Disaster victim identification is crucial for humanitarian and legal reasons. Forensic genetics plays an important role in these situations which often become a challenge for the different professionals involved due to their complexity. The establishment of guidelines and recommendations makes it easier to follow standardised protocols that make it possible to guarantee the reliability of the identification final result. Likewise, advances in forensic genetics contribute to speeding up the response, providing new analysis strategies and bioinformatic tools. This article aims to provide an overview of how forensic genetics and its advances can contribute in these situations, as well as some keys to understanding the work of forensic genetics laboratories in the identification of corpses in events with multiple victims.
La identificación de los afectados por un suceso con víctimas múltiples es una prioridad por razones humanitarias y legales. La genética forense juega un importante papel en estas situaciones que, por su complejidad, a menudo se convierten en un reto para los distintos profesionales implicados. El establecimiento de guías y recomendaciones facilita el seguimiento de protocolos estandarizados que permiten garantizar la fiabilidad del resultado final de la identificación. Así mismo, los avances en la genética forense contribuyen a agilizar la respuesta, aportando nuevas estrategias de análisis y herramientas de tipo bioinformático. Con este artículo, se pretende dar una visión general de cómo la genética forense y sus avances pueden contribuir en estas situaciones, así como algunas claves para entender la labor de los laboratorios de genética forense en la identificación de cadáveres en sucesos con víctimas múltiples.
The reliable identification of bodies in mass fatality incidents (MFI) is a primary objective for humanitarian and legal reasons. As outline in the Spanish National Technical Commission's Disaster Victim Identification Guide,1 “In the case of an event with a large number of victims, the recovery and identification of mortal remains is a priority so that they can be returned to their families in the shortest possible time”.
There are diverse mechanisms that can cause this type of disaster, ranging from natural phenomena to human action (acts of terrorism or accidents). The nature of these mechanisms, and the magnitude of the incident have a different impact on the bodies of the victims and, therefore, on their integrity, which affects their identification. Sometimes, the bodies are damaged to such an extent that the use of classic forensic identification procedures (anthropometric, fingerprint, or dental) is compromised; this is when genetic analysis for identification purposes can be of decisive value.
The success of the identification process by genetic analysis is determined by certain guidelines that are essential in this type of action. In this regard, different bodies and standardisation groups worldwide have issued guidelines and recommendations, including those published by Interpol,2 the International Society for Forensic Genetics, ISFG,3 the International Red Cross,4,5 or Royal Decree 32/2009 of 16 January, approving the Spanish National Protocol for Forensic Medical and Scientific Police Action in Multiple Victim Events,6 whose scientific standards with respect to genetic identification were reviewed by Vallejo and Alonso in 2009.7 All highlight the multidisciplinary nature of the process, in which different groups and professionals intervene in the identification process, in a coordinated and structured manner.
The field of forensic genetics now has new analysis strategies and bioinformatics tools that provide greater reliability and speed in the process of the genetic identification of victims of MFI. In this article, we provide a breakdown of the different recommendations issued by the ISFG in this area and review some methodological and technological advances, which promise to be of great use in the identification of corpses. Finally, we review the problems associated with the final step of the identification process and the usefulness of bioinformatics tools in this process.
Recommendations of the International Society of Forensic Genetics for the use of genetic markers in mass fatality incidentsIn 2007, the DNA Commission of the ISFG published 12 recommendations on the role of forensic genetics in the investigation process,3 which are divided into 4 main blocks. The first (Recommendations 1 and 2) refer to the importance of planning the different aspects related to intervention in the incident. In this planning, genetic identification laboratories should be actively involved in operational aspects, especially those related to the most appropriate choice of samples and conditions of submission and preservation. Furthermore, participating laboratories need to address throughput capacity, in terms of agility of response, and define the personnel responsible for the different phases of the process.
