Situated in West Central Morocco, the Doukkala region carries the distinction of being one of North Africa's oldest human settlement zones. Nonetheless, it has been notably understudied in the realm of population genetics. Through allele frequency analysis and integration of forensic parameters, the research aims to gain insights into the genetic structure and neighboring affiliations of the Doukkala population.
MethodsThis study employed the AmpFlSTR Identifiler PCR system to assess the allelic frequencies and forensic parameters of 15 autosomal STRs in a cohort of 134 unrelated, healthy individuals from the Doukkala region who identify as Arab-speakers. Additionally, we explored the genetic relationships between the Doukkala population and other reference groups, considering both our dataset and previously published population data.
ResultsA total of 180 alleles were observed in the study population. With a count of 19 alleles, D18S51 proved to be the most diverse marker in the study. After Bonferroni's correction, 3 loci (FGA, TH01, and TPOX) deviated from Hardy–Weinberg equilibrium. The combined power of discrimination (PD) was 0.99999999999999999526 and the combined probability of exclusion (PE) was 0.99999664790900144592. The Arabic-speaking population of Rabat-Salé–Zemmour-Zaer and Southern population from Morocco exhibit the shortest genetic distance from population of Doukkala. No significant difference was observed between the Arabic-speaking population of Doukkala and all North African populations at all loci, except for the Egyptian population (North-East Africa), where a difference was observed at 2 specific loci (CSF1PO and TH01).
ConclusionsThese results indicate that the diversity found in populations from North Africa transcends geographic and linguistic barriers. The dataset's relevance to this research could stem from its capacity to offer valuable reference data for forensic, anthropological, and genetic investigations.
Situada en el centro-oeste de Marruecos, la región de Doukkala se distingue por ser una de las zonas de asentamiento humano más antiguas del norte de África. Sin embargo, ha sido muy poco estudiada en el ámbito de la genética de poblaciones. Mediante el análisis de las frecuencias alélicas y la integración de parámetros forenses, la investigación pretende profundizar en la estructura genética y las afiliaciones vecinales de la población de Doukkala.
MétodosEn este estudio se empleó el sistema de PCR AmpFlSTR Identifiler para evaluar las frecuencias alélicas y los parámetros forenses de 15 STR autosómicos en una cohorte de 134 individuos no emparentados y sanos de la región de Doukkala que se identifican como hablantes de árabe. Además, exploramos las relaciones genéticas entre la población de Doukkala y otros grupos de referencia, considerando tanto nuestro conjunto de datos como los datos poblacionales publicados previamente.
ResultadosSe observó un total de 180 alelos en la población estudiada. Con un recuento de 19 alelos, D18S51 resultó ser el marcador más diverso del estudio. Tras la corrección de Bonferroni, tres loci (FGA, TH01 y TPOX) se desviaron del equilibrio de Hardy–Weinberg. El poder de discriminación (PD) combinado fue de 0,99999999999999999526 y la probabilidad de exclusión (PE) combinada fue de 0,99999664790900144592. La población arabófona de Rabat-Salé–Zemmour-Zaer y la población meridional de Marruecos presentan la menor distancia genética con la población de Doukkala. No se observaron diferencias significativas entre la población arabófona de Doukkala y todas las poblaciones norteafricanas en todos los loci, excepto en el caso de la población egipcia (África nororiental), donde se observó una diferencia en dos loci específicos (CSF1PO y TH01).
ConclusionesEstos resultados indican que la diversidad encontrada en las poblaciones del norte de África trasciende las barreras geográficas y lingüísticas. La relevancia del conjunto de datos para esta investigación podría derivarse de su capacidad para ofrecer valiosos datos de referencia para investigaciones forenses, antropológicas y genéticas.
Short tandem repeats (STRs) are sequences of nucleotides discovered in eukaryotic DNA in the early 1970s. These STRs are widespread in the genomes of all living organisms, from bacteria to humans, and play vital roles in DNA structure, chromatin organization, and gene expression. The length of STRs generally ranges from 2 to 6 base pairs.1 They are abundant in the human genome, appearing approximately every 10 000 base pairs on average. They constitute up to 75% of the human genome.2 The number of repeats at a specific STR site can vary between individuals, even between individuals of the same sex or between monozygotic twins. This diversity arises from random mutations taking place during DNA replication. STRs find significant applications in both forensic genetics and population genetics.3,4 They serve as invaluable markers for distinguishing individuals based on their unique DNA profiles, facilitating precise identification and aiding in criminal investigations.5
The study of a population's short tandem repeats (STRs) provides valuable forensic data for individual identification, determination of familial relationships, analysis of genetic diversity, and support for cold case investigations. This analytical process is fundamental within the realm of forensic genetics.
