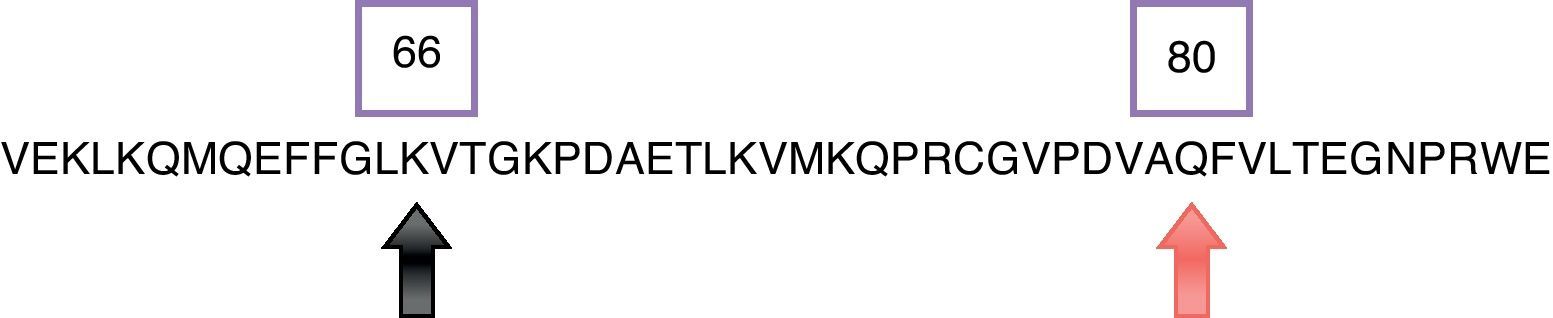The objective of this study is to demonstrate the molecular action of Porphyromonas gingivalis cysteine proteases such as gingipains (R1, R2 and K) upon human molecules.
Materials and methodsUsing the information on protein structure and function available in international databases (UniProtKB and Merops Database), the molecular interactions already described between gingipains and host molecules were clarified.
ResultsPossible cleavage sites were identified in host-produced elastase inhibitors and in pro-Matrix MetalloProteinase (MMP)1. Analysis of the results leads to the suggestion that the elastase inhibitor alpha1-antitrypsin is also degraded by interpain A, a cystein protease of Prevotella intermedia sharing a high homology with the PrtT and periodontain of P. gingivalis.
ConclusionThe information obtained suggests a synergistic molecular mechanism by which cysteine proteases of different bacteria can be responsible for the clinical manifestations of periodontal disease, and illustrates the use of bioinformatics to establish and predict molecular mechanisms.
O objetivo deste trabalho é verificar o detalhe molecular da ação de proteases de cisteína de Porphyromonas gingivalis como as gingipains (R1, R2 e K) em moléculas do hospedeiro.
Material e métodosUtilizando a informação disponível sobre estrutura e função de proteínas nas bases de dados internacionais (UniProtKB e Merops Database) as interações já descritas entre gingipains e moléculas do hospedeiro são clarificadas.
ResultadosSão identificados possíveis locais de corte das gingipains em inibidores naturais das elastases e a identificação molecular de um local de corte na pro MetaloProteinase da Matriz (MMP) 1. A análise dos resultados sugere que a interpain A, uma protease de cisteína de Prevotella intermedia com elevada homologia estrutural com as proteases de cisteína PrtT e periodontain de P. gingivalis, também degrada o inibidor de elastases, alfa1antitripsina.
ConclusãoCom a integração da informação obtida, sugere-se um possível mecanismo molecular da sinergia criada entre proteinases de cisteína no sentido de promover a doença periodontal. Este estudo ilustra a forma como as ferramentas bioinformáticas podem ser úteis no esclarecimento dos mecanismos moleculares e na previsão de resultados experimentais, melhorando o desenho de estudos laboratoriais.