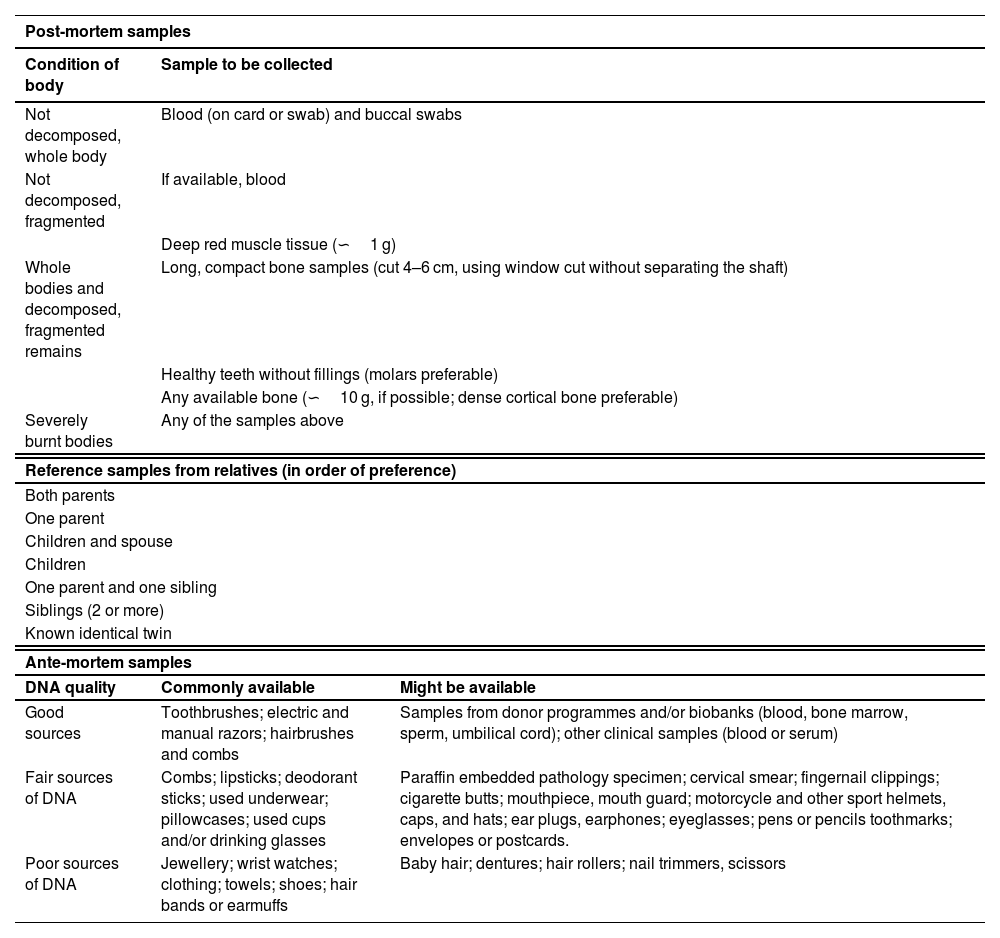
The second block of recommendations is dedicated to the selection of biological samples of interest (Table 1). The speed in recovering post-mortem samples, their correct preservation and appropriate selection (Recommendation 3) are critical to the success of the genetic identification process. In this regard, the correct identification and traceability of the samples must be guaranteed throughout the process. Blood is preferable as a post-mortem sample, when the corpse or the remains to be identified are preserved in appearance. Conversely, when signs of degradation are visible on the remains, analysis of long bone fragment (e.g., femur, humerus) or molar in a good state of preservation is recommended (Table 1). It is recommended to collect post-mortem biological samples even when the body has been identified by other means.
