A prospective study was designed to assess the performance of the new CT OligoGen kit and the cobas 4800 assay for detection of Chlamydia trachomatis.
MethodsA set of samples that included urine samples (n=212), endocervical (n=167), rectal (n=53), pharyngeal (n=7) and urethral swabs (n=3). The samples were sent from a regional sexually transmitted diseases (STD) clinic in Seville, Spain, and were collected from 261 men and 181 women. Discordant results were re-analyzed and clinical data and other tests were reviewed in order to resolve them.
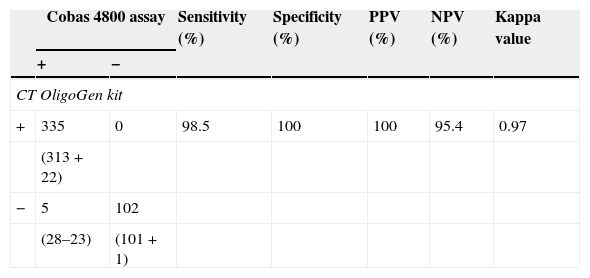
ResultsSensitivity, specificity, positive predicative value (PPV), negative predictive value (NPV) and kappa value for C. trachomatis detection using the CT OligoGen kit were 98.5%, 100%, 100%, 95.4% and 0.97, respectively.
ConclusionsThis new kit had a high sensitivity, specificity, PPV and NPV for C. trachomatis, therefore the performance profile confirms the usefulness and reliable results of this new assay.
Se diseñó un estudio prospectivo para evaluar las características del nuevo kit CT OligoGen en comparación con el test cobas 4800 para la detección de Chlamydia trachomatis.
MétodosSe analizaron una serie de muestras que incluían orinas (n=212), exudados endocervicales (n=167), rectales (n=53), faríngeos (n=7) y uretrales (n=3). Estas muestras provenían de un centro de infecciones de transmisión sexual (Sevilla) y pertenecían a 261 hombres y 181 mujeres. Los resultados discordantes se reanalizaron y revisaron historias clínicas y otras pruebas para resolverlas.
ResultadosLos valores de sensibilidad, especificidad, valor predictivo positivo (VPP) y negativo (VPN) y valor kappa para el kit CT OligoGen fue 98,5%, 100%, 100%, 95,4% and 0,97, respectivamente.
ConclusionesEste nuevo kit tuvo una alta sensibilidad, especificidad, VPP y VPN para la detección de C. trachomatis, por lo que esta evaluación confirma su utilidad y fiabilidad.
Chlamydia trachomatis is the most common bacterial sexually transmitted infection. According to the World Health Organization (WHO), 101 million chlamydial infections are detected annually worldwide, and women carry the major burden of the disease.1 Several complications because of undetected genital infections may evolve to ectopic pregnancy, pelvic inflammatory disease and infertility in female patients2 and, as other STIs, can also facilitate the transmission of human immunodeficiency virus infection3 or as a cofactor for endocervical cancer progress among females coinfected with the human papillomavirus.4 In males, epididymitis, urethritis, and prostatitis are among the main diseases.5–8
Nowadays, we can find several methods for C. trachomatis detection including culture method, serology, ELISA, direct fluorescence assay and nucleic acid amplification tests (NAATs). Polymerase chain reaction (PCR) assays are more sensitive and specific compared to other diagnostic methods available8 and allow the use of non-invasive procedures for detection of asymptomatic C. trachomatis infection. Urine specimens and urogenital, anorectal or oropharyngeal swabs can be used for reliable STI detection with cost-saving test strategies.9,10 One of them is the cobas 4800 CT/NG assay (Roche Diagnostics, Mannheim, Germany) for testing a substantial number of samples simultaneously. For a few samples, retesting results or cheaper alternatives, other assays, as the new CT OligoGen kit (Operon-Inmuno & Molecular Diagnostics, Zaragoza, Spain), can be used.
PCR assays, sometimes, can be an unavailable method because of their high cost or the equipment being under routine use in developing countries, so there is an urgent need for the development of a rapid, highly sensitive and cost-effective detection method. In the present study, we have evaluated a pre-commercial kit for detection of C. trachomatis against the diagnostic method currently used in our hospital (cobas 4800) in order to value its specificity and sensibility with several types of obtained samples from a STI clinic.
