing and optimizing amplification conditions, as well as replicating the amplification in independent runs. This helps in the selection of reliable and consistent loci, eliminating those that are prone to scoring error (see Zwartjes, 1999). Thus, RAPD markers have been successfully used to reliably detect avian population structure (Haig et al., 1994; Haig et al., 1996), provided strict amplification protocols ensuring band consistency are followed, such as the ones used in this study.
In light of the limited understanding of the evolutionary and ecological forces shaping the genetic structure of Mexican frigatebird breeding colonies, in this study we integrate published and unpublished ecological (reproductive and dispersal) and genetic (nuclear and mitochondrial) data to test with increased power the influence of non-physical barriers in the genetic structure of the species. Specifically, integrating mitochondrial (control region sequences) and nuclear (RAPDs) markers will allow us to use uni- and bi-parental inheritance group-structured models that explicitly incorporate breeding ecological variables to infer relevant population genetic parameters such as female and male dispersal (Chesser, 1991b; Chesser, 1991a; Chesser and Baker, 1996; Scribner and Chesser, 2001). We propose that the evidence of genetic structure at different spatial scales, not only between ocean basins, but also among Pacific breeding colonies, will strengthen the hypothesis that behavior and not physical barriers, such as distance or land masses, will be the prime force limiting gene flow among reproductive colonies of this species.
Materials and methodsFrigatebird tagging and blood collectionAs part of this and other studies (Madsen, 2005; González and de la Cueva, 2007; Madsen et al., 2007a; Madsen et al., 2007b; González-Jaramillo et al., 2010; González-Jaramillo and Rocha-Olivares, 2011), almost 2 800 frigatebirds nesting in Isabel Island were banded on the left wing (numbered nylon yellow canvas tags 20×8cm and 10.8gr) and ringed using numbered metal rings on the left tarsus from 1998 to 2003. Six satellite radio-transmitters were fitted on the backs of 3 banded males and 3 banded females in 1999 (V. Madsen pers. comm.). A record of the tagged and tracked frigatebirds at Isabel Island was kept during the following years from 2000 through 2005 breeding seasons (see González and de la Cueva, 2007; González-Jaramillo et al., 2010). In addition, a record of opportunistic resightings of banded frigatebirds reported by several birdwatchers along the Mexican Pacific was kept in order to document dispersal patterns from Isabel island. The other colonies were visited once during the 2004 breeding season in search of banded breeding frigatebirds; during April in Santa Margarita and Tunitas Islands in the Mexican Eastern Pacific and during March in Contoy Island in the Mexican Caribbean. At that time, blood samples (n×99) were collected by extracting blood from the brachial vein of adult female (nf×14) and male (nm×15) frigatebirds nesting in Santa Margarita, Tunitas (nf×13, nm×14) and Isabel (nf×8, nm×9) islands, as well as adults (nf×10, nm×8) and juveniles (nj×8) on Contoy island (Fig. 1).
DNA isolation and genetic analysesThis study is based on the same blood samples used in González-Jaramillo and Rocha-Olivares (2011), where methodological details about DNA extraction can be found. Mitochondrial sequences from the second half of conserved domain II and the entire highly variable domain III of the mitochondrial control region were produced as part of our previous study (González-Jaramillo and Rocha-Olivares, 2011). For RAPD marker development, one organism from each breeding colony was assayed with 20 primers (Kit C, Operon Technologies, Inc., www.eurofinsdna.com) alone and in combinations in order to identify their ability to amplify bright, distinct and easily scorable bands. Reactions were optimized for DNA template amount and thermal cycling. RAPD amplifications (25μl) included 60ng of genomic DNA,10mM Tris-HCl, 50mM KCl, 1.5mM MgCl2,0.25mM each dNTPs, 0.12μM each primer (Operon), 0.001% gelatin and 0.75 U Taq Hot start DNA polymerase (New England Biolabs). We used 2 different amplification cycles: (1) 3min at 94° C and 45 cycles of 15s at 94° C, 30s at 30° C, 90s at 72° C, and 7min at 72° C for individual primers; and (2) 3min at 94° C and 65 cycles of 1min at 94° C, 1min at 33° C, 2min at 72° C, and 7min at 75° C for primer combinations. Amplifications were screened in 1.5% agarose gels stained with ethidium bromide, run for 3h at 4V.cm−1 (0.5X TBE) with a negative control reaction and using a DNA molecular weight marker (Hi-Lo, Bionexus Inc.).
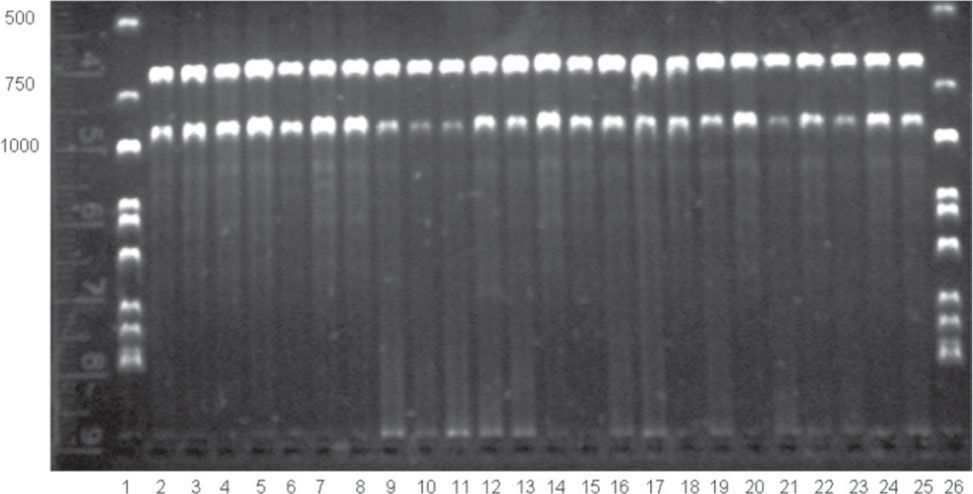
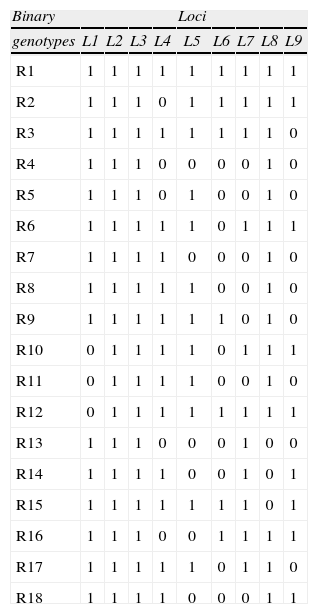
One individual primer (OPC-9 [5' CTC ACC GTC C-3']) and 2 combinations (OPC-1 [5'-TTC GAG CCA G-3']+OPC-2 [5' GTG AGG CGT C-3']; and OPC-19 [5'-GTT GCC AGC C-3']+OPC-20 [5'-ACT TCG CCA C-3']) produced 9 consistent bands: OPC-9=2;OPC-1+OPC-2=2; and OPC-19+OPC-20=5 (Fig. 2). We based our quality control protocol on Zwartjes (1999) to improve the stringency and the consistency of our genotyping: 1) DNA samples of the 99 frigatebirds were independently amplified twice with each primer and primer combinations and fragments of the same molecular weight were identified using a constant reference ladder; 2) loci from different replicates were scored in separate gels as present (visible dominant allele 1) or absent (null allele 0) per individual per locus and results were compared between replicates; 3) repeatability was checked by considering as repeatable only bands that amplified consistently in independent replicate amplifications, hence 100% of the alleles analyzed were deemed repeatable and reliable by this criterion (Table 1).