Samples recommended for analysis in mass fatality incidents according to type: post-mortem samples, reference samples from relatives, and ante-mortem samples.
| Post-mortem samples | ||
|---|---|---|
| Condition of body | Sample to be collected | |
| Not decomposed, whole body | Blood (on card or swab) and buccal swabs | |
| Not decomposed, fragmented | If available, blood | |
| Deep red muscle tissue (∽1 g) | ||
| Whole bodies and decomposed, fragmented remains | Long, compact bone samples (cut 4–6 cm, using window cut without separating the shaft) | |
| Healthy teeth without fillings (molars preferable) | ||
| Any available bone (∽10 g, if possible; dense cortical bone preferable) | ||
| Severely burnt bodies | Any of the samples above | |
| Reference samples from relatives (in order of preference) | ||
| Both parents | ||
| One parent | ||
| Children and spouse | ||
| Children | ||
| One parent and one sibling | ||
| Siblings (2 or more) | ||
| Known identical twin | ||
| Ante-mortem samples | ||
| DNA quality | Commonly available | Might be available |
| Good sources | Toothbrushes; electric and manual razors; hairbrushes and combs | Samples from donor programmes and/or biobanks (blood, bone marrow, sperm, umbilical cord); other clinical samples (blood or serum) |
| Fair sources of DNA | Combs; lipsticks; deodorant sticks; used underwear; pillowcases; used cups and/or drinking glasses | Paraffin embedded pathology specimen; cervical smear; fingernail clippings; cigarette butts; mouthpiece, mouth guard; motorcycle and other sport helmets, caps, and hats; ear plugs, earphones; eyeglasses; pens or pencils toothmarks; envelopes or postcards. |
| Poor sources of DNA | Jewellery; wrist watches; clothing; towels; shoes; hair bands or earmuffs | Baby hair; dentures; hair rollers; nail trimmers, scissors |
Adapted from Prinz et al.3 (2007).
The appropriate choice of reference samples (Recommendation 4) depends on the available relatives, with preference given to relatives with a direct genetic link (e.g., parent(s) or first-degree descendant(s)) (Table 1). It is recommended that samples are taken from as many relatives as possible. In all cases, an informed consent form is necessary that includes the information and data, in line with international and local standards. Occasionally, as an alternative or complement, ante-mortem samples consisting of objects used by the victim or biological samples taken from the victim (e.g., hospital samples), are used) (Table 1). It is essential that their authenticity is ensured for them to be used as attributed samples.
Forensic geneticists play a key role in guiding the choice of the most suitable ante- and post-mortem samples, which helps to speed up and secure the results.
The special complexity involved in the analysis of post-mortem samples makes it advisable that the qualification of the laboratories involved in the analysis of this type of samples be demonstrated (Recommendation 5). The high discriminatory power provided by the analysis of autosomal STR markers makes them the first analytical choice. At least 12 STRs should be used and, where appropriate, these should be agreed with the countries involved in the incident (Recommendation 6). Currently, such commercial kits usually include a larger number of STRs agreed by the international scientific community, which enables an adequate response to this recommendation.
The nature of these types of post-mortem samples, often affected by degradation or the presence of possible inhibitors, can lead to certain artefacts in the profile after genetic analysis (e.g., allelic losses), which can result in the assignment of incorrect genetic profiles and thus in errors during the identification process. Therefore, Recommendation 7 refers to the need to establish technical protocols to ensure the allelic assignment of the analysed sample. Sometimes, the degree of involvement of the sample or the availability of non-direct relatives prevents the use of autosomal STR markers. Thus, Recommendation 8 proposes the use of single nucleotide polymorphisms (SNPs), Y chromosome STR markers, or mitochondrial DNA (mtDNA) analysis, although the latter two types of genetic markers have less discriminatory power due to their single-parent inheritance pattern.
The last block of recommendations addresses data processing aspects. Recommendation 9 refers to the need for centralised databases and the importance of the electronic uploading of genetic profiles to avoid transcription errors. There are now different bioinformatics tools that allow us to respond to this important need, which we shall discuss in more detail in the last section of this article. Recommendation 10 refers to the importance of having other complementary identification tools (e.g., anthropometric studies), especially in situations where there are several members of the same family among the victims.