Material and methodsClinical specimens includedA total of 4305 clinical samples, submitted for C. trachomatis and Neisseria gonorrhoeae detection, were sent from a regional sexually transmitted infection (STI) clinic from Seville (Spain) to our laboratory. These patients were enrolled in this study following institutional ethical committee clearance and informed oral consent. The specimens were collected from December 2012 to December 2013. These samples were collected in cobas PCR media Urine Sample kit or cobas PCR media Female swab sample kit (Roche Diagnostics GmbH, Mannheim, Germany) according manufacturer's instructions and analyzed by cobas 4800 assay. Original samples were stored at room temperature for up to 3 days before being tested by cobas 4800 assay and at −80°C for up to 30 days before being analyzed by CT OligoGen kit following manufacturer's instructions. After obtaining testing results with cobas 4800 assay and reviewing clinical data, a total of 442 clinical specimens were selected for CT OligoGen kit evaluation: 102 (23.1%) negative for both chlamydia and gonococcus, 329 (74.4%) positive for chlamydia, and 11 (2.5%) positive samples for both chlamydia and gonococcus.
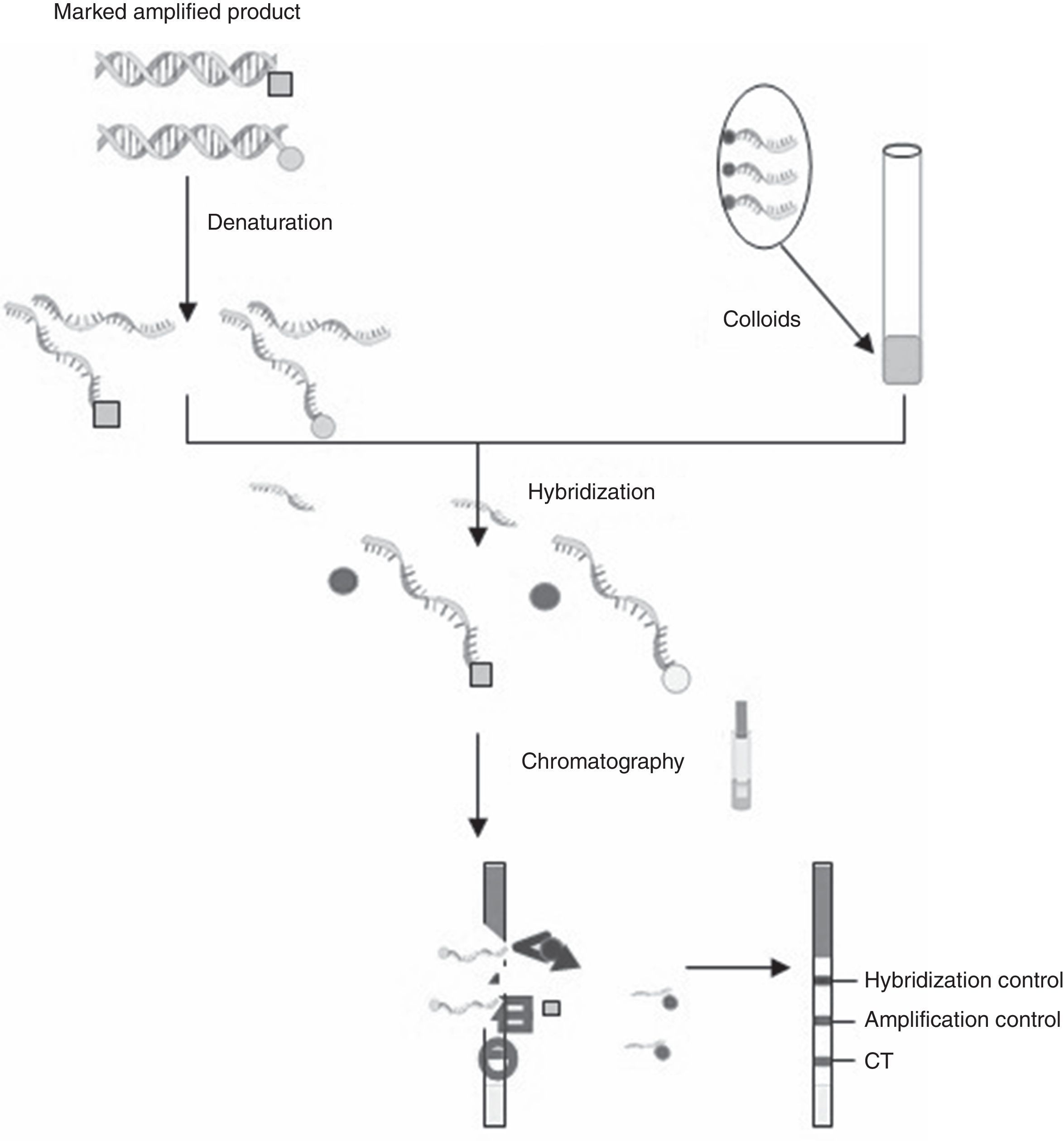
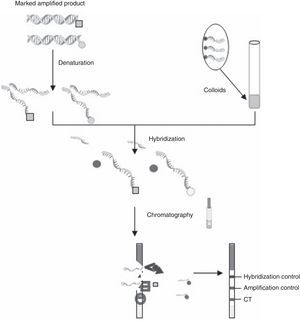
C. trachomatis testing, samples selection and DNA isolationC. trachomatis and N. gonorrhoeae DNA isolation was performed by cobas X 480 (Roche Diagnostics, Mannheim, Germany). Original specimens and DNA were stored at −80°C up to the analysis with CT OligoGen kit. Two qualified technicians performed the tests and were blinded for the results. Briefly, CT OligoGen test is able to detect and identify infections caused by CT, with high sensitivity and specificity, by means of PCR amplification and subsequent detection by oligo-immunochromatography. This is done by amplifying two independent CT DNA regions, one in the cryptic plasmid and the other in the CT genome (omp gene). As an internal PCR control, a human control gene (GAPDH) is also amplified. Amplification was performed in a final reaction volume of 50μl containing 38μl of master mix, 5μl of primer mix and 2μl of Taq DNA polymerase. Amplification conditions consisted of initial polymerase activation at 96°C for 5min, 40 cycles of 96°C for 60s, 59°C for 60s and 72°C for 60s and a final elongation step at 72°C for 5min. A negative water control is included in each run. During this PCR, different