RAPD amplification using OP-9 primer of DNA frigatebird Fregata magnificens samples from Tunitas breeding colony, Mexican Eastern Pacific. Lanes 1 and 26 contain a DNA ladder, ladder molecular weight and assigned numbers of frigatebird samples are indicated in the horizontal and vertical axes respectively.
RAPD binary genotypes of Mexican frigatebirds Fregata magnificens. Band patterns were scored as present (visible allele 1) or absent (null allele 0) per locus (L) per individual
| Binary | Loci | ||||||||
| genotypes | L1 | L2 | L3 | L4 | L5 | L6 | L7 | L8 | L9 |
| R1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 |
| R2 | 1 | 1 | 1 | 0 | 1 | 1 | 1 | 1 | 1 |
| R3 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 0 |
| R4 | 1 | 1 | 1 | 0 | 0 | 0 | 0 | 1 | 0 |
| R5 | 1 | 1 | 1 | 0 | 1 | 0 | 0 | 1 | 0 |
| R6 | 1 | 1 | 1 | 1 | 1 | 0 | 1 | 1 | 1 |
| R7 | 1 | 1 | 1 | 1 | 0 | 0 | 0 | 1 | 0 |
| R8 | 1 | 1 | 1 | 1 | 1 | 0 | 0 | 1 | 0 |
| R9 | 1 | 1 | 1 | 1 | 1 | 1 | 0 | 1 | 0 |
| R10 | 0 | 1 | 1 | 1 | 1 | 0 | 1 | 1 | 1 |
| R11 | 0 | 1 | 1 | 1 | 1 | 0 | 0 | 1 | 0 |
| R12 | 0 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 |
| R13 | 1 | 1 | 1 | 0 | 0 | 0 | 1 | 0 | 0 |
| R14 | 1 | 1 | 1 | 1 | 0 | 0 | 1 | 0 | 1 |
| R15 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 0 | 1 |
| R16 | 1 | 1 | 1 | 0 | 0 | 1 | 1 | 1 | 1 |
| R17 | 1 | 1 | 1 | 1 | 1 | 0 | 1 | 1 | 0 |
| R18 | 1 | 1 | 1 | 1 | 0 | 0 | 0 | 1 | 1 |
We estimated the following breeding parameters for the group-structured model (Chesser and Baker, 1996; Chesser, 1998): (b) mean number of mates per male (b=1.0, in case of frigatebirds given their socially consecutive monogamy breeding system –i.e., males mate with only 1 female each breeding season, even though they may differ between breeding seasons) –; (k) mean progeny per female that survives to reproduce, assuming a minimum breeding female lifespan of 35 years and a life span >40 years (Madsen et al., 2007a), and including all the possible female productivities (0, 0.1, 0.2, 0.3, 0.4 and 0.5 chicks per year) multiplied by the nesting success (0.17, Osorno, 1996) and by the fledging survival rate up to breeding age (φˆ=0.62; González and de la Cueva, 2007); (σk2) variance of k;mean number of females (n) and variance (σn2), and mean number of males (m) based on census at each breeding colony (Fig. 1); and (l) mean number of male mates per female (l=1, assuming single paternity; Chesser and Baker 1996).
Genetic structure analysesIn order to assess the level of genetic structure using RAPDs, we first used the program RAPDistance v1.04 (Armstrong et al., 1995) to compute a matrix of genetic distances between binary genotypes from all populations. We estimated Excoffier et al. (1992) inter-genotypic distance metric: d=n*(1-(n11/n)), where n is the total number of RAPD loci in 2 genotypes being compared, and n11 is the number of bands present (scored 1) in both genotypes. A distance-based approach allows the quantitative assessment of these data in the presence of dominance without the assumption of Hardy-Weinberg or linkage equilibria. We computed ΦST, which is a molecular analog of FST, as implemented in the program Arlequin v3.5.1.2 (Excoffier et al., 2005), using the matrix of inter-genotype Excoffier et al. (1992) distances as Euclidian distances. Two AMOVAs were initially performed to estimate the following molecular variance components: i) among the 4 breeding colonies (populations): Santa Margarita, Tunitas, Isabel and Contoy to test for overall panmixia (ΦSTALL); and ii) a nested AMOVA decomposing the molecular variance between 2 “groups”: the Eastern Pacific group (Santa Margarita, Tunitas and Isabel) and the Caribbean sample (Contoy); among populations within groups and within populations. In addition, pairwise ΦST were also computed between colonies. The Benjamini and Yekutieli False Discovery Rate method (B-Y FDR, Benjamini and Yekutieli, 2001) was used to adjust significance for multiple comparisons. In order to complement AMOVA ΦST analyses, which are not specifically designed for dominant markers such as RAPDs, we adopted 2 model-based approaches based on Bayesian inference and Markov Chain Monte Carlo (MCMC) simulations to assess the level of genetic structure. These specifically incorporate the uncertainty in the identifiability of genotypes and allele frequencies introduced by dominance. First, we computed the statistic θB to assess differentiation at the same hierarchical levels of the AMOVAs above. θB is an FST analog derived from a Bayesian hierarchical model appropriate for dominant markers, given that it incorporates the uncertainty of the magnitude of inbreeding into FST estimates (Holsinger, 1999; Holsinger et al., 2002). We ran the f-free model, which is independent of assumptions regarding the inbreeding coefficient (f), whose estimation is unreliable from dominant markers. We report values of θ(II), which is a θB-statistic better suited in estimating the proportion of genetic diversity due to differences among contemporaneous populations. Simulations were carried out with the program Hickory v1.1 (Holsinger and Lewis, 2007) with default parameters (burn-in= 5 000, sample= 100 000, thin= 20) and each run was independently replicated (n=10) to assess consistency and convergence of the sampler. Second, we used the model-based Bayesian clustering assignment method implemented in the program STRUCTURE v2.3.4 (Pritchard et al., 2000), in which a number of population clusters (K) of individuals are delineated on the basis solely of their multilocus genotypes using a Bayesian approach. The model accounts for the existence of linkage and Hardy-Weinberg equilibria by introducing population structure and attempts to find the groupings (i.e., clusters) that maximize linkage and genotypic equilibria (Pritchard et al., 2000). We used the statistic ΔK of Evanno et al. (2005) to estimate the most likely number of clusters inherent in our data using values of K=1−6. MCMC simulations were carried out using the STRUCTURE model extended to dominant markers that incorporates a new update step into the algorithm accounting for the uncertainty introduced by dominance in the ascertainment of genotypes by assessing likelihoods based on the probabilities of all possible genotypes (Falush et al., 2007). All MCMC simulations of the correlated allele-frequencies and admixture model sampler were independently replicated (n=10) and run for a length of 106 steps with the first 104 discarded as burn-in to assure independence from initial conditions.
