Prior to modeling the potential distribution of a species it is recommended to carry out analyses to reduce errors in the model, especially those caused by the spatial autocorrelation of presence data or the multi-collinearity of the environmental predictors used. This paper proposes statistical methods to solve drawbacks frequently disregarded when such models are built. We use spatial records of 3 species characteristic of the Mexican humid mountain forest and 2 sets of original variables. The selection of presence-only records with no autocorrelation was made by applying both randomness and pattern analyses. Through principal component analysis (PCA) the 2 sets of original variables were transformed into 4 different sets to produce the species distribution models with the modeling application in Maxent. Model precision was higher than 90% applying a binomial test and was always higher than 0.9 with the area under the curve (AUC) and with the partial receiver operating characteristic (ROC). The results show that the records selected with the randomness method proposed here and the use of the PCA to select the environmental predictors generated more parsimonious predictive models, with a precision higher than 95%, and in addition, the response variables show no spatial autocorrelation.
Cuando se modela la distribución potencial de una especie es deseable efectuar algunos análisis previos para reducir errores en el modelo resultante, especialmente los ocasionados por la autocorrelación espacial de los registros de presencia y la correlación entre los predictores ambientales utilizados. En este trabajo se proponen métodos estadísticos que sirven para resolver estos inconvenientes que con frecuencia se presentan al elaborar los modelos de distribución potencial. Se emplearon los registros de presencia de 3 especies características del bosque húmedo de montaña de México y 2 conjuntos de variables originales. A los datos de presencia se les aplicó un análisis de aleatoriedad y de patrones para seleccionar registros no autocorrelacionados. Mediante análisis de componentes principales (PCA), los 2 conjuntos de variables originales se transformaron en 4 conjuntos distintos para generar los modelos de distribución de especies utilizando el algoritmo Maxent. La precisión de los modelos fue mayor al 90% con una prueba binomial y mayor de 0.9 del área bajo la curva (AUC) con la característica operativa del receptor parcial (ROC). Los resultados muestran que la selección de registros por el método de aleatoriedad propuesto y el uso de componentes principales como predictores ambientales generan modelos predictivos más parsimoniosos, con una precisión mayor al 95%, además de que sus variables predictivas no presentan autocorrelación espacial.
The models that predict the potential distribution of species through the combination of presence-only records and digital layers of environmental variables are of great interest in both theoretical and applied disciplines (Guisan and Thuiller, 2005; Elith and Leathwick, 2009a; Peterson et al., 2011). Such models use the association between environmental variables, presumably of predictive value, and the species occurrence records; thus are identified the environmental conditions where a species could survive indefinitely (Pulliam, 2000; Guisan and Thuiller, 2005; Elith and Leathwick, 2009b). This approach is especially important to produce basic information for such disciplines as biogeography, conservation biology, ecology, evolutionary biology, and others (Guisan and Zimmermann, 2000; Guisan and Thuiller, 2005; Elith and Leathwick, 2009b; Peterson et al., 2011).
Species distribution models implicitly suppose that the geographical data points for species records are independent, although this is not necessarily true. In addition, the environmental layers used as hypothetical predictive variables and associated to the geographical records of species also show problems of spatial autocorrelation. The spatial autocorrelation is the degree of dependency of variables in geographical space (Cressie, 1991; Legendre, 1993; Anselin et al., 2004); accordingly, disparity among variable values is strongly influenced by the distances among geographical data points where a species has been observed (Anselin et al., 2004; Segurado et al., 2006). Spatial autocorrelation represents an intrinsic characteristic in most of the geospatial data (Legendre, 1993; Segurado et al., 2006) and it can be an important bias in most geospatial analyses (Anselin et al., 2004). Spatial autocorrelation inflates type I errors of traditional statistics and it can affect the estimated parameters in model selection (Lennon, 2002).
The species distribution models obtained from a large data set of associated environmental covariates often inherently result in multi-collinearity, a statistical problem defined as a high degree of correlation among covariates. Multi-collinearity is a serious statistical problem in non-experimental situations, where the researcher has no control of the risk associated to hypothetical factors related to independent variables. Multi-collinearity is found, for instance, when many covariates are used as predictor variables to model selection and several of them measure similar phenomena. This is so because in most cases the researcher does not have a priori knowledge on which predictive environmental variables should be included in the model. However, the researcher must have a model in mind that usually includes a large number of predictive variables and hopes that using an appropriate statistical analysis will provide him/her with a correct model. It should be taken into account that multi-collinearity does not violate the assumptions that underlie to the statistical analysis, i.e., its presence does not affect the estimate of the dependent variable. In other words, estimation values for the dependent variable are the best unbiased estimates from the conditional population average. However, the existence of multi-collinearity tends to inflate both the variances of predicted values of the response variable and the variances of the estimated parameters. Therefore, if one considers that multi-collinearity is present in a dataset, it is important to know how the linear relationships are among the predictive environmental variables. For these reasons, it is critical both to the researcher as to the research to be sure that those environmental predictive variables are orthogonal to each other, that is, they are mutually independent.