The Doukkala region (Fig. 1), which is the subject of this study, is situated to the south of the central plain on the Atlantic coast of Morocco, and covers an area of about 7–106 km2, with a population around 458 000 inhabitants.6 In Doukkala, previous studies extensively investigated GM immunoglobulin allotype, Y chromosome polymorphism, and class I and II HLA polymorphism.7–9 These studies have significantly contributed to our understanding of genetic diversity within the Doukkala population.
This research employs 15 autosomal STRs to perform genotyping on individuals from the Doukkala region who speak Arabic. The study aims to examine the Doukkala population's genetic structure, relationships with neighboring populations, and affiliations by analyzing allele frequencies and incorporating forensic parameters.
Materials and methodsSamples collection and DNA extractionThe sampling was conducted by the Higher Institute of Health Sciences, Hassan First University of Settat, Morocco. Informed consent was managed following ethical guidelines from the Biomedical Research Ethics Committee (CERBC) of Casablanca, Morocco.
Buccal swabs were collected using a sterile cotton swab from 134 randomly selected, healthy adult individuals, comprising 71 women and 63 men. All participants are native Arabic-speakers, hailing from families that have resided in the region for a minimum of 3 generations. It is worth noting that there are no blood ties among them. All participants were properly informed of their rights and granted written informed permission to participate in the study, which was carried out with the closest respect to ethical standards. DNA was extracted using the Chelex® 100 method.10 The concentration and quality of the DNA were measured using a NanoDrop TM 8000 spectrophotometer (Thermo Scientific) according to manufacturer's guidelines.
PCR amplification, capillary electrophoresis, and quality controlThe AmpFlSTR® Identifiler® kit (Applied Biosystems, Foster City, CA, USA) was employed for PCR co-amplification of 15 STR markers (CSF1PO, D13S317, D16S539, D18S51, D19S433, D21S11, D2S1338, D3S1358, D5S818, D7S820, D8S1179, FGA, TH01, TPOX, and vWA). According to the manufacturer's recommendations, 1 ng of DNA was amplified for each sample examined in a final reaction volume of 25 μl using a 2720 Thermal Cycler (Applied Biosystems, USA). Genotypes were assigned using the GeneMapper® ID-X v1.1 software by comparison with reference allelic ladders (Applied Biosystems). The ABI 3500 genetic analyzer was utilized to detect and separate the amplified products (Applied Bio-systems, Foster City, CA, USA). The laboratory's internal control standards and kit controls were adhered to during all experimental procedures.
Statistical analysisThe allele frequencies and statistical parameters such as the Polymorphic Information Content (PIC), Matching Probability (MP), Power of Discrimination (PD), Power of Exclusion (PE), and Typical Paternity Index (TPI) were evaluated by using STRAF online program v1.0.5.11 After applying the Bonferroni correction (0.05/15 = .0033), only P-values below .0033 were considered statistically significant for adjusted exact test of Hardy–Weinberg equilibrium.12
Pairwise uncorrected Fst distances were calculated between the 11 populations to perform a standard non-metric MDS analysis. The MDS plot was generated using Past v4.03 software13 to visualize the multivariate genetic data. To calculate these uncorrected genetic distance metrics (Fst), we utilized the Poptree2 software.14 Arlequin software v3.5.2.215 computed locus-by-locus Fst and corresponding P-values for 15 STRs, comparing Doukkala to reference populations.