In order to assess the influence of geographic distance among breeding colonies on the genetic structure of frigatebirds, we tested for isolation by distance by performing a Mantel test correlating the matrix of pairwise ΦST values and the corresponding geographic linear distances between pairs of colonies using Arlequin v3.5.1.2 (Excoffier et al., 2005).
Gene flow estimationsWe estimated gene flow (Nm) levels using different approaches. A first-order estimation was obtained assuming Sewall Wright's island model of population structure and using ΦST and θB as FST analogs in: Nm=[(1/FST)-1]/4, where: FST is the probability that 2 alleles chosen at random are identical by descent within the same population, and Nm is gene flow in number of effective migrants per generation (see Slatkin and Barton, 1989).This approach, however, has been questioned on the grounds that the assumptions behind the island model are seldom met in natural populations (Whitlock and McCauley, 1999). Hence, we adopted an alternative approach (Bielawski and Pumo, 1997) modified from Slatkin's (1981) method to graphically estimate lower and upper bounds of gene flow for frigatebirds. The graphic method involves plotting the observed conditional average marker frequencies p(i) as a function of i/d, where i refers to the number of demes, which total d, in which an allele is present (i.e., occupancy number in Slatkin, 1981) and comparing them to theoretical curves numerically simulated by the model. Maximum qualitative estimates were based on null markers whereas minimum estimates were determined by ignoring null markers and treating all rare dominant markers as homozygous, thereby overestimating rare marker frequencies (Bielawski and Pumo, 1997).
Integrative group-structured population modelWe used recently derived theory developed for group-structured populations to integrate ecological (reproductive) and genetic (nuclear and mitochondrial) data into a model allowing us to estimate male (dm) and female (df) dispersal rates from uni- and bi-parentally inherited markers (Chesser and Baker, 1996; Chesser, 1998). The model analyzes the accumulation of coancestry among individuals within reproductive groups as affected by the mating system, the mean and variance in the number of progeny produced in each mating, and the movement of individuals among groups. The relevant parameters are the probability that a randomly selected pair of mates would come from the same population:
where σn2 is the variance in the mean number mates per group (n) and s is the number of groups (Chesser and Baker, 1996, Eq. 5) and the probabilities that a random pair of progeny are born from the same female ψf (Chesser and Baker, 1996, Eq. 1) or from the same male ψm (Chesser and Baker, 1996, Eq. 2-4), which in case of monogamy and single paternity reduce to ψ=ψm=ψf, where: ψ=[σk2+k(k−1)]/k(ks−1), where σk2 is the variance in the mean progeny per female that reproduces (k) and s is the number of groups (Chesser and Baker, 1996, Eq. 1). With this information exact equations are available for the degree of matrilineal genetic structure (ΦSTmtDNA) and the female effective population size (NemtDNA) as a function of df, ψ, and η (Eqs. 3 and 4, Scribner et al., 2001). The expression of matrilineal genetic structure can be used to solve for df as: df= ψ(1 - ΦSTmtDNA)/ΦSTmtDNA [1- ψ (1 - η) - ΦSTmtDNA η] (Chesser and Baker, 1996, Eq. 16) and the female effective population size is NemtDNA= 1/(df η ΦSTmtDNA) (Chesser and Baker, 1996, Eq. 12). Hence, empirical mtDNA data can be used to derive an explicit estimate of female dispersal accounting for the observed ΦST. No analogous exact equation is available to express dm as a function of reproductive parameters using data derived from nuclear biparental genetic markers (i.e., RAPDs in our case); instead, a numerical simulation approach is necessary to solve for dm and diploid effective population size (Nenuclear). Simulations involve numerically iterating the transition equations of Chesser and Baker (1996, Eqs. 27–28) to simulate inbreeding (Ft+1), coancestry (θt+1), and gene correlation among breeding groups (αt+1) as a function of (Ft, θt, αt), where t is time in generations. We followed Scribner and collaborators (Scribner and Chesser, 2001; Scribner et al., 2001) to implement simulations using increasingly higher dm values from 0 in 0.01 increments. For each dm, asymptotic values of FST and Nenuclear were derived after 10 000 generations, where Nenuclear ≈ [4 − η (1 + 3 FIS)] / 6η (FST-FIS), where FIS=(F−θ) / (1−θ) and FST=(θ−α) / (1−α) at asymptotic equilibrium and η is as defined above. After each of the dm iterations, the simulated FST was compared to the empirical value to assess convergence or departure. Transition equations were used until simulated estimates converged on the observed FST (i.e., ΦST or θB fromRAPDs). Initial conditions for all simulations were F0=θ0=α0=0.ResultsDispersal and breeding ecologyOf the almost 2 800 banded frigatebirds on Isabel island, a resighting record was obtained for 870 (males, n=506; females, n=364) from 1998 to 2003, of which 64% (n=324) of the banded males and 49% (n=178) of the females returned to the breeding colony at least 1 year after they were tagged (González and de la Cueva, 2007). No banded breeding frigatebirds from the Isabel colony were found breeding on Santa Margarita, Tunitas, or Contoy islands during our fieldwork in 2004. Nevertheless, 3 banded females were observed breeding in Bahía Santa María, adjacent to the Tunitas colony (R. S. Quintero-Félix and A. Castillo pers. comm.), but details about resighting data were unavailable. From 1995 to 2012, 24 banded frigatebirds (21 males and 3 females) have been reported flying over the Mexican Eastern Pacific coast: in the upper Gulf of California during 1995 and from 2001 to 2003 (16 males and 3 females; J. C. Pérez y S. Jauregui, pers. comm.); and on the coasts of Oaxaca in 2004 (1 male; T. Trilar, pers. comm.); Sonora in 2010 (1 male; J. Cruz-Nieto, pers. comm.) and Nayarit in 2010 (1 male; C. Contreras, pers. comm.) and in 2012 (1 female; K. Fry and C. Contreras, pers. comm). Two of the 3 post-breeding satellite tracked males banded in 1999 on Isabel island, which were recorded flying over the Eastern Pacific coast as far as Nicaragua (Lee and Osorno, unp. data), returned to the Isabel breeding colony at least 1 year after being marked (2000–2001 breeding season) and in the next breeding season (2001–2002) (Fig. 1). These birds were no longer recorded on the island from 2002 to 2005. The third satellite-tracked male could only be tracked for a few days because of failure of the transmitter, during which time it remained close to the colony (Lee and Osorno, unp. data). The 3 satellite tracked females moved over Nayarit's coast during chick rearing and after chick fledging (Lee and Osorno, unp. data), and were not resighted on Isabel island from 2000 to 2005 breeding seasons.