When there is a genetic match between post-mortem fragments or compatibility of the post-mortem remains with a specific relative or family group, it is necessary to establish a minimum value at which it is statistically possible to establish the identity as reliable. The use of the likelihood ratio (LR) is recommended (Recommendation 11). This threshold value will depend on several factors, e.g., the number of victims involved in the incident, increasing as the number of victims increases,8 or other circumstances (e.g., closed versus open population). A large amount of information, records, samples, or DNA extracts are generated during the preparation and execution of the identification process. Therefore, laboratories should have described in their working procedures how to manage this information and derived data, the policy for notifying relatives of results, and provide for storage, delivery, or disposal of recovered biological material (Recommendation 12).
Advances and new tools in forensic geneticsDNA polymorphism analysis has become the gold standard for victim identification both in MFI and in forensic cases where human remains are highly fragmented and/or degraded.9,10 This is mainly due to the high degree of discrimination that DNA-based identification can provide. There are numerous challenges from a genetic perspective; these include, among others, the high number of victims, the destruction mechanisms, the degree of fragmentation, the availability of relatives, or the existence of ante mortem samples, which will be crucial for the final identification process. In the last 20 years, forensic genetics has undergone important technical and technological advances that can help in victim identification in MFI. We shall give an overview below of these major developments from different perspectives: DNA source, DNA recovery, genetic markers, and methods of analysis.
DNA sourceThe Spanish National Protocol for Forensic Medical and Scientific Police Action in Mass Fatality Incidents (Annex VII.1)6 and the Rules for the preparation and submission of samples for analysis by the INTCF (arts. 30 and 32)11 detail the type of samples required (Table 1) and the submission conditions for these types of MFI identification studies.
Apart from the most suitable type of samples identified in these references, recent studies have shown the availability of DNA from certain tissues and organs, not previously considered for genetic identification purposes, with promising results. These DNA sources may also offer savings in time and resources for MFI teams compared to time-consuming sample collection, preparation, and extraction of DNA from traditionally used samples (e.g., the femur). Thus, it has been found that, from human remains buried for periods of between 4 and 42 years, DNA/RNA is more stable in certain organs, such as brain or heart, obtaining good quality genetic profiles with fewer signs of degradation than from other organs.12 Small spongy bones (e.g., bones of the hands, feet, or ankles) have also been found to have higher DNA concentrations per unit mass than dense cortical bones such as the femur.13
DNA recoveryDNA retrieval (extraction) methods are increasingly optimised and automated,14,15 using high-throughput techniques using paramagnetic particles, including prior demineralisation steps (in the case of skeletal remains),16,17 which minimise human intervention and potential risk of contamination or handling errors.
There are also some current solutions, especially indicated for reference samples, but which have also been tested on cadaveric samples, which further reduce extraction times, integrating in a single step even the process of obtaining the profile in just over 90 min. These are the Rapid DNA systems,18 which are completely portable to the scene and do not require extensive technical training. Their usefulness and effectiveness have been demonstrated, for example, in the case of the French Gendarmerie's mobile laboratory (mobil'DNA), taken to the site of Germanwings flight 9525 incident in 2015,19 or to Nice in 2016, and Beirut in 2020, on the occasion of the respective attacks, and the French government has recently provided a unit to Ukraine for the identification of victims of the current armed conflict. However, these systems do not have high sensitivity, and therefore they would not be appropriate in cases where samples are affected by degradation.
The Direct-to-PCR20 techniques may also be of interest in these types of incidents. They consist of adding biological samples directly to the PCR tube. This type of technique considerably reduces the time taken to obtain results by eliminating the DNA extraction step. However, the success and quality of the recovered profiles will obviously depend on the nature of the sampled material and the presence of PCR inhibitors, which are usually removed during the DNA extraction process. In combination with this technique, preservatives have also been described that capture DNA from the recovered tissue and retain it in a solution that can be used directly in the PCR process.21
Genetic markersAutosomal STRS are the genetic markers that are normally used in this type of incident, which have a high discriminatory power.9 In some cases, depending on the relatives available (e.g., only male siblings), Y chromosome STR markers can also be useful, which allow paternal lineages to be established,22 or X chromosome STRs.23 There are numerous commercial kits for these 3 types of markers from different companies, which are highly standardised and internally validated in most forensic genetics laboratories.14 The mtDNA kit, maternal inheritance, is also often very useful in the forensic field, especially in cases of missing persons and unidentified remains, when there is not enough nuclear DNA available, due to the high number of copies of mtDNA per cell and its circular conformation, which makes it more resistant to degradation,24 and to confirm/rule out maternal relationships with relatives of second degree or higher. However, mtDNA has very low discriminatory power, which makes it, despite its advantages with degraded samples, of limited resolution in MFI.