Periodontitis is an inflammatory and infectious disease affecting the supporting tissues of the tooth. Despite the fact that the microflora associated with periodontal disease has been extensively studied,1 no species by itself could be identified as the etiologic agent.2P. gingivalis has been identified as one of the major organisms associated with destructive adult periodontitis. The virulence of this microorganism is particularly related to its proteolytic activity, and the cysteine peptidases gingipains have been extensively studied and characterized.3,4 Gingipains have been described and studied as being particularly efficient and versatile enzymes, capable of proteolysis of several host molecules associated with physiological processes such as host immune defense, cell adhesion and vascular permeability.5–9 Studies demonstrating the “in vitro” cleavage of host molecules by gingipains often just report a digestion10 and sometimes identify the resulting fragments either by size11 or by antibody5 based techniques, but lack information on the molecular details of the interaction between the gingipains and the host molecules.
Public databases accumulate an enormous quantity of sequences and structural data and several bioinformatics tools have been developed. Some of the tools are integrated in the databases and repositories (e.g. Blast search and ClustalW2 alignment algorithms) while others are developed independently such as PyMol.12,13 In either case, the objective is the same: to extract biological significance of the accumulated data. This knowledge may then be used to direct further scientific research prompting new experimental studies or, in other instances, be applied in the development of chairside innovative tools and/or procedures.
The purpose of this work is to show how the structural information available in public databases such as UniProtKB14 can be explored with bioinformatics tools such as PyMol12 and Psipred13 to produce information on the interactions between host molecules and gingipains. This analysis clarifies the molecular detail of these interactions and proposes new experimental approaches to elucidate other interactions between microbial proteases and host molecules. The knowledge of the molecular interactions is fundamental not only in the development of specific inhibitors for the action of gingipains and other microbial proteases,15 but also in the development of additional diagnostic tools such as the recently described by Kaman et al.,16 and therefore provides a basis for improvement and innovation in the development of new therapeutic approaches and accurate diagnosis of periodontal diseases.
Materials and methodsThe bioinformatics analysis of the interactions between the gingipains and host molecules was performed using freeware tools and information available in international databases. This study focused on host molecules whose interaction was described in the literature7 and for which crystallographic structures were available. Because the modeling of membrane proteins still has limitations, we used only molecules which are soluble and not integrated in the membrane. To analyze the interactions the steps followed are described below.
Characterization of gingipainsThe amino acid sequence of proteins used in this study was obtained from the UniProtKB database14 (http://www.uniprot.org/). The 3D model of Gingipain R2 was obtained from crystallographic data available at http://www.pdb.org,18 with code 1cvr. The remaining models of gingipains were generated through the program MODWEB19 (http://modbase.compbio.ucsf.edu/ModWeb20-html/modweb.html) and recorded in the program of structural analysis PyMol12 (http://www.PyMol.org/).
Characterization of host substratesThe information on the host substrates was obtained from the UniProtKB14 and Merops17 database by searching the name of the protein. For each of the host substrates we searched at http://www.pdb.org18 for the crystallographic information available. In many cases the available crystallographic structure was that of the molecule associated to an inhibitor. When this was the case, MODWEB19 was used to produce the 3D structure of the host substrate.
Whenever the secondary structure of a protein was necessary the Protein Structure Prediction Server (PSIPRED) was used. This is a highly accurate method for protein secondary structure prediction freely available on-line.13
ResultsTo clarify the molecular details of the cleavage of Secretory Leukocyte Protease Inhibitor (SLPI) by gingipains, a determination of the secondary structure of this molecule was done and is presented in Fig. 1. All arginine (R) residues of the molecule are in areas of the corresponding to beta sheets and therefore possible cleavage sites. The analysis of Fig. 1 also reveals that there are 4 lysine (K) residues on SLPI which could be cleavage sites for gingipain R2.