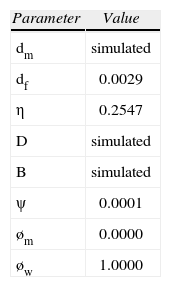
Estimated breeding parameters and breeding probabilities were: i) mean progeny per female that survive to reproduce (k=0.9225) and variance of k(σk2=0.476); ii) mean number of females (n=8 997) and variance((σn2=1.5335 ×106)), and mean number of males (m=8 826); iii) the probability that a randomly selected pair of mates would come from the same population was: η=0.255, and iv) the probability that a random pair of progeny were born from the same female and the same male was: ψ=5.30¿10−5. Finally, using Chesser and Baker's (1996) model and a global ΦSTmtDNA= 0.0182, obtained from our control region data (González-Jaramillo and Rocha-Olivares, 2011), we estimated the female rate of dispersal as df=0.0029 (Table 2).
Breeding ecology parameter values used in the group- structured genetic model of Chesser and Baker (1996). See text for details
| Parameter | Value |
| dm | simulated |
| df | 0.0029 |
| η | 0.2547 |
| D | simulated |
| B | simulated |
| ψ | 0.0001 |
| øm | 0.0000 |
| øw | 1.0000 |
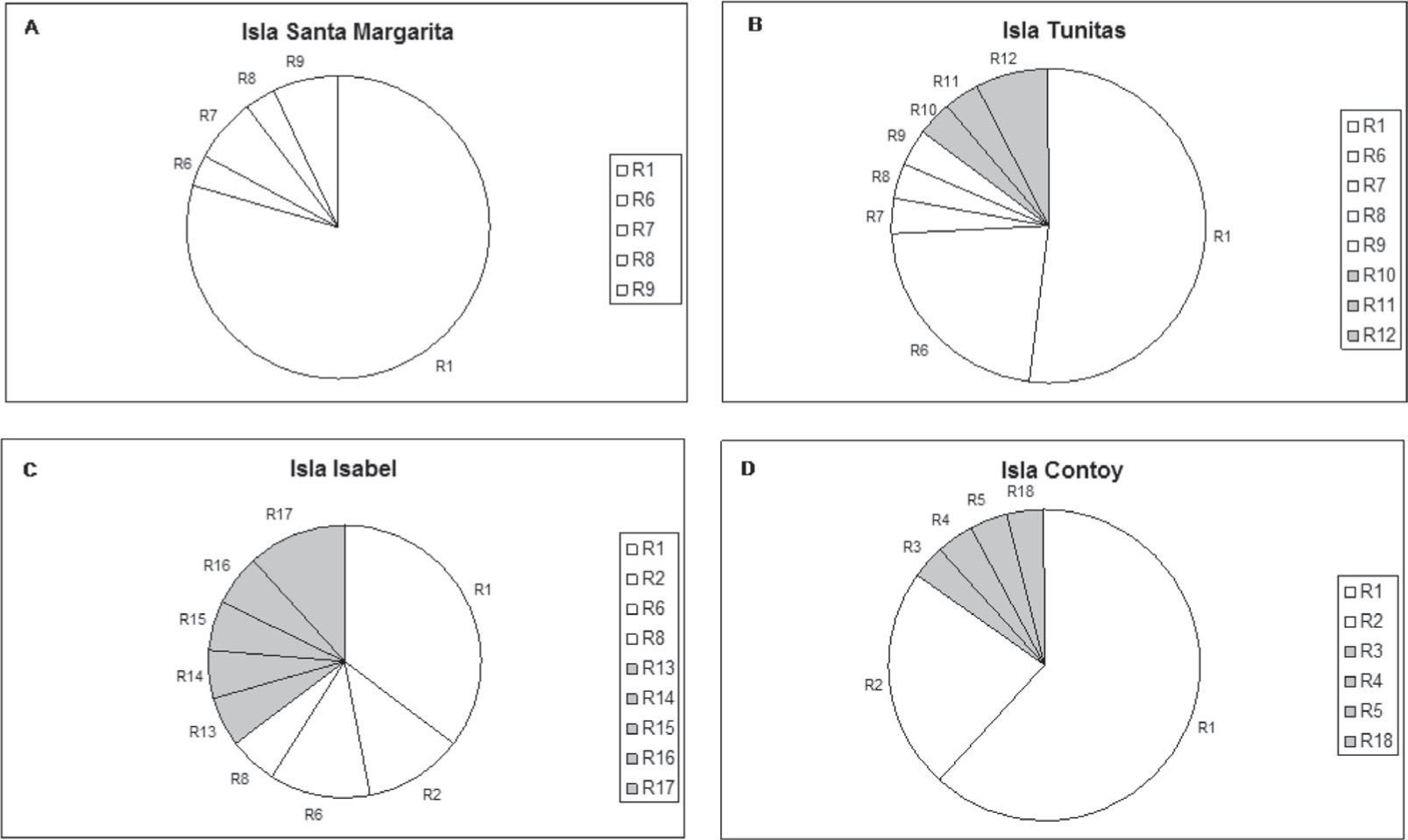
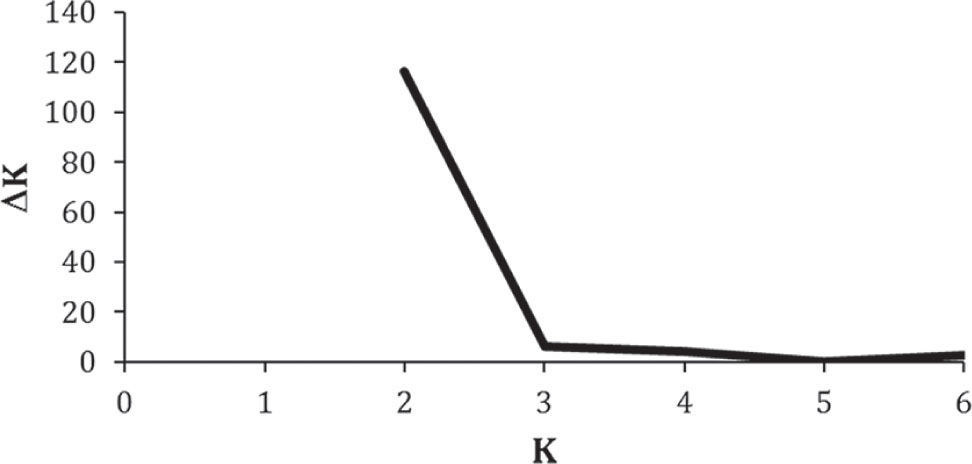
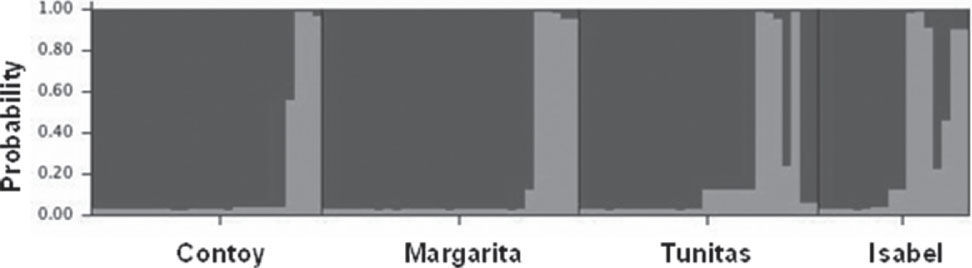
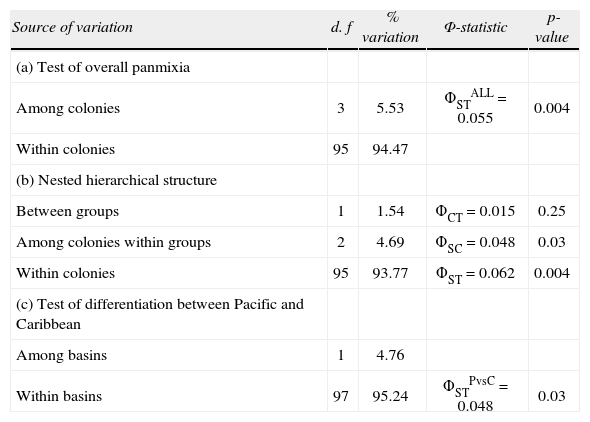
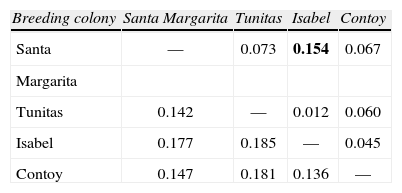
A total of 18 binary genotypes involving 9 loci were scored among 99 frigatebirds, of which genotype R1 was the most abundant in all localities. Six genotypes were shared among colonies (R1, R2, R6, R7, R8, R9; Fig. 2). Except for R1, genotypes found on Santa Margarita were only shared with the rest of Eastern Pacific colonies. On the other hand, except for R2 shared with Isabel island, the rest of Contoy genotypes were private. However, the proportion of private genotypes decreased from the Caribbean to the Pacific Contoy (66%), Isabel (55%), and Tunitas (37%) (Fig. 3). Fixation indices yielded largely congruent results regarding the patterns of genetic structure; however, the Bayesian model-based θ(II) yielded values twice or larger than ΦST obtained from the AMOVAs. Depending on the statistic, a small (i.e., <10%) but significant 5.5 to 16% of the molecular variance was accounted for by differentiation among the 4 frigatebird breeding colonies (ΦSTALL= 0.055, p= 0.004; θ(II)ALL= 0.163, p< 0.05, Table 3a). A hierarchical nested AMOVA confirmed this finding and provided support for the genetic distinction among the 3 Pacific colonies (ΦSC=0048, p=0.03; θ(II)=0.175, p<0.05, Table 3b). However, grouping