Species distribution models are not explicitly spatial (Franklin, 2009); they suppose that the geographical occurrences of records are mutually independent. However, this violates a fundamental principle of the spatial geography establishing that spatially proximate objects are similar and proximate localities tend to have similar values due to the possibility they reciprocally influence each other, or both are influenced by the same pattern that generates geographical processes (Franklin, 2009). Disregard and not avoid spatial autocorrelation has consequences, for example: a) it can increase the probability of incurring in type I errors or incorrectly rejecting the null hypothesis of no effect, b) variable selection may be predisposed toward more strongly auto-correlated predictors (Lennon, 2002), c) coarse scale predictors may be better selected against more locally influencing predictors, and d) model selection based on the Akaike information criterion will tend to model with larger number of predictive variables due to the committed residual variance structure. In summary, if spatial autocorrelation is present and ignored or not resolved, one may be incurring in a biased selection of variables or model coefficients.
Among the statistical procedures proposed to solve or to reduce autocorrelation, principal component analysis (PCA), ridge-regression, and latent-root regression have been mentioned (Mason and Gunst, 1985; Afifi et al., 2012). The advantages of PCA compared with the other 2 procedures is the availability of an exact theory on estimate distributions, that is, the term or the error of the regression and the estimates are normally distributed (Gunst and Mason, 1977) and the principal components (PCs) are useful exploratory tools to detect and quantify mutual relationships among variables (Afifi et al., 2012).
The reduction of dimensionality is among the many applications of the PCA, that is, the reduction to a number of predictive variables that retain a high proportion of the original information (Tabachnick and Fidell, 2007). The PCs obtained are placed hierarchically according to their variance size; consequently, the first PC explains the maximum variance recorded in the predictive variables, the second PC explains the maximum of the residual variance and so forth, until the last PC which explains the remainder variance (Tabachnick and Fidell, 2007). Since the first PCs are those that retain the highest proportion of information, the dimensionality reduction is attained by choosing those first PCs, which explain a high percentage of variation recorded in the original data. The selection of the number of PCs is a function of the variance percentage that satisfies the standards of the research carried out. Another important property of obtained PCs is their independency of each other, that is, they are orthogonal. Accordingly, obtaining the PCs permits them to be used as independent, non-correlated variables in analyses of modeling potential species distribution.
This contribution seeks to evaluate and propose statistical methods to resolve or minimize spatial autocorrelation, from both presence species records and environmental variables used in species distribution modeling. The methods proposed are then tested to model the potential distribution of 3 species characteristic of the humid mountain forest of Mexico (Cruz-Cárdenas et al., 2012). A contrasting number of environmental variables and number of presence records are used to validate the resulting models and to select the best one for each species using as criteria the reduction of spatial autocorrelation, the precision of the predicted potential distribution, the parsimony principle, and the surface of simulated occurrence.
Materials and methodsPredictive environmental variablesTable 1 shows the 58 presumably predictive environmental variables (pixel size 1 km2) used in the analysis, which include: 1) 19 climatic variables taken from WorldClim data base (Hijmans et al., 2005; http://www.worldclim.org/current.htm); 2) 7 seasonal climatic variables calculated from WorldClim data; 3) 9 variables of soil properties (Cruz-Cárdenas et al., 2014); 4) 8 topographic variables generated from a digital elevation model extracted from the GTOPO data base (http://edc.usgs.gov/products/elevation/gtopo30/gtopo30.html), and 5) 14 normalized vegetation indices, one for each month of the year 2009 obtained from remote sensing data (MODIS), and one normalized vegetation for the dry months, and another for the humid months.