Locus-by-locus allele frequency-based Analysis of Molecular Variance (AMOVA) was conducted using Arlequin v.3.5.2.2,15 exploring 3 distinct grouping approaches: Firstly, populations were grouped based on spoken language, resulting in Group 1 encompassing Arab-speaking populations (Arab-speakers of Doukkala, Arab-speakers of Rabat-Salé–Zemmour-Zaer, Arab-speakers of southern Morocco, Libya, Egypt), and Group 2 comprising Berber-speaking populations (Berber-speakers of Asni, Berber-speakers of Azrou, Berber-speakers of Béjaïa in Algeria). Secondly, populations were grouped by geographical distribution, yielding Group 1 representing Western North African populations (Arab-speakers of Doukkala, Arab-speakers of Rabat-Salé–Zemmour-Zaer, Berber-speakers of Asni, Berber-speakers of Azrou, Arab-speakers of southern Morocco, Berber-speakers of Béjaïa in Algeria), and Group 2 representing Eastern North African populations (Libya, Egypt).
Comparative population dataIn this study, we included 10 populations with previously published allelic frequencies to compare them with the population of Doukkala (DK). These reference populations encompassed a diverse range of regions and genetic backgrounds, including: North African population: Arabic-speaking population of Rabat-Salé–Zemmour-Zaer (RZ),16 Berber-speaking from Asni (AS),4 Berber-speaking population of Azrou (AZ),17 Arabic-Speaking population in Southern Morocco (SO),3 Berber population of Bejaia (DZ),18 Libyan Arabs (LY),19 Arab Egyptians (EG);20 and Sub-Saharan African population: Uganda (UG),21 Namibia (NB),22 and Tanzania (TZ).23
Ethical approvalThis study were approved by the Biomedical Research Ethics Committee (CERBC) of Casablanca, Morocco. The Ethics Committee is based on the Declaration of Helsinki 2008. Registration number: 06/19.
Results and discussionAllele frequency distribution and forensic parametersA total of 180 alleles were observed in the Arabic-speaking population of Doukkala in Morocco (Supplementary Table 1). Three of the 15 studied markers (FGA, TH01, and TPOX) present a deviation from Hardy–Weinberg equilibrium after Bonferroni correction (P < .05/15 = .0033). Multiple factors can contribute to a deviation from Hardy–Weinberg equilibrium (HWE), which encompasses aspects such as population size, consanguinity, population stratification, migration, genetic mutation, and the impact of natural selection.24 Approximately, a quarter of all matrimonial alliances in the Doukkala population were comprised of consanguineous unions, accounting for 26.56% of the total.25
With 19 alleles, D18S51 exhibited the most pronounced autosomal STR polymorphism. The alleles 16, 14, 12, and 17 were the most prevalent, representing 14.6%, 13.8%, 13.1%, and 12.3%, respectively. On the other hand, alleles 14.2, 15.2, 16.2, 22, and 26 were the rarest, each accounting for a mere 0.4%. D13S317, D8S1179, and TPOX exhibited the least polymorphism, with 8 alleles each. In contrast, D13S317, D8S1179, and TPOX demonstrated the lowest level of polymorphism, each showcasing only 8 alleles.
Supplementary Table 1 provides a comprehensive overview of the statistical parameters associated with the 15 markers that were examined. The study population exhibited a spectrum of heterozygosity (Obs.Het), with values ranging from 0.694 to 0.851. Heterozygosity values are at their lowest point, respectively, at the TPOX (Obs.Het = 0.694) and D5S818 (Obs.Het = 0.709) loci, this includes the alleles that have the highest frequencies (0.399 for allele 8 and 0.444 for allele 12, respectively). This clarifies the lowest levels of power of exclusion (PE = 0.419 for TPOX and PE = 0.442 for D5S818) and typical paternity index (TPI = 1.634 for TPOX and TPI = 1.718 for D5S818). The highest heterozygosity value is observed at the loci, D21S11 (Obs.Het = 0.851) and D2S1338 (Obs.Het = 0.851) which has the highest values of power of exclusion (PE = 0.696) and of typical paternity index (TPI = 3.35), respectively. The examined 15 STRs reveal a significant level of polymorphism, displaying a high powers of discrimination (PD > 0.870) and exclusion (PE > 0.418). The combined power of discrimination (CPD) demonstrates an exceptionally high precision, measuring 0.99999999999999999526. Similarly, the combined power of exclusion (CPE) value is also quite high, reaching 0.99999664790900144592. These findings provide strong evidence for the significance of these 15 genetic markers in forensic cases involving the Doukkala population. These loci hold considerable significance in applications such as individual identification and the determination of paternity.