the 4 colonies into 2 unbalanced groups (a Pacific group composed of 3 colonies and a Caribbean group including 1 colony) produced a smaller and non-significant inter- group fixation index (ΦCT=0.015, p=0.25, Table 3b). This relates to the very small power of the test given that the significance for this variance component is computed by shuffling at random entire colonies between groups (Excoffier et al., 1992). Indeed, with 4 samples and 2 groups there are only 7 possible unordered combinations, thus any random outcome has a probability of occurrence of 0.14, which represents the smallest p-value. In order to address that problem, we computed inter-basin differentiation by grouping Pacific genotypes into one sample and computing a ΦST between both samples (i.e., Pacific vs. Caribbean). This produced a small but significant fixation index accounting for 4.8% of the molecular variance between basins (ΦSTPvsC= 0.048, p= 0.03, Table 3c), a differentiation confirmed by the considerably larger index derived from Bayesian simulations (θ(II)PvsC=0.140, p<0.05). Pairwise ΦST values ranged from 0.012 between Isabel and Tunitas to 0.154 between Santa Margarita and Isabel, and most were significant individually but only the latter remained so (p=0.003) after applying the B-Y FDR correction. On the other hand, pairwise θ(II) were all larger than 0.13 and none of the Bayesian credibility intervals overlapped zero. In general, they varied proportionally to ΦST, except for the comparisons of Tunitas with Isabel and with Contoy that were much larger than predicted by their ΦST counterpart (Table 4). According to Evano's ΔK, 2 were the most likely number of clusters fulfilling the linkage and HW equilibria of Bayesian clustering, of which one was predominant (dark grey, Fig. 4). The vast majority of birds had probability of ancestry traceable to one of the clusters and very few appeared to be of mixed descent. Notably, putative hybrids with 20-50% of the “rare” cluster (lighter grey, Fig. 4) were found both in the Pacific (2 in Isabel and 1 in Tunitas) and the Caribbean (1 in Contoy). Both clusters were present in all breeding colonies and the less abundant increased in frequency in Isabel Island followed by Tunitas (Fig. 4). These results are consistent with those of the Bayesian Hickory analyses and the distance-based AMOVA, which point to Santa Margarita and Isabel as the most differentiated colonies, respectively (Table 4).