labels are added to the DNA amplified fragments. Amplification products are detected by oligo-immunochromatography. Firstly, denatured PCR fragments are recognized by specific probes covalently bounded to latex colored particles (colloids) (hybridization process). Secondly, these DNA+colloid complexes migrate through a membrane (immunochromatography) and are captured by specific antibodies that recognize the labels added during the PCR, resulting in the appearance of a colored bands pattern (Fig. 1). This kit allows the detection and identification of infections caused by different serotypes of CT with high sensitivity and specificity.
Discordant results reanalysisSamples identified as C. trachomatis positive or C. trachomatis negative with both CT OligoGen kit and the cobas 4800 assay were defined as true positives and true negatives, respectively. Discordant results were retested as follows:-Retest the discordant DNA samples with both CT OligoGen kit and cobas 4800 assay to confirm previous result and/or dismiss DNA degradation.-New DNA isolation from discordant samples to dismiss DNA degradation. For urine specimens, Urine Bacterial Concentration and DNA Isolation Kit (Norgen Biotek Corp, Ontario, Canada) were used. For swab samples QIAamp DNA mini kit (Qiagen GmbH, Hilden, Germany) was used. These DNA samples were stored at −70°C to be retested with CT OligoGen kit.
Statistical analysisWe used the cobas 4800 results as reference assay to calculate the sensitivity, specificity, positive predictive value (PPV) and the negative predictive value (NPV) that were calculated with 95% confidence intervals (CI) to test the significance of the estimates. Agreement between the 2 assays was determined by unweighted kappa (κ) statistics and 95% CIs. The measure of agreement was generally interpreted as follows: less than 0.20, poor agreement; 0.21–0.40, fair agreement; 0.41–0.60, moderate agreement; 0.61–0.80, good agreement; and more than 0.80, excellent agreement. All statistical analyses were performed with the SPSS Statistics program v. 20.0 (IBM Ibérica, Madrid, Spain).
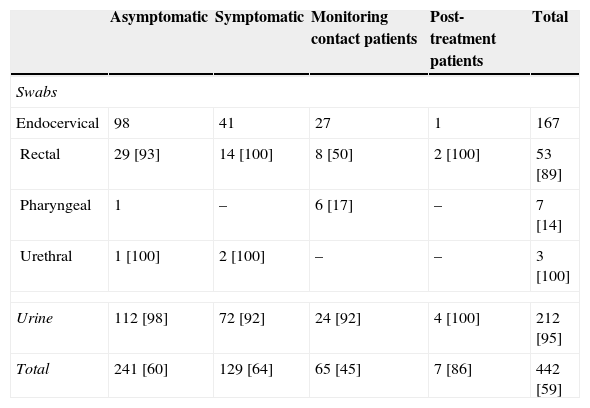
ResultsSamples included in this study comprised 212 (48.0%) urine specimens, 167 (37.8%) endocervical, 53 (12.0%) rectal, 7 (1.6%) pharyngeal and 3 (0.7%) urethral swabs. These patients were mainly men (59.0%) with a median age of 28 years [Q1–Q3: 23.8–33.0]. A summary of the specimens is included in Table 1.