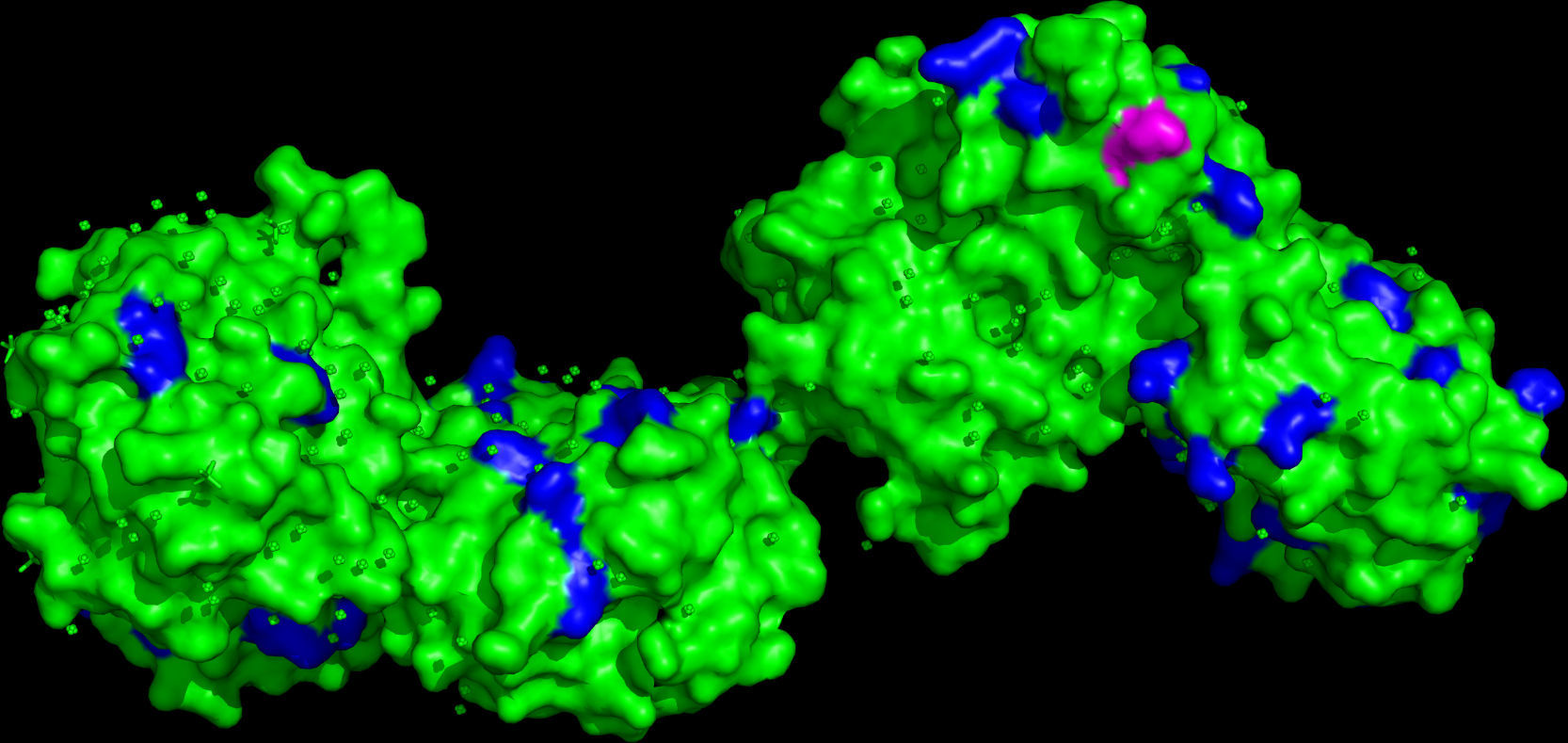
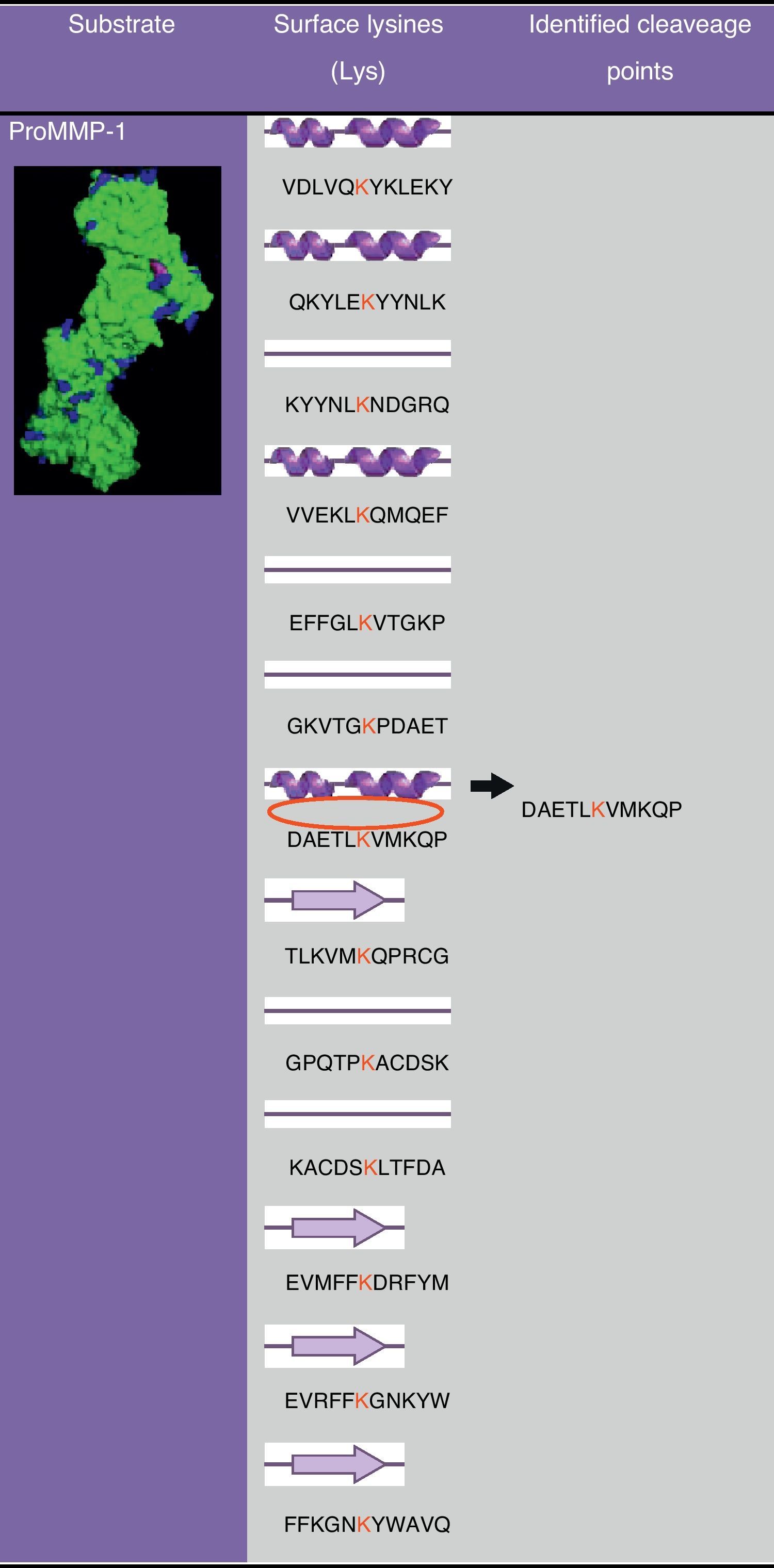
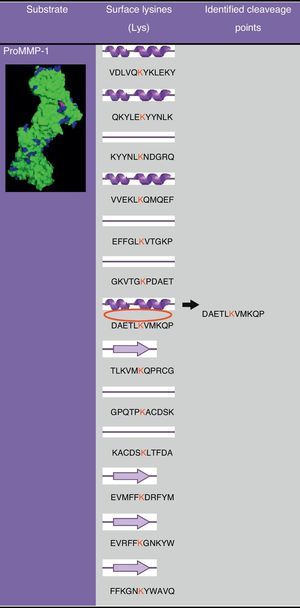
In Fig. 2 a model of the 3D structure of pro-MMP1 is presented showing the surface arginine and lysine residues which may be cleaved by gingipain R and K, respectively. The secondary structure of pro-MMP1 is presented in Fig. 3 where the K residue described previously as the cleavage site is marked in red. In Fig. 4 the location of the cleavage site identified for gingipain K in this study (black arrow) is compared with the cleavage site described in the literature (blues arrow) in Fig. 4.