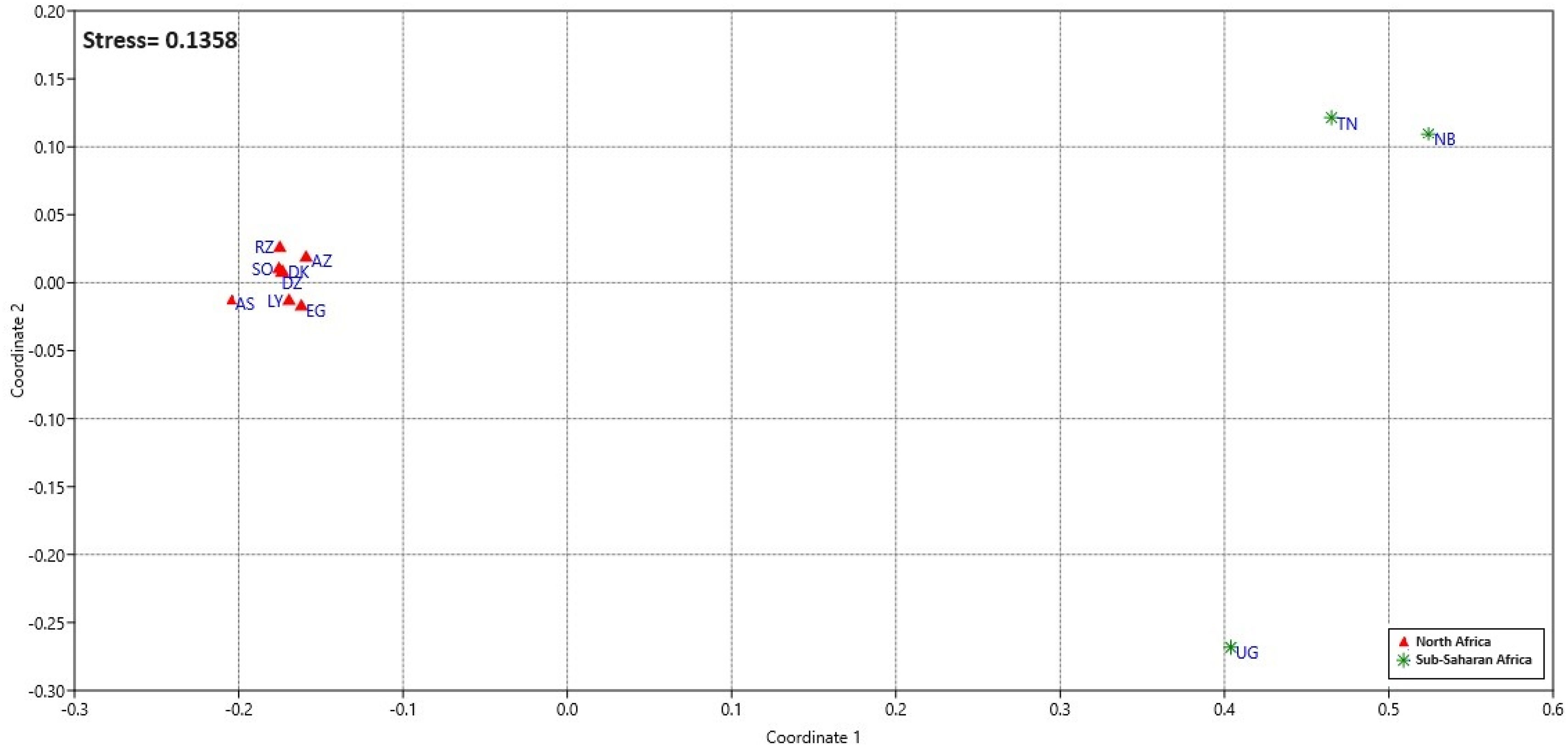
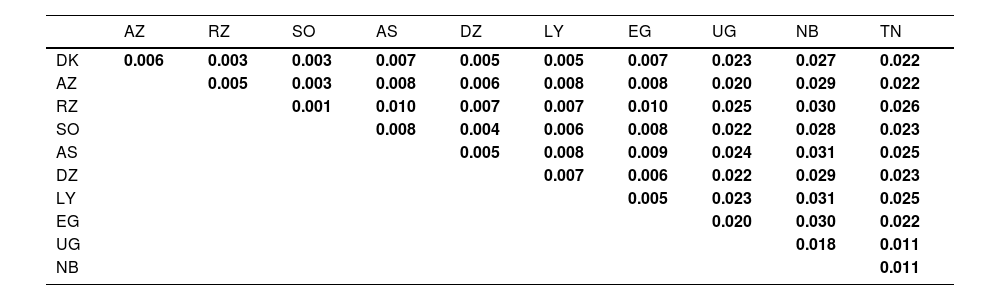
Interpopulation comparisons: The MDS plot, Locus Fst, and AMOVA analysisThe Fst distances (Table 1) reveal that Arabic-speaking population of Doukkala is closest to Arabic-speaking populations of Rabat-Salé–Zemmour-Zaer (RZ) and Southern population (SO) from Morocco, both having an Fst value of 0.003. On the other hand, the most distant populations from North Africa are Berber-speaking from Asni (AS) from Morocco and Egypt population (EG), both having an Fst value of 0.007. Furthermore, the furthest population from the Arabic-speaking population of Doukkala within Africa is Namibian population (NB), with an Fst value of 0.027, representing a Sub-Saharan population.
The Fst genetic distances among the populations under study.
| AZ | RZ | SO | AS | DZ | LY | EG | UG | NB | TN | |
|---|---|---|---|---|---|---|---|---|---|---|
| DK | 0.006 | 0.003 | 0.003 | 0.007 | 0.005 | 0.005 | 0.007 | 0.023 | 0.027 | 0.022 |
| AZ | 0.005 | 0.003 | 0.008 | 0.006 | 0.008 | 0.008 | 0.020 | 0.029 | 0.022 | |
| RZ | 0.001 | 0.010 | 0.007 | 0.007 | 0.010 | 0.025 | 0.030 | 0.026 | ||
| SO | 0.008 | 0.004 | 0.006 | 0.008 | 0.022 | 0.028 | 0.023 | |||
| AS | 0.005 | 0.008 | 0.009 | 0.024 | 0.031 | 0.025 | ||||
| DZ | 0.007 | 0.006 | 0.022 | 0.029 | 0.023 | |||||
| LY | 0.005 | 0.023 | 0.031 | 0.025 | ||||||
| EG | 0.020 | 0.030 | 0.022 | |||||||
| UG | 0.018 | 0.011 | ||||||||
| NB | 0.011 |