Original environmental predictive variables used in species distribution modeling. In bold, the a priori selected variables are indicated
| A priori selected | |
| a) Climatic variables | |
| bio1=Annual mean temperature | X |
| bio2=Mean diurnal range (Tmax - Tmin) | |
| bio3=Isothermality (bio1/bio7) × 100 | X |
| bio4=Temperature seasonality (standard deviation × 100) | X |
| bio5=Maximum temperature of warmest month | |
| bio6=Minimum temperature of coldest month | |
| bio7=Temperature anual range (bio5–bio6) | |
| bio8=Mean temperature of wettest quarter | |
| bio9=Mean temperature of driest quarter | |
| bio10=Mean temperature of warmest quarter | |
| bio11=Mean temperature of coldest quarter | |
| bio12=Annual precipitation | X |
| bio13=Precipitation of wettest month | |
| bio14=Precipitation of driest month | |
| bio15=Precipitation seasonality | X |
| bio16=Precipitation of wettest quarter | |
| bio17=Precipitation of driest quarter | |
| bio18=Precipitation of warmest quarter | |
| bio19=Precipitation of coldest quarter | |
| b) Seasonality climatic variables | |
| ETRA=Real annual evapotranspiration | X |
| ETRAH=Real evapotranspiration of the humid months (may to october) | X |
| ETRAS Real evapotranspiration of the dry months (november to abril) | X |
| PPH=Precipitation of the humid months | X |
| PPS=Precipitation of the dry months | X |
| TH=Mean temperature of the humid months | X |
| TS=Mean temperature of the dry months | X |
| c) Soil properties | |
| Ca=Calcium | |
| CE=Electric conductivity | X |
| CO=Organic carbon | |
| K=Potasium | |
| MO=Organic material | X |
| Mg=Magnesium | |
| Na=Sodium | |
| pH=Hydrogen potential | X |
| RAS=Sodium absorption relationship | |
| d) Topografic attributes | |
| Aspect | X |
| Anisotropic heating | |
| Elevation | X |
| Runoff | |
| Convergence index | |
| Topographic humidity index | |
| Terrain rugosity index | |
| Vector rugosity measurement | |
| Slope | X |
| e) Normalized vegetation indices for 2009 | |
| IVNENE=January normalized index+ | |
| IVNFEB=February normalized index | |
| IVNMAR=March normalized index | |
| IVNABR=April normalized index | |
| IVNMAY=May normalized index | |
| IVNJUN=June normalized index | |
| IVNJUL=July normalized index | |
| IVNAGO=August normalized index | |
| IVNSEP =September normalized index | |
| IVNOCT=October normalized index | |
| IVNNOV=November normalized index | |
| IVNDIC=December normalized index | |
| IVNH=Humid months of year normalized | X |
| index | |
| IVNS=Dry months of year normalized index | X |
Evapotranspiration was calculated using the Turc’s model (Turc, 1954): ETRA=P / [0.9 + (P/L)2]½, where, P=total annual precipitation (mm), L=300 + 25T +0.05T3, and T=average annual temperature (° C).
Considering studies by García (1965) and Mociño and García (1974), we selected from WorldClim data those variables that account for the distribution of the vegetation types in Mexico. For instance, based on conclusions by these authors, the distribution patterns of rainfall and temperature in Mexico are strongly influenced by orography, and by the atmospheric circulation at both low and high elevations. Rainfall in Mexico is heterogeneous throughout the year; it increases latitudinally from north to south, and is also strongly influenced by the presence of the Gulf of Mexico and the Pacific Ocean (García, 1965), and by the link with the El Niño Southern Oscillation (Cavazos and Hastenrath, 1990). García (1965) and Mociño and García (1974) point out that about 70% of the rainfall in Mexico is recorded from May to October, and the remaining 30% between November and April, which define the humid and dry seasons, respectively. During the dry season, a proportion of species lose their foliage, producing physiognomic differences that characterize the different vegetation types of Mexico. The presence or absence of leaves during the humid or dry seasons should be reflected on the monthly normalized vegetation indices. Based on the works of the above authors, we made an a priori selection from the 58 variables listed in Table 1 discarding 38 and retaining 20 variables that presumably influence the potential distribution of plant species. These 20 variables are marked in Table 1 under the header a priori.
Species presence recordsThree species considered characteristic of the humid mountain forest of Mexico (Villaseñor, 2010) were selected for analysis: Liquidambar styraciflua L., Quercus rubramenta Trel., and Roldana robinsoniana (Greenm.) H. Rob. and Brettell. These species were selected based on the criteria by Cruz-Cárdenas et al. (2012) in which the species are restricted to the HMF geographic polygon besides that the literature has also referred them as charismatic of that biome. The localities of occurrence were obtained from specimens housed in the Herbario Nacional de México (MEXU) at Instituto de Biología, UNAM. Data for 193 collecting localities were transformed to geographical coordinates, 142 of them represented L. styraciflua, 41 Q. rubramenta, and 10 R. robinsoniana.