A model of the 3D structure of pro-MMP1, in which the cleavage point (in purple) for gingipain K is highlighted and other lysines are marked (blue) on the surface of the molecule. This three-dimensional model was obtained from crystallographic data27 and modified in PyMol.
The sequence of interpain A from Prevotella intermedia, identified by A9J7N5, PrtT (P43158) and periodontain (B2RJ74) from P. gingivalis was compared and is presented in Fig. 5. From this image it can be seen that the sequences for the catalytic site (TGCVATA) of the proteins is very similar.
DiscussionProteolytic activity of gingipainsSeveral in vitro studies showed the cleavage of many substrates related to periodontitis by gingipains R1, R2 and K.7,20–25 These substrates include complement proteins, kallikrein, cytokines, fibrinogen and matrix metalloproteases.8 Using bioinformatics and modeling tools we set to verify if the analysis of the crystallographic information available for each substrate supported the “in vitro” digestion results. Many of the host molecules described as being cleaved by gingipains have not had their crystallographic structure determined and therefore could not be submitted for this analysis. For instance, important host substrates such as Tumor Necrosis Factor alfa and protrombin have only had part of their crystallographic structure deposited in the PDB database.18 In other cases, the existing crystallographic structure is from a non-human version of the molecule. This is why we restricted our study to two molecules that are known not to be integral membrane proteins and for which digestion in vitro had been reported: Secretory Leukocyte Protease Inhibitor (SLPI) and the pro-Matrix Metalloprotease 1 (pro-MMP1).
SLPIUsing purified gingipain R1 and SLPI, Into et al.11 demonstrated, by immunoblot, the cleavage of SLPI by gingipain R1, originating 3 fragments, one with 12kDa and two smaller ones (≥6kDa, <12kDa).
When the SLPI molecule was analyzed relative to its surface we found 5 arginine residues in positions 45, 62, 83, 84 and 113, present in 2 domains of this molecule which represent potential cleavage sites for gingipain R. This would lead to the prediction of 6 fragments after digestion by gingipain. However, it is known that structures such as helices or beta sheets indicate areas where the cleavage will be less probable so, we further considered the secondary structure of SLPI26 (Fig. 1).
From Fig. 1 we conclude that all R residues present on the surface of SLPI are possible cleavage sites for gingipain R, but the size of the fragments obtained experimentally11 suggests that most likely the cleavage occurs at R residues 62 and 83/84. In this case the bioinformatics analysis of SLPI structure corroborates the experimental results obtained previously.11
A similar analysis of SLPI surface regarding K residues leads to the conclusion that there are 4 possible cleavage sites for gingipain K. These putative cleavage sites allow us to predict that if the cleavage occurs on residue 85, fragments very similar to those obtained by the digestion of gingipain R1 (12 and 6kDa) will be obtained; the cut in residue 99 is not likely due to the fact that this K is inserted in a beta-sheet and therefore inaccessible to gingipain K; if the cleavage occurs at residue 112 two fragments will be originated one over 12kDa and a smaller one; finally a cut near the end of the SLPI molecule is possible on residue 112 originating a much smaller fragment and a larger one almost the size of the uncut molecule. Most likely, there will be three fragments originating from the digestion of SLPI by gingipain K all below 12kDa. The literature lacks experimental data for this digestion and in this case, the bioinformatics tools allow the prediction of the experimental results.
MMP-1DeCarlo et al.10 demonstrated the cleavage and consequent activation of pro-MMP-1 by gingipain K, with SDS-PAGE, Western blot and sequencing of fragments obtained. The cleavage of pro-MMP-1 occurs at lysine residue 66. We analyzed the structure of MMP-1 in order to verify if this residue lies at the surface of the molecule and therefore the structural and experimental data are in concordance.
The analysis of ProMMP-1 models (Fig. 2) showed that there are several loops with lysine residues, representing putative cleavage points for gingipain K. The cleavage site identified experimentally by DeCarlo et al.10 is inserted into a helix (Fig. 3), which surprised us, because cleavage in helices is not very common. In order to obtain more data on the secondary structure of ProMMP-1 we ran the sequence of amino acids in PSIPRED program13.