Populations studied: DK: Arabic-speaking from Doukkala (Present study); AZ: Berber-speaking from Azrou; RZ: Arabic-speaking from Rabat-Salé–Zemmour-Zaer; SO: Arabic-speaking from southern Morocco; AS: Berber-speaking from Asni; DZ: Berber-speaking from Béjaïa in Algeria; LY: Libya; EG: Egypt; UG: Uganda; NB: Namibia; TZ: Tanzania.
In order to gain a thorough understanding of the genetic variations between populations, standard non-metric MDS analysis was conducted based on the pairwise genetic distances provided above.
The non-metric Multidimensional scaling (MDS) plot (Fig. 2) based on Fst distances succinctly groups North African populations: Arabic-speaking from Doukkala (DK), AZ: Berber-speaking from Azrou (AZ), RZ: Arabic-speaking from Rabat-Salé–Zemmour-Zaer (RZ), Arabic-speaking from southern Morocco (SO), Berber-speaking from Asni (AS), Berber-speaking from Béjaïa in Algeria (DZ), Libya (LY), Egypt (EG); revealing genetic affinities within this region. Uganda's population (UG) distinguishes itself from other sub-Saharan groups: Namibia (NB), Tanzania (TZ); with possible factors including historical genetic divergence, geographical isolation, population migrations, or natural selection.26
Non-metric MDS visualization of interpopulation relationships using uncorrected Fst distances (11 Populations).
Populations studied: DK: Arabic-speaking from Doukkala (Present study); AZ: Berber-speaking from Azrou; RZ: Arabic-speaking from Rabat-Salé–Zemmour-Zaer; SO: Arabic-speaking from southern Morocco; AS: Berber-speaking from Asni; DZ: Berber-speaking from Béjaïa in Algeria; LY: Libya; EG: Egypt; UG: Uganda; NB: Namibia; TZ: Tanzania.