Species distribution modelingFigure 1 illustrates the procedure to generate the species distribution models. Below, it is described in detail:
1) A randomness test was applied to the spatial records for each species (Bivand et al., 2008). If positive, 75% of the records were used for training the model and 25% for model validation. If negative, a pattern analysis was applied to the records by estimating the distance at which it is possible to find a single species record with a maximum probability. The pattern analysis was performed by using the public domain ILWIS 3.7 (http://52north.org/ilwis). Pattern analysis is similar to estimating the distance (or the range value of a variogram) for which the records do not show spatial autocorrelation (Hengl, 2007). The study area is divided in a grid cell system whose sides are the estimated distance value expressed in degrees; such a grid cells system is obtained by using the public Quantum GIS 1.7.4 software (http://qgis.osgeo.org). Finally, after the species records are randomly selected, a single record per grid cell is used to train the model.
2) The environmental layers were 7 principal components (PCs) obtained from the PCA for 20 environmental variables selected a priori for Mexico (Table 1). Such PCs explained more than 95% of variance of the original variables (Set I).
3) The 7 PCs and the training records were used for modeling the potential species distribution with the Maxent algorithm (http://www.cs.princeton.edu~schapire maxent/). Maxent requires presence-only records and a set of predictive environmental variables (Phillips et al., 2006); because it is a nonparametric algorithm, it incorporates interactions among variables, produces continuous maps of suitability or compatibility (Phillips et al., 2006), and performs better than other methods that estimate potential species distributions with presence-only data (Elith et al., 2006). Maxent configuration was used by default (Phillips and Dudik, 2008), except for the “Extrapolate” and “Do clamping” modules that were deactivated; the output format was logistic.
4) Models obtained with Maxent were transformed to boolean layers (presence-absence) with a cutoff threshold equal to 10% omission errors (Pearson et al., 2007).
5) A binomial test was used for model validation to assess whether it was better than any other model randomly obtained (p > 0.5). The number of successes was obtained quantifying the number of records with logistic values above the cutoff threshold.
6) A partial Receiver Operating Characteristic (ROC) test was applied as an additional precision analysis (Peterson et al., 2008). This test estimates the relationship between the Area Under the Curve (AUC) and the null expectation of bootstrap repetitions; a Z test is the decision rule to determine if such a relationship is less than 1 (considered a good model). Additionally, the number of bootstrap replicas with AUC≤ 1 was counted. As a more appropriate alternative, we used partial ROC approaches that weight omission error over commission error, such that the model evaluation is more appropriate to the challenge of predicting species’ distributions from incomplete, presence-only biodiversity data (Peterson et al., 2008).
Species distribution models were also obtained with other 5 sets of environmental variables. The second set (Set II) consisted of 7 PCs that explained 95% or more of data variance and obtained through the PCA applied to 58 environmental variables (Table 1). The third set (Set III) included 7 PCs obtained with the nonspatial data for 20 variables selected a priori (Table 1). The fourth set (Set IV) was prepared with 58 variables and nonspatial data, but the PCA was elaborated from a data matrix with records selected with step 1 (above described) as rows and the environmental variables as columns, that is, the matrix consisted of records selected for training the model with step 1 as rows and the environmental variables as columns. All PCA analyses were carried out with R software (R Development Core Team, 2012).
In each case, the variables selected were significantly correlated with the PC (95% confidence level). Thus, the linear combinations were made with the loading values of the selected variables, and for each of the 7 PCs their respective rasters were created. The sets V (20 variables) and VI (58 variables) included the original environmental predictors. Steps 3 (species distribution modeling), 4 (boolean layer built-up), 5 (binomial test), and 6 (partial ROC test) of the proposed methodology were similar for the 6 sets of environmental predictors. In brief, environmental predictors integrating sets I to III are 7 PCs generated with 20 original variables, sets II and IV are likewise integrated by 7 PCs generated with 58 original variables, and the sets V and VI included 20 and 58 original variables respectively (Table 1).