The lysine residue in this representation is in the transition between a helix and a loop which increases the likelihood of access of the gingipain active site. In this case, the structural analysis of the ProMMP-1 molecule does not strongly support the experimental data available and raises the question of why are superficial lysine residues not cleaved by gingipain K? Are there other factors affecting gingipain K specificity? Is the structure of ProMMP-1 altered “in vivo”? These and other suggestions should be targeted in further studies to elucidate the molecular behavior of gingipain K and enhance the development of more efficient inhibitors.
Comparing the proximity in the cleavage of pro-MMP1 by gingipain K and the usual cleavage point that leads to its activation “in vivo”, we found that these sites are nearby, which supports the suggestion of DeCarlo et al.10 that the cleavage by gingipain K leads to the activation of pro-MMP1 (Fig. 4). Other MMPs have been described as being activated by gingipains even though the molecular mechanisms of the cleavage have not been described in detail.28,29 For example, MMP-9 seems to be activated by the cleavage of the 92kDa proMMP-9 zymogen to the 82kDa active form by P. gingivalis.30 In another in vitro study Grayson et al.31 found that latent MMP-2 in the presence of gingipain R1 was activated.
Proposal of proteolitic activity by other proteasesThe effect of microbial proteases on elastase inhibitors seems to play an important role in the virulence mechanisms associated with periodontitis. The degradation of alpha 1 antitrypsin by PrtT and periodontain32 had already been described. These two cysteine proteases of P. gingivalis are included in C1017 family along with streptopain and interpain A. The fact that the C10 protease family includes two proteases from P. gingivalis and one from another bacteria (P. intermedia) that is commonly associated with periodontal disease, led us to search for the evidence of structural identity between these proteases making the proposal that, if there is structural homology, there may be functional similarities.
Interpain A is encoded by the locus PIN0048 and secreted as a zymogen of 868 aa including a signal peptide of 44 aa, a pro-domain (Ala1-Asn111), a catalytic domain (Val112-Pro359) and a C-terminal with possible regulatory and secretory functions; however, their specific targets and functions are yet to be clarified.33 The alignment of interpain A sequence and PrtT and periodontain (Fig. 5) shows great similarities especially at the active site of the enzymes. Consequently, we suggest that interpain A, produced by P. intermedia, also cleaves alpha 1 antiproteinases, preventing the inhibition of elastases produced by neutrophils. This suggestion supports the polymicrobial nature of periodontitis, showing virulence factors of different microorganisms with identical molecular effects. In fact, Potempa et al.34 have shown the degradation of complement factor 3 by interpain A from P. intermedia in vitro which is one of the molecules also degraded by the gingipains.8
ConclusionThe use of bioinformatics tools such as PyMol, Psired and other structure analysis tools, coupled with the structural information available in several public databases (UniprotKB, MEROPS, PDB) enabled the verification of the molecular detail of in vitro reactions (gingipains digestion of SLPI and MMP1). The results of SLPI digestion by gingipain R1 were supported by the structural analysis, and the activation of MMP1 by gingipain R1 was partially supported. Predictions of the fragments most likely to be obtained from the digestion of SLP by gingipain K are also made. Furthermore, the fact that experimentally less fragments are observed than what might be expected from the theoretical prevision suggests that other factors may be affecting enzyme specificity and/or substrate conformation “in vivo”.
The prediction of new interactions was based on the comparison of the structural similarities shared by the members of the C10 protease family. We suggest that, based on those similarities, interpain A (produced by P. intermedia) cleaves alfa 1 antitripsin proteinase supporting a synergistic action with the proteases of P. gingivalis promoting host tissue destruction.
Ethical disclosuresProtection of human and animal subjectsThe authors declare that no experiments were performed on humans or animals for this investigation.
Confidentiality of dataThe authors declare that no patient data appear in this article.
Right to privacy and informed consentThe authors declare that no patient data appear in this article.
Conflicts of interestThe authors have no conflicts of interest to declare.