To gain deeper insights into the genetic relationships, a detailed analysis was conducted by comparing the Arabic-speaking population of Doukkala with African populations, enabling a locus-by-locus comparison. The Fst values and corresponding P-values (Supplementary Table 2) provide additional validation for the genetic affinities that Doukkala shares with the North African populations, as confirmed by standard non-metric MDS analysis. In Morocco, no significant differences in the 15 genetic systems studied were observed between the Arab-speaking population of Doukkala and the other Moroccan populations, whether they are Arab- or Berber-speakers (Arab-speakers of Rabat-Salé–Zemmour-Zaer, Berber-speakers of Asni, Berber-speakers of Azrou, and Arab-speakers of Southern Morocco). The lack of genetic variation may be attributed to the genetic structure of the overall population in North Africa. Prior research by Flores et al. analyzing the Y chromosome polymorphism, reveals genetic convergence between Arabic- and Berber-speakers in Morocco, despite distinct cultural and linguistic backgrounds.8 In addition, other studies examining North African populations have consistently revealed the absence of genetic disparities between Arabs and Berbers when scrutinizing individual genetic markers.27–30
Based on the FST comparison test (Supplementary Table 2), no significant difference was observed between the Arabic-speaking population of Doukkala and all North African populations at all loci, except for the Egyptian population (North-East Africa), where a difference was observed at 2 specific loci (CSF1PO and TH01). The findings support AbdEl-Hafez et al.20 study on Egyptian autosomal STRs. They reveal significant differences in 12 loci between Egyptian and Moroccan population.20 Fadhlaoui-Zid et al. work highlights the existence of historical and ongoing gene flow from Arabian Peninsula and Egypt with Eurasians, rather than with North-West Africans,28 confirming divergence from the Egyptian population.
The Moroccan population of Doukkala shows a significant difference from Sub-Saharan African populations (Uganda: CSF1PO, D2S1338, D8S1179, D18S51, D21S11, FGA, TH01, TPOX, and vWA; Namibia: D2S1338, D3S1358, D5S818, D7S820, D8S1179, D13S317, D16S539, D18S51, D19S433, D21S11, FGA, and TH01; Tanzania: CSF1PO, D2S1338, D3S1358, D7S820, D8S1179, D13S317, D16S539, D18S51, D21S11, FGA, TH01, and TPOX). Our results corroborate and align with several previous studies31 that explored the divergence between the Doukkala population and the Sub-Saharan population using autosomal RFLP markers. Similarly, the study by González-Pérez et al. based on genetic markers such as Alu polymorphisms and STRs confirmed the presence of significant differences between the North-West African population and the Sub-Saharan African population.32 The genetic differentiation observed between the Doukkala population and the Sub-Saharan population indicates that the Sahara desert acted as a stronger barrier to gene flow.31
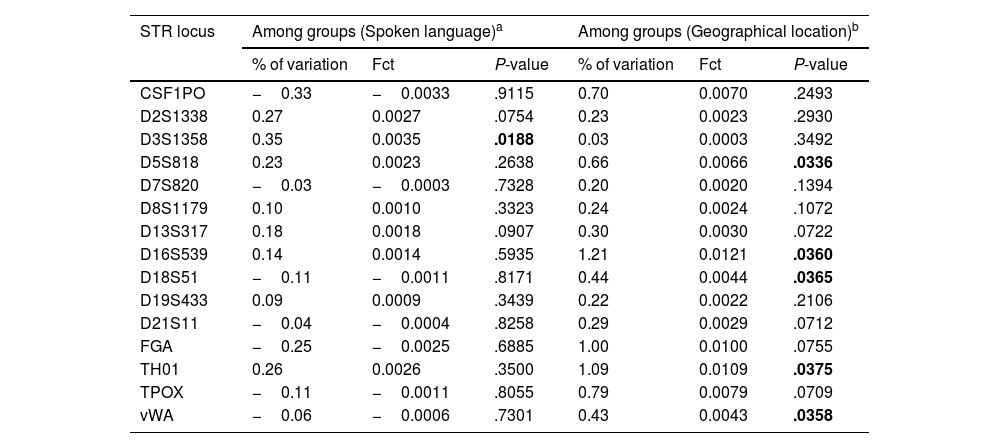
In order to better understand the structure and relationships between Arabic- and Berber-speaking groups in North Africa, an AMOVA analysis was used (Table 2). The first analysis focused on language spoken, while the second focused on geographical location. AMOVA results indicate significant genetic structuring within North African populations. Analysis based on spoken language revealed that only one autosomal STR marker (D3S1358) has a P-value <.05. Fct values ranged from −0.0033 to 0.0035 and percentages of variation from 0.03 to 0.35%. In addition, AMOVA based on geographic location showed 5 significant autosomal STR markers (D5S818, D16S539, D18S51, TH01, and vWA). Fct values ranging from 0.0003 to 0.0121 and percentages of variation from 0.03% to 1.21%, indicating genetic variation between populations in Morocco (Arab-speakers of Doukkala, Arab-speakers of Rabat-Salé–Zemmour-Zaer, Berber-speakers of Asni, Berber-speakers of Azrou, Arab-speakers of southern Morocco), Algeria (Berber-speakers of Béjaïa), Libya, and Egypt. These results highlight the influence of language and geography on genetic diversity in this region.