Frequency of RAPDs genotypes in the 4 largest Mexican breeding colonies of frigatebirds Fregata magnificens: Isla Santa Margarita, Isla Tunitas and Isla Isabel (Eastern Pacific Ocean), and Isla Contoy (Caribbean Sea). White slices are shared genotypes and grey slices are private genotypes.
Analyses of molecular variance (AMOVA) based on RAPDs of 4 major nesting colonies of Mexican Frigatebirds from the Mexican Pacific (Isla Santa Margarita, Isla Tunitas, and Isla Isabel) and the Mexican Caribbean (Isla Contoy)
| Source of variation | d. f | % variation | Φ-statistic | p-value |
| (a) Test of overall panmixia | ||||
| Among colonies | 3 | 5.53 | ΦSTALL=0.055 | 0.004 |
| Within colonies | 95 | 94.47 | ||
| (b) Nested hierarchical structure | ||||
| Between groups | 1 | 1.54 | ΦCT=0.015 | 0.25 |
| Among colonies within groups | 2 | 4.69 | ΦSC=0.048 | 0.03 |
| Within colonies | 95 | 93.77 | ΦST=0.062 | 0.004 |
| (c) Test of differentiation between Pacific and Caribbean | ||||
| Among basins | 1 | 4.76 | ||
| Within basins | 97 | 95.24 | ΦSTPvsC=0.048 | 0.03 |
RAPDs pairwise comparisons of genetic differentiation ΦST (above diagonal) and θ(II) (below diagonal) among the 4 largest Mexican breeding colonies of frigatebirds Fregata magnificens. Significant ΦST value at table-wide α=0.05 after B-Y FDR correction indicated in bold. None of the 95% Bayesian credibility intervals of θ(II) values overlapped zero
| Breeding colony | Santa Margarita | Tunitas | Isabel | Contoy |
| Santa | — | 0.073 | 0.154 | 0.067 |
| Margarita | ||||
| Tunitas | 0.142 | — | 0.012 | 0.060 |
| Isabel | 0.177 | 0.185 | — | 0.045 |
| Contoy | 0.147 | 0.181 | 0.136 | — |
Plot of Evano's et al. (2005) statistic ΔK as a function of the number of clusters (K) used to infer the most likely number of genetic groups inferred from Bayesian clustering using the program STRUCTURE (Pritchard et al., 2000).
The hypothesis of isolation by distance was rejected in the absence of a significant correlation between genetic and geographic distances (Mantel test, p=0.55) among the 4 colonies of frigatebirds.
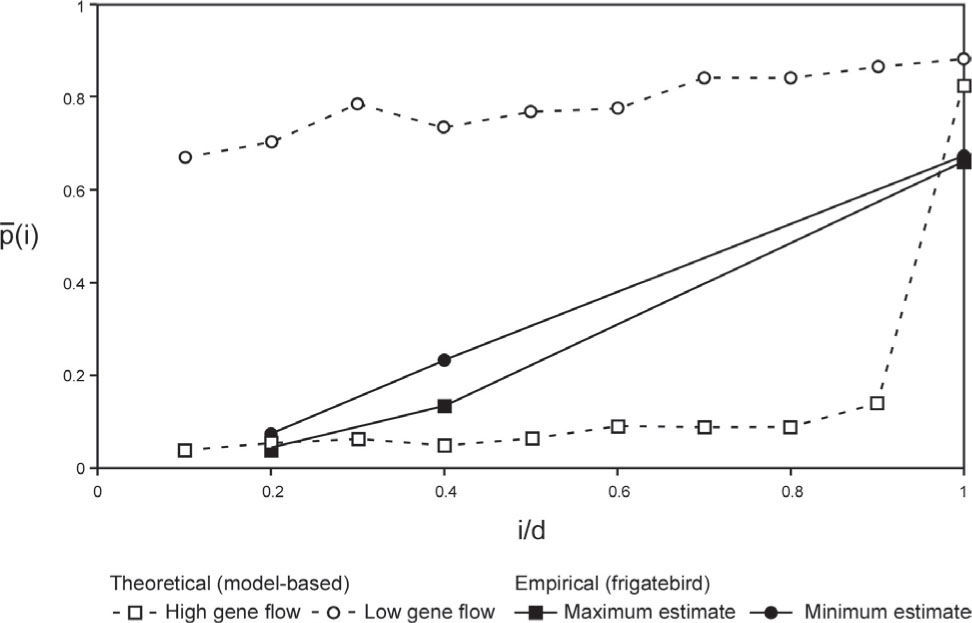
Gene flowA first order estimation of gene flow levels based on AMOVA fixation indices produced values between 4 and 5 effective migrants per generation overall and between regions of interest (ΦSTALL= 0.059, p< 0.001; Nm= 3.99; ΦSC= ΦSTPvsC= 0.048, p= 0.03; Nm= 4.96). Corresponding gene flow values estimated from the higher-valued θ(II) statistics were consequentially smaller (1.1<Nm<1.54). The small number of studied populations did not allow having a large number of occupancy values for RAPD markers to compare with Slatkin's (1981) theoretical model. Nevertheless, the lower and upper bound limits of the empirical conditional average marker frequencies were clearly biased towards the theoretical expectations of a high gene flow species (Fig. 6).
Conditional average frequencies p¯(i) as a function of standardized occupancy numbers (i/d) used to estimate gene flow levels relative to high- and low-gene flow theoretical expectations based on numerical simulations of Slatkin's (1981) gene flow model.