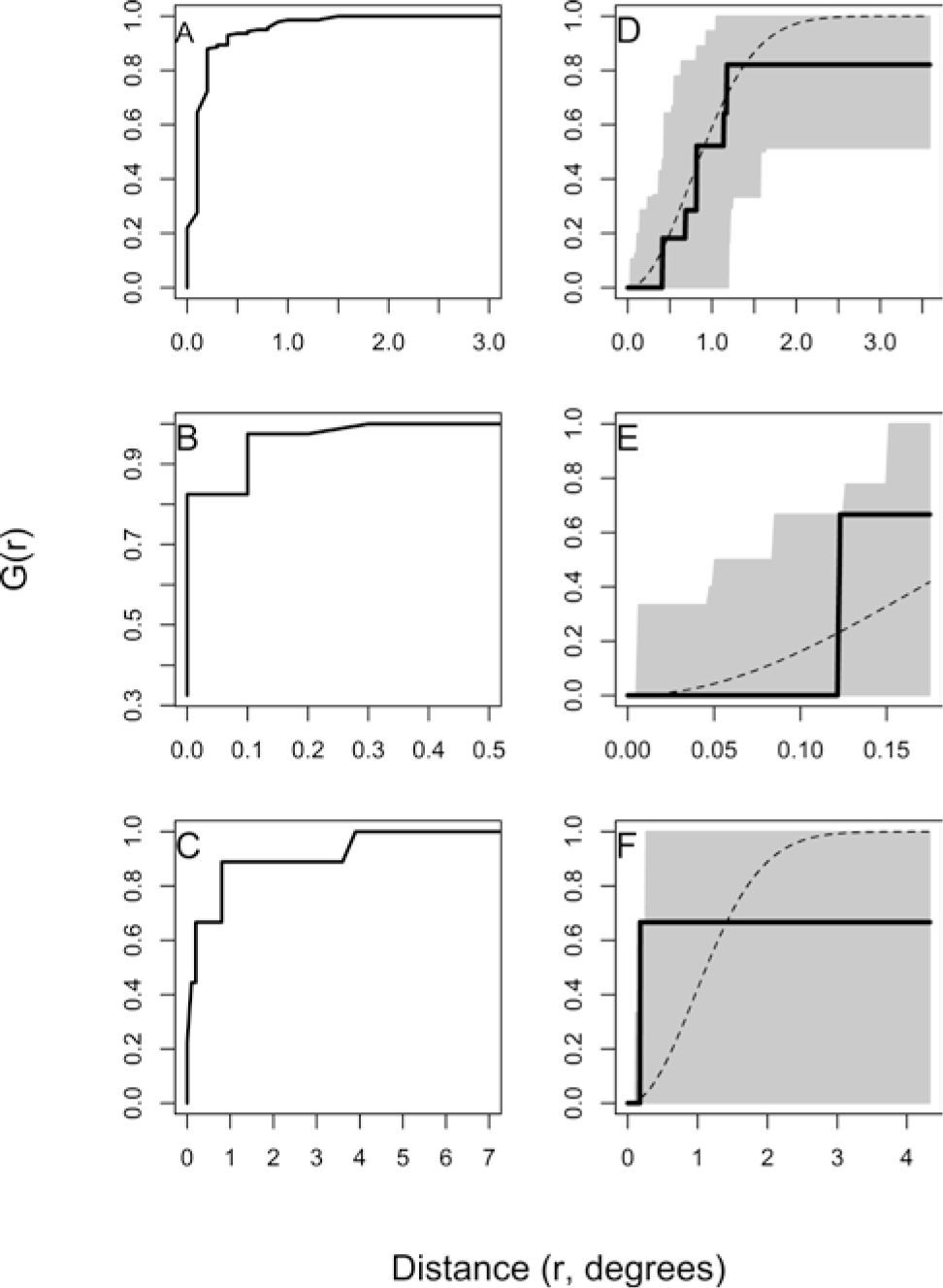
ResultsFigure 2 shows the results of the randomness test applied to the records for each species. The analyses indicate that the records for the 3 species show aggregated spatial patterns, since the observed values (continuous line) are distributed up and out of the confidence bands estimated to any distance (r). Records of Q. rubramenta (Fig. 2B) display a strong aggregation since at distances lower than 6.5km (0.06 degrees) the probability of finding a record is high (G(r) ≈ 0.9). In contrast, the records of R. robinsoniana (Fig. 2C) show a weak aggregation because at distances of about 77km (< 0.4 degrees), the probability of finding a record is lower (G(r) ≈ 0.7). On the other hand, records of L. styraciflua (Fig. 2A) display an aggregated distribution at short distances and a random distribution pattern at intermediate distances, with a maximum probability (G(r) ≈ 0.9) of finding a record at distances of about 16km (G(r) ≈ 0.15).
Randomness test for recorded sites of Liquidambar styraciflua (A), Quercus rubramenta (B), and Roldana robinsoniana (C). G(r)=average number of records inside a radius r, equivalent to distance in degrees. The continuous line corresponds to observed values, the discontinuous line to the theoretical values, and the gray area to the confidence band.
Figure 3 shows the pattern analyses for the records of the 3 species. Based on these analyses, cutoff distances resulted with maximum likelihood of species randomly distributed were 1.5, 0.3, and 3.9 degrees for L. styraciflua, Q. rubramenta, and R. robinsoniana respectively. Records selected by this process were 16 for L. styraciflua, 7 for Q. rubramenta, and 5 for R. robinsoniana, which when applied, satisfied their randomness test. However, it is important to point out that the records selected with this method for R. robinsoniana were only 2; therefore 3 additional records were randomly selected to reach the minimum number of records (n=5). Figure 3 (D, E y F) shows the observed values distributed into the confidence bands, which ensures the randomly distribution pattern of records for the 3 species. Finally, the number of records used to validate the distribution models were 35 for L. styraciflua, 10 for Q. rubramenta, and 5 for R. robinsoniana.