More recently, for MFI cases in which the recovered DNA is degraded and/or inhibitory, some markers less susceptible to degradation have become available, which can help in genotyping.15 This is the case of SNPs, which are single-base sequence variations, whose amplicon (DNA fragment resulting from amplification) is usually much shorter, which offers an advantage over degraded samples.25 Validated commercial panels of up to 90 autosomal SNPs are now available, with better random match probabilities than for the set of STR markers typically used in the forensic field.26 But their potential goes even further, with the implementation of markers and analysis methodologies in the burgeoning field of investigative genetic genealogy,27 where more than 600,000 SNPs could be analysed. However, a commercial panel with 10,230 SNPs has recently been developed to be used in the forensic field.28 Thanks to this large number of markers, it is potentially possible to establish more distant kinship relationships (e.g., third cousins), which would facilitate the study in MFI cases where more direct relatives (e.g., parents, children, or siblings) are not available.
Furthermore, while DNA analysis in the context of forensic identification has traditionally been strictly based on comparison with relatives or ante mortem samples, recent advances are allowing highly informative DNA markers to be used to predict externally visible characteristics (EVC) and biogeographical ancestry (BGA)29 in certain specific cases. Within this field are phenotype informative SNPs (piSNPs) and ancestry informative SNPs (aiSNPs), the analysis of which can provide valuable information such as eye, hair, or skin colour, or clues to the biogeographical ancestry of the DNA donor, data that can be useful for cadaver identification when no reference samples are available for comparison. In this respect, the international VISAGE project30 for the development of SNP panels and bioinformatics tools is of great importance.
Methods of analysisCurrently, most forensic genetics laboratories obtain their DNA profiles by capillary electrophoresis (CE), a standardised method based on the separation of DNA fragments according to their charge/size.31
However, new technologies such as massively parallel sequencing (MPS) are already upcoming. MPS allows a substantial increase in throughput, because a large number of genetic markers and samples can be analysed simultaneously at increasingly affordable prices. Moreover, because it is simultaneous sequencing, not based on fragment size, panels have been designed with smaller amplicon sizes, making it an ideal technology for degraded samples, such as those found in MFI cases or in the identification of missing persons.32
Using MPS, commonly used forensic STR markers are more informative (with higher discriminatory power), as they allow the detection of SNPs, undetectable by CE, both within the repetitive regions of the STRs and in the flanking regions.32 Along with the STR identification information, large panels of SNPs can also be sequenced to increase the power of discrimination with autosomal SNPs, as well as adding information on the ancestry and phenotype of the donor of the sample.32 With regard to the analysis of mitochondrial DNA, it is possible to further reduce the size of the amplicons, making them overlap, and increasing the region sequenced, which can increase the discriminatory power of this marker. But it also simplifies the complex mtDNA analysis by Sanger-based sequencing, allowing higher throughput with little operator intervention.32
Bioinformatics tools for comparisonThe final stage in the process of the genetic identification of victims of an MFI is to compare between the genetic profiles obtained from post-mortem samples and the genetic profiles of samples attributed to the missing persons or from samples taken from their relatives (Table 1). Logically, there are differences in the way this comparison is made depending on the origin of the reference samples. In the case of attributed (ante mortem) samples, full matches with the genetic profiles of the post-mortem samples will be sought. When matching against the genetic profiles of relatives, the aim of the search is to find a relative or family group that is compatible for the kinship relationship that is being investigated. Thus, if first-degree relatives (direct ascendants or descendants) and genetic profiles of autosomal STR markers are available, the search aims to locate the genetic profile that shares at least one allele in all the markers analysed (unless a mutation occurs). When the kinship relationship is more distant, it is advisable to additionally analyse other types of markers, e.g., haplotypic markers, which allow us to establish kinship relationships through the paternal (Y chromosome STR marker haplotypes) or maternal route (mtDNA haplotypes) and whose information will allow us to reinforce compatibility, or to exclude a kinship relationship in cases of chance compatibility with the autosomal markers. In the event that the bodies of the victims are fragmented, it is advisable to reassemble all the fragments of the same body beforehand and select the most complete genetic profile available for this set of fragments to be used as a representative profile of the victim in question, since reducing the number of profiles involved in the comparisons will facilitate the processing of the data.