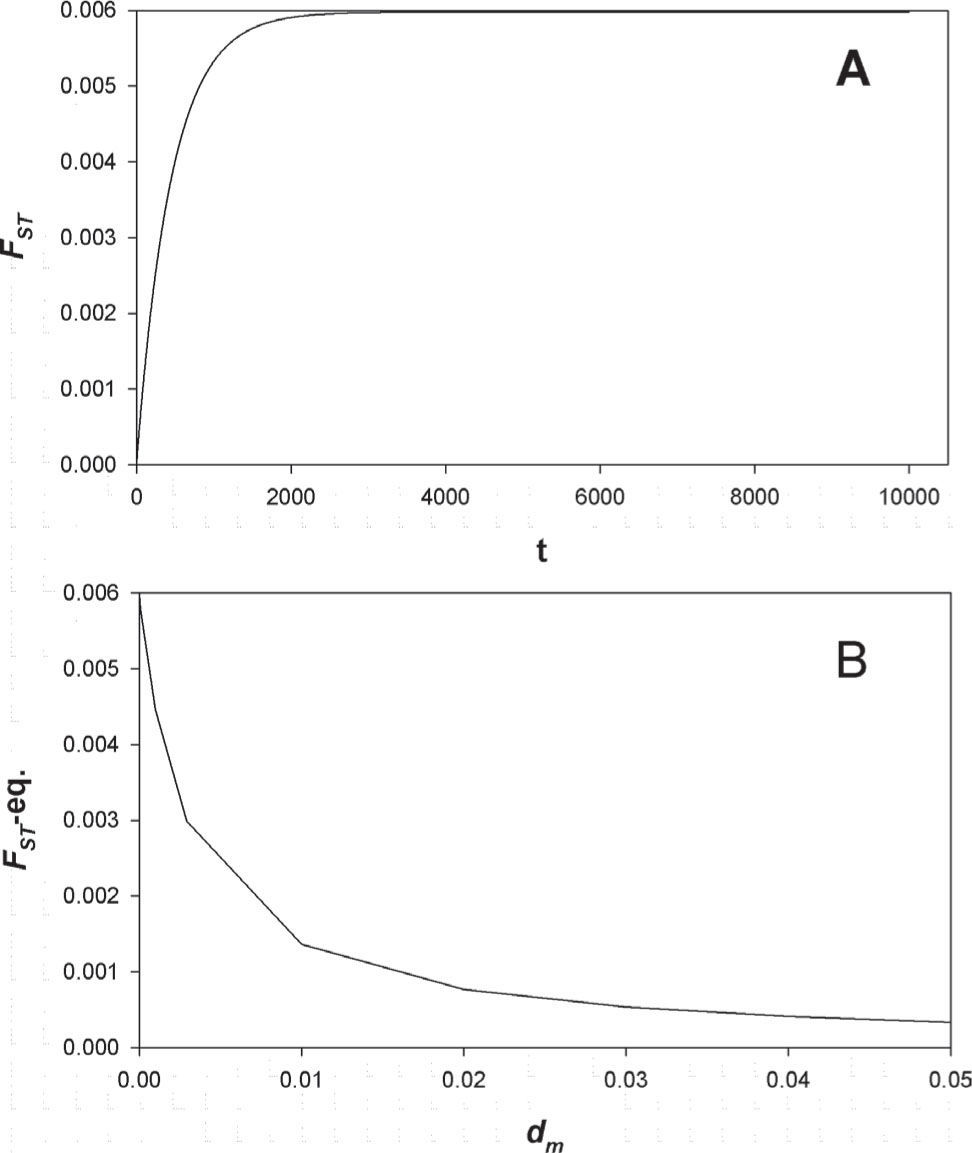
The starting simulation at dm=0 (zero male dispersal equivalent to 100% reproductive philopatry) produced an FST=0.006 at equilibrium. In all simulations, equilibrium was reached at 3 000 generations or before (Fig. 7A). This estimate represents an upper bound for FST in the absence of male dispersal; hence, as expected, increasing dm produced a hyperbolic reduction in the level of genetic structure resulting from higher levels of gene flow. At dm=df=0.0029 the level of predicted genetic structure was FST=0.006 (Fig. 7B). Given that our biparental (RAPDs) estimates of genetic structure are at least an order of magnitude higher among all colonies (ΦSTALL= 0.059; θ(II)ALL= 0.163), model results suggest that our mtDNA-based estimate of female dispersal is positively biased. Using numerical simulations and dm=0 to solve for a female dispersal accounting for at least FST=0.059 yields df=0.0021×10−5, which is 5 orders of magnitude smaller than the mitochondrial-based estimate.
Results of the group-structured model of Chesser and Baker (1996). A) Forward in time simulation of the level of genetic structure during 10 000 generations using transition equations, parameters in Table 2, and dm=0. B) Values of FST at equilibrium after 10 000 generations with increasing levels of male dispersal (dm).
In this paper we integrated genetic and dispersal ecological data to address the influence of non-physical barriers to gene flow among Mexican frigatebird colonies. We found that ecological mark and recapture, as well as genetic data point to high levels of dispersal and gene flow in these birds. However, gene flow is highly biased and insufficient for complete homogenization of gene pools. Group-structured population modeling allowed us to assess sex-biased dispersal parameters from maternally (mtDNA) and biparentally (RAPDs) inherited markers, which were consistent with the high degree of male-biased breeding philopatry and female mate choice leading to sequential monogamy observed in the field. These non-physical behavioral forces appear to be responsible for breeding isolation promoting nuclear genetic differentiation at different geographic scales. On the other hand, the absence of a genetic pattern of isolation by distance suggested that geographic distance does not play a significant role in the genetic distinction of breeding colonies.
Patterns of dispersal and genetic differentiationSighting data of banded and tracked frigatebirds suggest that female-biased breeding dispersal is mainly restricted to adjacent colonies, as field observations support the movement westward from Isabel to Bahía Santa María, Sinaloa, among Eastern Pacific colonies, but not from the Pacific to the Caribbean. On the other hand, males may spend the non-breeding season mainly in the upper Gulf of California (M. González-Jaramillo unpublished data) or flying south as far as Nicaragua (Lee and Osorno, unpublished data), traveling up to ∼750km away from Isabel. However, evidence indicates that these males return to Isabel for breeding, hence manifesting breeding philopatry (González and de la Cueva, 2007) and site fidelity (Ososrno, 1996) (Fig. 1).
The increasing percentage of private genotypes from the northeast Pacific to the Caribbean (Santa Margarita 0% < Tunitas 37.5 % < Isabel 55% < Contoy 66%), with Contoy having the largest fraction of private genotypes (4/6), could be consistent with a hypothetical unidirectional westward gene flow from the Caribbean to the Pacific (Fig. 3). The number of private alleles among populations has been used to infer gene flow trends among some source-sink bird populations (e. g., Lanius collurio,Pustjens et al., 2004), with the expectation of a decreasing fraction of private alleles in receiving rather than in source populations. Furthermore, the presence of shared genotypes (R1 and R2, Fig. 3) between Isabel, in the Eastern Pacific, and Contoy, in the Caribbean Sea, (separated by 1 977km) suggests the action of sequential founder events in island colonization of frigatebirds, as proposed for other seabird species (Friesen et al., 1996; Patirana et al., 2002; Olson and Hearty, 2003; Steeves et al., 2003; Steeves et al., 2005b).