Figure 4 shows the species distribution models and the areas of occupancy predicted by the models. Predicted areas are low, intermediate, and high. Models that predicted low geographical areas were those that used 20 and 58 variables as environmental predictors corresponding to sets V y VI (Fig. 4). In contrast, models that predict wide geographical areas were those whose simulations derived from the use of 7 PC, that is, set I to IV (Fig. 4). Set I predicted the smaller area as compared with the others.
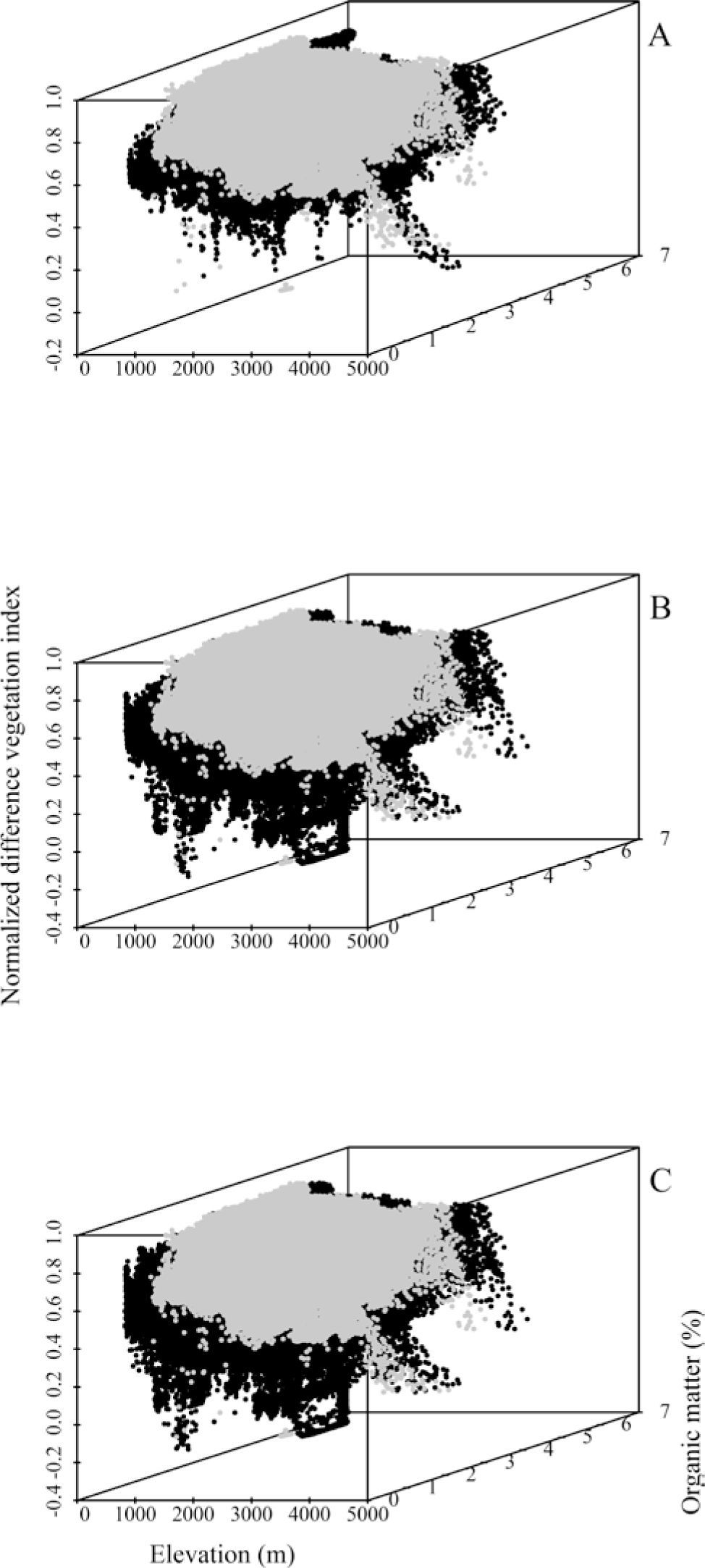
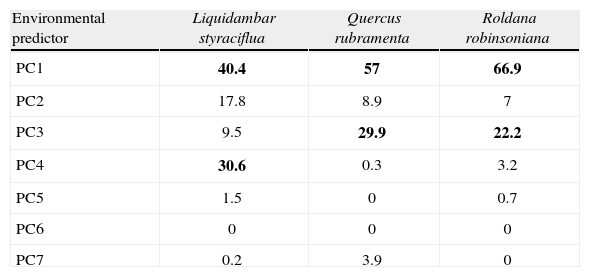
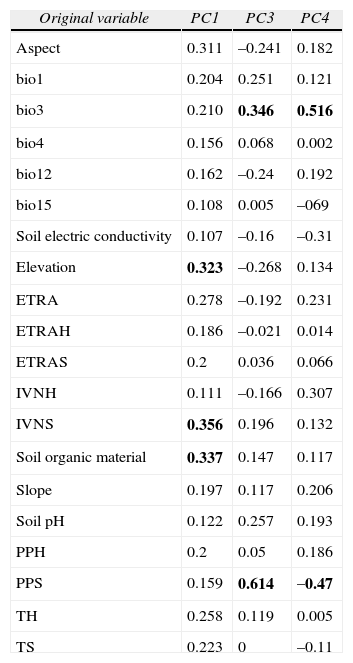
The relative contribution of environmental predictors, represented by the orthogonal arrangement of PCs were: PC1 and PC4 for L. styraciflua (Table 2) that explain 71% of total variance registered in the original variables, and PC1 and PC3 both for Q. rubramenta and for R. robinsoniana, which contributed to explain 86.9% and 89.1%, respectively. The most important original variables included in the PCs of each species were selected based on their highest loading values (Table 3). Elevation, normalized vegetation index of dry months, organic matter, isothermality, and rainfall for the dry months of the year turned out to be the most important variables to explain the potential distribution for the 3 species. The first 3 variables were used to generate the environmental space (Fig. 5) of the occurrence of the 3 species in the humid mountain forest of Mexico (Cruz-Cárdenas et al., 2012).
Number of principal components retained and their relative contribution to modeling distribution of species resulting from Maxent outputs. The most important principal components for each species are highlighted in bold face
| Environmental predictor | Liquidambar styraciflua | Quercus rubramenta | Roldana robinsoniana |
| PC1 | 40.4 | 57 | 66.9 |
| PC2 | 17.8 | 8.9 | 7 |
| PC3 | 9.5 | 29.9 | 22.2 |
| PC4 | 30.6 | 0.3 | 3.2 |
| PC5 | 1.5 | 0 | 0.7 |
| PC6 | 0 | 0 | 0 |