There are multiple factors that influence the phase of comparison of genetic profiles. One important aspect is whether, in terms of prior knowledge of the number and identity of the deceased, the incident is open (e.g., natural disaster) or closed (e.g., plane crash, where in principle, the list of possible victims is available). Logically, in the latter case it will be easier to access all necessary reference samples (either ante mortem or from relatives) than in the former case. However, the magnitude of the incident, the number of victims, the completeness or dismemberment of the bodies, and certain circumstances of the incident that may facilitate the degradation of human remains (e.g., flood or fire) and thus make it difficult to obtain complete genetic profiles, will also influence the complexity of the matching and the final result. Accessibility to reference samples may also be hampered in the case of incidents involving international victims, bearing in mind that, in certain cases, it may also be necessary to resort to different population genetic databases than the one usually used. Likewise, the time elapsed between the incident and performing the genetic analysis and matching is a factor that works against obtaining the maximum genetic information from the remains, because it affects both the preservation of the unidentified human remains and, therefore, the possibility of obtaining the maximum genetic information from them, and the possibility of obtaining suitable reference samples (attributed samples or samples from direct relatives). The project of the national DNA bank of victims of the Spanish Civil War and dictatorship (art. 23, Law 20/2022, of 19 October),33 is an illustrative example of this last factor in Spain, which aims to identify victims of events that took place up to 87 years ago, whose main challenges are 1) the degradation of the DNA of the remains, which will be highly variable depending on the conditions of conservation, and 2) the absence of direct relatives, which will force us to resort to second, third, or fourth degree relatives (grandchildren, nieces/nephews, cousins) and, therefore, to the design of complex pedigrees. Also, in this type of situation, there is the added difficulty that many bodies may be in common graves with the consequent possibility of the remains of different individuals being mixed, or there being individuals not related to the event in question.
The complexity of the process of comparison between multiple samples and the different variables mentioned above makes it essential that the process is carried out using advanced bioinformatics tools to optimise the detection of potential coincidences/compatibilities, as well as their corresponding statistical evaluation.34
In Spain, the national database of identifiers obtained from DNA (regulated in LO 10/2007, of 8 October)35 is currently hosted in the CODIS (Combined DNA Index System) application, provided free of charge by the FBI (US Department of Justice) to the Ministry of the Interior, whose State Secretariat for Security is responsible for the national administration of this database. This database is fed with the genetic profiles obtained in the forensic genetics laboratories of the security forces and police corps, both national (National Police and Civil Guard) and regional (Mossos d'Esquadra, Ertzaintza, and Policía Foral de Navarra), and the National Institute of Toxicology and Forensic Sciences, which depends on the Ministry of Justice. In this repository, apart from the index of genetic profiles of criminal interest (in which the proven and unproven genetic profiles of all samples collected or taken in relation to the investigation of the crimes specified in the law itself are stored and compared), there is an index of social interest, which includes both the ante mortem and unproven genetic profiles of the relatives of missing persons and the genetic profiles obtained from corpses or unidentified human remains, for the sole purpose of identifying the latter.
As mentioned above, successful identification will depend to a large extent on the quality of the genetic profiles obtained from post-mortem remains and the availability of suitable reference samples (see ISFG3 recommendations3). If it is not possible to obtain samples from any of the recommended relatives, or reliable ante mortem samples, other more distant relatives can be used, but in this case, it is highly recommended to have as many relatives as possible.