Mexican frigatebird breeding colonies were not found to be panmictic since nuclear genetic structure was found between ocean basins, in consistence with our previous work (González-Jaramillo and Rocha-Olivares, 2011), and among Eastern Pacific breeding colonies. The degree of genetic structure among Eastern Pacific frigatebird colonies (ΦSC=0.048) was higher than that found in Leach's Storm-petrels (Oceanodroma leucorhoa), another seabird with high dispersal capability (ΦSC=0.026, Paterson and Snyder, 1999). The patterns and strength of nuclear genetic structure of frigatebirds varies among methodological approaches but they were consistent with significant differentiation and with the presence of 2 non- geographically restricted genetic groups among Mexican colonies, as inferred from Bayesian clustering using STRUCTURE (Pritchard et al., 2000). The presence of a genetic dichotomy in RAPDs genotypes is noteworthy given the dimorphic nature of mitochondrial variation found in our earlier study (González-Jaramillo and Rocha-Olivares, 2011); however, the mitochondrial haplotype frequencies were highly biased in Pacific and Caribbean birds. In RAPDs, shared genotypes suggest that gene flow is likely taking place between adjacent islands as Isabel and Tunitas (420km far apart) and between Tunitas and Santa Margarita (305km far apart). Unexpectedly, gene flow among the 4 colonies, as well as among the Eastern Pacific colonies was lower than between ocean basins: Nm≈4 vs. 5 migrants per generation respectively. Furthermore, absence of a significant correlation between genetic and geographic distance reflects the fact that genetic structure among frigatebird breeding colonies is not mediated by geographic distance, which is highly consistent with additional evidence pointing to the weak role of distance as a barrier to dispersal in other seabirds (see Friesen et al., 2007).
Our combined RAPDs and mtDNA results reveal patterns of differentiation that could be at odds with those of Hailer et al. (2011). In that study, involving a larger geographical area, Galapagos frigatebirds showed significant reproductive isolation from other colonies in the Pacific and Caribbean, but no genetic distinction was found between Eastern Pacific and Atlantic breeding colonies. In order to reconcile these findings, several factors must be considered (González-Jaramillo and Rocha-Olivares, 2011): 1) genetic data from Mexican frigatebirds analyzed by Hailer et al. (2011) consisted only of mtDNA sequences from 5 historical individuals among the 36 from the Eastern Pacific; 2) relative to the colonies studied by Hailer et al. (2011) closer to the center of abundance in the Eastern Pacific, the breeding colonies that we analyzed are peripheral and hence may experience different demographic and connectivity patterns (edge effects); 3) the distance between Eastern Pacific and Caribbean/Atlantic breeding colonies in this study were much larger (minimum 2800km vs. maximum 1900km in Hailer et al., 2011), and 4) differences in the relative rates of evolution and diversification of the molecular markers. These non-mutually exclusive factors may account for the differences among studies.
Non-physical barriers to reproductionAlthough frigatebirds have a high dispersal capability (Weimerskirch et al., 2003; Hailer et al., 2011), their monogamous mating system along with male-biased philopatry (González and de la Cueva, 2007) and complex female mate choice behavior (Madsen et al., 2004), are likely influencing their population structure among Eastern Pacific colonies. Sex-specific philopatry and mating tactics have direct influence in gene dynamics by increasing coancestry (Sugg et. al., 1996). This results from the increased probability of mating with related individuals, relative to random mating, produced by the reproductive behavior. Hence, assortative mating significantly influences gene frequencies, as well as genotype distributions and proportions relative to those expected with panmixia (Chesser, 1991b; Chesser, 1991a; Sugg et al., 1996). To our knowledge, this is the first time reproductive ecological parameters and genetic data from maternally and biparentally inherited genetic markers are integrated into a group-structured model for a pelagic seabird. Most notably, this integrative approach provided irrefutable evidence of the influence of male- based philopatry in shaping the genetic patterns observed, even if the absolute magnitudes of dispersal estimates did not entirely coincide with model expectations. However, we cannot discard the possibility that other processes, in addition to behavior, are influencing the genetic differences between some Eastern Pacific colonies. Ecological factors such as differences in ocean productivity (Kessler, 2006), community and flock composition, as well as differences in local predation regimes (Drummond et al., 2000; Wolf et al., 2006) may be exerting differential selective pressures, as suggested in the Great Frigatebird (Fregata minor; Dearborn et al., 2003).
Overall, our results concur with previous genetic studies on Magnificent Frigatebirds from North America (González-Jaramillo and Rocha-Olivares, 2011; Hailer et al., 2011) citing the prime influence of non-physical barriers such as behavioral characters, instead of physical barriers such as the Panama Isthmus, in the population genetic structure of frigatebirds. Furthermore, population genetic structure within ocean basins related to behavioral traits has also been found in several pelagic seabirds such as the Red-legged Kittiwake (Rissa brevirostris; Patirana et al., 2002), the Leach's Storm-petrel (Oceanodroma leucorhoa; Paterson and Snyder, 1999), the Masked booby (Sula dactylatra; Steeves et al., 2005a), and the Black-browned (Thalassarche melanomorphis; Burg and Croxall, 2001) and Wandering (Diomedea spp.; Burg and Croxall, 2004) albatrosses around Antarctica.
Evidence of historical and contemporaneous evolutionary processes acting as barriers to dispersal revealed that mechanisms of population differentiation are complex in seabird species (Friesen, 2007; Friesen et al., 2007). Nevertheless, in frigatebirds behavior has a prime role in governing population genetics as a non-physical barrier to interbreeding. Hence, our results integrating ecological and genetic data have a considerable impact on the management plans and conservation strategies of this important seabird.
We thank Vinni Madsen, José Luis Osorno, Horacio de la Cueva, Marco Antonio González, Alfredo Castillo, Ignacio Barajas, and Donny Canul for facilitating blood samples, and Nancy Saavedra for helping in the laboratory. The managers of Isla Contoy and Isla Isabel National Parks generously provided all the facilities to visit and work at the islands. Pronatura, Sinaloa, provided logistic support and staff to visit Isla Tunitas; the Mexican Navy and the Comisión de Nacional de Áreas Naturales Protegidas provided logistic support to work at Isla Isabel. Dirección General de Vida Silvestre provided authorization for collecting samples (Oficio NUM/SGPA/DGVS/08778). Fishers from San Blas and Boca de Camichin were instrumental in the field and are gratefully acknowledged. This work was supported by a Centro de Investigación Científica y de Educación superior de Ensenada internal grant (ARO). The second author benefited from a Ph.D. scholarship from Conacyt during her studies in the Marine Ecology graduate program at CICESE. We thank two anonymous reviewers for their constructive comments.