Locus-by-locus AMOVA comparing North African populations by spoken language and geographic location. Significant P values, indicated in bold.
| STR locus | Among groups (Spoken language)a | Among groups (Geographical location)b | ||||
|---|---|---|---|---|---|---|
| % of variation | Fct | P-value | % of variation | Fct | P-value | |
| CSF1PO | −0.33 | −0.0033 | .9115 | 0.70 | 0.0070 | .2493 |
| D2S1338 | 0.27 | 0.0027 | .0754 | 0.23 | 0.0023 | .2930 |
| D3S1358 | 0.35 | 0.0035 | .0188 | 0.03 | 0.0003 | .3492 |
| D5S818 | 0.23 | 0.0023 | .2638 | 0.66 | 0.0066 | .0336 |
| D7S820 | −0.03 | −0.0003 | .7328 | 0.20 | 0.0020 | .1394 |
| D8S1179 | 0.10 | 0.0010 | .3323 | 0.24 | 0.0024 | .1072 |
| D13S317 | 0.18 | 0.0018 | .0907 | 0.30 | 0.0030 | .0722 |
| D16S539 | 0.14 | 0.0014 | .5935 | 1.21 | 0.0121 | .0360 |
| D18S51 | −0.11 | −0.0011 | .8171 | 0.44 | 0.0044 | .0365 |
| D19S433 | 0.09 | 0.0009 | .3439 | 0.22 | 0.0022 | .2106 |
| D21S11 | −0.04 | −0.0004 | .8258 | 0.29 | 0.0029 | .0712 |
| FGA | −0.25 | −0.0025 | .6885 | 1.00 | 0.0100 | .0755 |
| TH01 | 0.26 | 0.0026 | .3500 | 1.09 | 0.0109 | .0375 |
| TPOX | −0.11 | −0.0011 | .8055 | 0.79 | 0.0079 | .0709 |
| vWA | −0.06 | −0.0006 | .7301 | 0.43 | 0.0043 | .0358 |
In essence, the 15 STRs have been effectively employed to analyze the genetic structure of the Arabic-speaking population of Doukkala, providing valuable insights into its genetic relationship with African populations. Our findings unveiled a clear genetic affinity between the Doukkala population and North-West African populations, thus supporting the historical connections between these groups. Indeed, the 15 STRs loci exhibited a high level of informativeness, thereby establishing their efficacy in applications such as forensics, anthropology, and population genetic studies. To acquire a more comprehensive overview of Doukkala's genetic landscape, subsequent studies should consider the inclusion of supplementary genetic markers and the enlargement of the sample size.
Author contributionsAll authors contributed to the study conception and design. Material preparation, data collection, and analysis were performed by AE, KC, and ND. HE helped with the experimental work. The first draft of the manuscript was written by AE and all authors commented on previous versions of the manuscript. All authors read and approved the final manuscript. JT, AH, and HE created the initial concept for this study and guided through each step of the process.
Funding statementThis research did not receive any specific grant from funding agencies in the public, commercial, or not-for-profit sectors.