| PC7 | 0.2 | 3.9 | 0 |
Loading factors of the predictive environmental variables used in the principal component analysis
| Original variable | PC1 | PC3 | PC4 |
| Aspect | 0.311 | –0.241 | 0.182 |
| bio1 | 0.204 | 0.251 | 0.121 |
| bio3 | 0.210 | 0.346 | 0.516 |
| bio4 | 0.156 | 0.068 | 0.002 |
| bio12 | 0.162 | –0.24 | 0.192 |
| bio15 | 0.108 | 0.005 | –069 |
| Soil electric conductivity | 0.107 | –0.16 | –0.31 |
| Elevation | 0.323 | –0.268 | 0.134 |
| ETRA | 0.278 | –0.192 | 0.231 |
| ETRAH | 0.186 | –0.021 | 0.014 |
| ETRAS | 0.2 | 0.036 | 0.066 |
| IVNH | 0.111 | –0.166 | 0.307 |
| IVNS | 0.356 | 0.196 | 0.132 |
| Soil organic material | 0.337 | 0.147 | 0.117 |
| Slope | 0.197 | 0.117 | 0.206 |
| Soil pH | 0.122 | 0.257 | 0.193 |
| PPH | 0.2 | 0.05 | 0.186 |
| PPS | 0.159 | 0.614 | –0.47 |
| TH | 0.258 | 0.119 | 0.005 |
| TS | 0.223 | 0 | –0.11 |
PC=Number of principal components from PCA with set I of variables. The acronyms at first column are indicated in Table 1. Highest loading values are highlighted in bold face.
Environmental space of the 3 species built with the organic matter (%), normalized vegetation index (INV), and altitude (m) variables. Liquidambar styraciflua (A), Quercus rubramenta (B), Roldana robinsoniana (C). In black it is indicated the environmental space of the species and the gray color shows the environmental space of the humid mountain forest in Mexico.
The binomial test determined that all species distribution models were better that any others randomly obtained (p > 0.5). The precision of generated models for L. styraciflua with the set I of environmental layers (97%), set II (90%), and set IV (94%) were not statistically different; precision of these 3 sets was higher than that of sets III (77%), V (68%), and VI (88%). On the other hand, precision of distribution models for Q. rubramenta and R. robinsoniana with all the 6 sets of environmental predictors were statistically similar because they included 100% of success with validation records.