The computer programmes currently used to make comparisons have the option of constructing family trees (pedigrees) against which to carry out comparative searches, which makes it possible to extract as much information as possible from the family group and, therefore, maximise the possibilities of identification. The laboratories integrated in COMSIGENI (Regulatory and Coordinating Committee of the National. Management System for DNA-based identifiers) recently participated in a collaborative exercise consisting of an MFI simulation. The purpose of this exercise was for all the institutions to familiarise themselves with the pedigree management module available in CODIS, as well as to measure the limitations and problems that can be encountered in genetic identification for humanitarian purposes using this application. As a result of this exercise, which has been very positively evaluated by all the institutions, a guide for genetic identification using CODIS in mass fatality incidents (MFI) is currently being developed, which will serve as a reference manual in our country for the management of genetic profiles and their comparison using pedigrees in CODIS in relation to these types of incidents.
The importance of these types of collaborative exercises had already been highlighted in a previous initiative promoted by the ISFG Spanish and Portuguese Speaking Group (GHEP-ISFG, which organised two international collaborative exercises on the identification of victims in disasters (Disaster Victim Identification, DVI).36,37 The aim of these exercises was to explore the correct reassembly of remains, the identification of victims through kinship analysis, including related victims, the handling of mutations or an insufficient number of reference relatives, all working within a Bayesian framework, and to assess the correct use of the bioinformatics applications used.
Other bioinformatics applications are used in this field apart from CODIS, the most important of which are described below. It is worth mentioning the recent development of open-source applications, such as SmartRank, ForeStatistics, or GENis.15 Among these developments, the integration of the DVI Module,38 specific for the identification of victims of an MFI, in the free programme Familias,39 deserves special mention, which makes it possible to calculate probabilities and likelihoods in cases where we know the genetic profiles of some individuals, but their family relationships are in doubt, because it allows multiple alternative pedigrees to be taken into account and can compute which pedigree is more probable, and how much more probable it is with respect to the other pedigrees. A feature that sets Familias apart is its ability to deal with complex cases, where the possibility of mutations has to be considered, together with the possibility of handling different pedigrees simultaneously.
The M-FISys® (Mass Fatality Identification System) programme40 also has great potential in this field, developed by the US company Gene Codes Corp. for the identification of the victims of the World Trade Centre bombing in 2001. M-FISys® is capable of integrating and filtering a wide variety of data (such as anthropological descriptions, location where remains were recovered - including GPS or grid coordinates - and identifications made by fingerprints, dental or other methods, with access to supporting documents including images and PDF files associated with each sample). This software was the first to combine STR, mtDNA, and SNP marker data in an integrated manner, with progress reporting and workflow management, including administrative review tools to establish sample chain of custody and audit trails in critical operations. It includes quality control tools that allow the detection of inconsistencies that may be the result of mixed remains, sample changes, or contamination and greatly reduce the possibility of false identification.
Finally, Bonaparte41 is another highly versatile application (from the Dutch company SMART Research BV). Bonaparte has been validated in the forensic field in different international scenarios (NFI, Netherlands Forensic Institute; ACIC, Australian Criminal Intelligence Commission) and has recently been acquired by INTERPOL within the international I-Familia programme, for the identification of missing persons worldwide.42 This software allows the creation of complex pedigrees and uses an open mathematical algorithm based on a Bayesian inference model for the calculation of LR (Likelihood Ratio) adaptable to any type of family relationship investigated. It is also scalable to thousands of SNP markers, which are very useful in the analysis of degraded samples, and which, as mentioned above, when used in large numbers, make it possible to establish, kinship relationships with distant relatives, with a high degree of reliability.
Please cite this article as: Márquez MC, Barrio Caballero PA, Espuny MJF. Aportaciones y avances de la genética forense en los sucesos con víctimas múltiples. Revista Española de Medicina Legal. 2023. https://doi.org/10.1016/j.reml.2023.04.005.