Sample types and patient groups analyzed in this study. In brackets, percentage of men from samples.
| Asymptomatic | Symptomatic | Monitoring contact patients | Post-treatment patients | Total | |
|---|---|---|---|---|---|
| Swabs | |||||
| Endocervical | 98 | 41 | 27 | 1 | 167 |
| Rectal | 29 [93] | 14 [100] | 8 [50] | 2 [100] | 53 [89] |
| Pharyngeal | 1 | – | 6 [17] | – | 7 [14] |
| Urethral | 1 [100] | 2 [100] | – | – | 3 [100] |
| Urine | 112 [98] | 72 [92] | 24 [92] | 4 [100] | 212 [95] |
| Total | 241 [60] | 129 [64] | 65 [45] | 7 [86] | 442 [59] |
Of the 442 clinical specimens evaluated, 313 (70.8%) provided positive results by both assays (147 urine specimens, 126 endocervical, 34 rectal, 4 pharyngeal and 2 urethral swabs from 182 men and 130 women; cobas 4800 cycle threshold values ranging from 23.2 to 40.9 cycles, median 33.2 cycles) and 101 specimens provided negative results in both CT OligoGen and cobas 4800 assays. Twenty-eight samples (13 urine specimens, 8 endocervical, 5 rectal, 1 pharyngeal and 1 urethral swab) provided positive results by cobas 4800 but were negative by CT OligoGen. We did not find positive results by CT OligoGen and negative results by cobas 4800.
Discordant results were retested with the following results, after a new DNA extraction, 22 specimens were considered as positive results by cobas 4800 and CT OligoGen assays. This fact confirmed DNA degradation of the original sample. One sample was considered as negative result by both assays because the retesting provided negative results after retesting original sample and a new DNA extraction from original sample. Clinical data supported this result.
Finally, 5 specimens (2 urine specimens, 2 endocervical and 1 rectal swab) were considered as false-negative results by CT OligoGen. The retest with cobas 4800 assay provided again a positive result meanwhile a new DNA extraction from original sample provided a negative result by CT OligoGen kit. Clinical data of these patients supported cobas 4800 results (2 symptomatic and 3 monitoring contact patients with Chlamydia suspicion). Statistical data obtained for the cobas 4800 and CT OligoGen assays are provided in Table 2.
DiscussionRecent studies in Spain showed a C. trachomatis prevalence of 4.1% in global population11–12; meanwhile in a STI center like ours, we could find a major prevalence (10.2–10.5%, internal data from 2013 and 2014), higher data than another recently published study (8.1–8.5% in a STI in The Netherlands).13
The choice of a suitable method for detecting C. trachomatis could depend on various factors such as: hands-on time during specimen handling and processing, reagent preparation, number and type of samples to process them and time to test them. Some laboratories cannot get an automatic or semi-automatic system to detect this organism because of pricing or the limited number of samples to test, so alternative methods have to be considered.
This comparison between assays achieved an excellent agreement including all kinds of samples between both assays, with a concordance value of 98.9% (437/442) if we considered analyses altogether with every sample tested. The results showed high specificity, sensitivity, PPV and NPV for this test.
One major problem that we found during this study was a relative high number of discordant results, due to DNA quality from original samples. After retesting these discordant samples, the majority of them were resolved by a new DNA extraction from original samples. Data suggested that samples with a low DNA amount obtained from cobas X 480 could not be well amplified with CT OligoGen kit or DNA degradation from original samples. This matter can be easily resolved by a new DNA extraction with a manual extraction kit, so other commercial DNA extraction kits can be used without problems.
Another limitation of the present study was the limited number of pharyngeal and urethral swabs included and tested. Only a few studies about sensitivity and specificity with these samples in cobas 4800 assay have been published, especially with rectal swabs.14 Although more specimens should be tested to confirm this data, sensibility and specificity of CT OligoGen kit would not be compromised and reliable results are expected.
In this comparison, we could confirm that the new CT OligoGen kit is a suitable technology to detect and identify C. trachomatis nucleic acid in swab and urine specimens as well. This assay enables the analysis of an unlimited number of samples in 3.5–5h by a single staff.
Conflict of interestNone declared.
Part of this study has been funded by Operon-Inmuno & Molecular Diagnostics. CT OligoGen kits were supplied by Operon-Inmuno & Molecular Diagnostics. The authors thank Mr. Parma for the proofreading and translation he did for this work.