DiscussionThe evaluation of spatial distribution patterns of species records is important to avoid errors caused by spatial autocorrelation (Dormann, 2007). If the researcher does not avoid or minimize the spatial autocorrelation before the selection of variables, there may be negative consequences in the modeling analysis among them, and perhaps the most important, is the skewed selection of the environmental variables assumed as hypothetically predictive (Lennon, 2002). Our results show that randomness and spatial pattern analyses are good statistical methods to eliminate or minimize spatial autocorrelation shown in the species records. Following Phillips et al. (2006), relative contribution of environmental variables to species distribution should be taken carefully, especially if we are not certain that these variables are or not spatially correlated. In this respect, the strength of this contribution is that the environmental variables submitted to the PCA, resulted in PCs that constitute orthogonal projections of the transformed variables and consequently free of autocorrelation.
The models of distribution generated with raw environmental predictors generally predict a smaller occupancy surface. That is so, because these models must satisfy a large number of rules, so that, a given record can be classified as a true presence. It is evident that predicted occupancy surface may have problems when hypothetical environmental variables are correlated, as is the case of those climatic variables mostly generated from rainfall and temperature data.
The autocorrelation problem was notably reduced by using PCA, especially due to the property of orthogonal transformation of variables. As a result, each PC is a linear combination of the original predictive variables such that the new variables decrease in hierarchical order allowing the selection of a reduced number of PCs that explain a high percentage of variance of the variables. PCA used along with the highest values of the loading factors is not the single criterion for the selection of variables; others include the grouping criterion and the combination of correlation and the highest average of the loading factors (Al-Kandari and Jolliffe, 2012).
Distribution models obtained for the 3 species and their predicted occupancy area was consistently higher when sets of variables II and IV were used (except for L. styraciflua). This is because the environmental layers used are influenced by the linear combinations from which they were generated. Moreover, models generated for L. styraciflua and R. robinsoniana with sets I and II (resulted with) have the same statistical precision but are different from those obtained with sets III and IV. With the exception of the distribution model for R. robinsoniana, the use of set II predicts the species distribution in the Yucatán Peninsula; such projections are incorrect since there is no evidence that the humid mountain forest ever existed in that region.
Our results show that distribution models obtained with the set of predictors number I were the most parsimonious and suitable; their predicted variables lack spatial autocorrelation and although the predicted surface area includes patches of L. styraciflua extending into the Yucatán Peninsula (mostly south of Campeche and Quintana Roo), precision was always higher that 95%. The predicted occurrence of species in the Yucatán Peninsula perhaps may be explained by historical factors (Luna-Vega and Magallón, 2010); maybe its presence was factual in remote times when climate was mild and later the species reduced their geographical distribution due to recent climatic fluctuations.
The 3 selected species used in the distribution modeling analyses are preferably distributed in the humid mountain forest of Mexico, although they are not strictly restricted to this biome. Accordingly, the environmental space of this biome determines the realized niches of these 3 species, since it represents the suitable environmental, ecological, and biological conditions where they would be able to coexist indefinitely. In addition, the environmental space given by the range of tolerances to the environmental combinations that assure species long-term survival configure their fundamental niches. In consequence, it is evident that the fundamental niche of the 3 species is wider than their realized niche, such as it is expected (Soberón and Peterson, 2011).
The results obtained by using the randomness and spatial pattern analyses reduced or eliminated autocorrelation among species records, and suggest that they are adequate statistical methods to solve that problem. On the other hand, the use of PCA and variable selection based on the highest loading factors of each PC guarantee that this diminishes the autocorrelation of variables before modeling the distribution of species.
The validation of the resulting models as well as the analyses of the statistical similarities or differences among them demonstrate their relevance to help us to select the best model. In this respect, it was decided that the best models were those that met the principle of parsimony, namely the simplest, with the smaller number of explanatory variables, with either statistical or ecological significance, and with maximum feasibility of occupancy surface area. As a corollary, before modeling of both real and potential species distribution the researcher must decide whether the suggestions proposed in this paper should be put into practice.
We thank Pedro D. Maeda and Francisco Espinosa for the reading of a preliminary version of this paper and their suggestions to improve the manuscript. Claudio Delgadillo read the English version of the manuscript and his suggestions were of great help to improve it. Joselin Cadena redrew Figure 1. The first author thanks the UNAM for the fellowship granted in its DGAPA (Dirección General de Asuntos del Personal Académico) 2011 Postdoctoral Fellowships Program. The Institute of Biology, UNAM provided human and material resources to carry out this research.