Coagulase-negative staphylococci (CNS) are a common cause of bovine subclinical mastitis (SCM). The prevalence of CNS species causing SCM identified by genotyping varies among countries. Overall, the antimicrobial resistance in this group of organisms is increasing worldwide; however, little information exists about a CNS species resistant to antibiotics. The aim of the present study was to genotypically characterize CNS at species level and to determine the prevalence and antibiotic resistance profiles of CNS species isolated from bovine SCM in 51 dairy herds located in the central region of the province of Cordoba, Argentina. In this study, we identified 219 CNS isolates at species level by PCR-restriction fragment length polymorphism of the groEL gene. Staphylococcus chromogenes (46.6%) and Staphylococcus haemolyticus (32%) were the most prevalent species. A minimum of three different CNS species were present in 41.2% of the herds. S. chromogenes was isolated from most of the herds (86.3%), whereas S. haemolyticus was isolated from 66.7% of them. The broth microdilution method was used to test in vitro antimicrobial susceptibility. Resistance to a single compound or two related compounds was expressed in 43.8% of the isolates. S. chromogenes and S. haemolyticus showed a very high proportion of isolates resistant to penicillin. Resistance to two or more non-related antimicrobials was found in 30.6% of all CNS. S. haemolyticus exhibited a higher frequency of resistance to two or more non-related antimicrobials than S. chromogenes.
Los estafilococos coagulasa negativos (ECN) son una causa frecuente de mastitis subclínica (MSC) en bovinos. La prevalencia de especies de ECN causantes de MSC identificadas por métodos genotípicos varía entre países. La resistencia antimicrobiana en este grupo de organismos se está incrementando en el mundo; sin embargo, existe poca información acerca de las especies de ECN resistentes a antibióticos. Los objetivos del presente estudio fueron caracterizar genotípicamente los ECN a nivel de especie y determinar la prevalencia y los perfiles de resistencia a antibióticos de las especies de ECN aisladas de MSC en bovinos de 51 rodeos situados en la provincia de Córdoba, Argentina. Mediante polimorfismos de los fragmentos de restricción del gen groEL identificamos 219 aislamientos de ECN a nivel de especie. Staphylococcus chromogenes (46,6%) y Staphylococcus haemolyticus (32%) fueron las especies más prevalentes. Un mínimo de 3 especies diferentes de ECN estuvieron presentes en el 41,2% de los tambos. S. chromogenes fue aislado en la mayoría de los tambos (86,3%), mientras que S. haemolyticus fue aislado en el 66,7% de aquellos. Para el análisis de sensibilidad a los antimicrobianos in vitro se usó el método de microdilución en caldo. La resistencia a un único compuesto o a 2 compuestos relacionados fue expresada en el 43,8% de los aislamientos. S. chromogenes y S. haemolyticus mostraron una muy elevada proporción de aislamientos resistentes a penicilina. La resistencia a 2 o más antimicrobianos no relacionados fue hallada en el 30,6% de los ECN. S. haemolyticus exhibió una frecuencia de resistencia a 2 o más antimicrobianos no relacionados más elevada que S. chromogenes.
Bovine mastitis is one of the most costly and complex diseases of the dairy industry3,9. The complexity is reflected in the numerous causative pathogens, the variety and magnitude of the physiological responses to these pathogens and the variation in efficacy of control measures for different causative organisms4,7,17,18,23,32. Coagulase-negative staphylococci (CNS) have been traditionally considered minor pathogens. However, their importance has increased because they have become the most frequently isolated group of species from bovine milk in many areas around the world16,29,33. CNS usually cause subclinical mastitis (SCM), resulting in an increase in the somatic cell count (SCC) and a decrease in milk quality with the resulting economic losses that this implies30. The understanding and control of CNS mastitis are complicated by the heterogeneity of this group of bacteria. So far, based on Jean Euzéby's List of Procaryotic names with Standing in Nomenclature (http://www.bacterio.net/s/staphylococcus.html), the genus Staphylococcus contains 47 species and 24 subspecies. Even though, the prevalence of CNS species varies among the studies; six species have been reported to be commonly involved in intramammary infections (IMI)20,30. Recently, it has been found that CNS speciation based on the biochemical profile is not accurate enough for bovine CNS identification26. Hence, molecular methods have become an important diagnostic tool to improve CNS species differentiation5,15,39. CNS tend to be more resistant to antibiotics than Staphylococcus aureus and easily develop multiresistance2,27,32. In addition, limited information is available regarding differences in antimicrobial susceptibility among CNS species identified by genotyping27,38. As yet, nothing has been reported about the prevalence and susceptibility antibiotics of CNS species of dairy cows in Argentina. Recently, a research conducted in our laboratory8 showed that CNS represent the most prevalent bacterial group of minor pathogens isolated from the subclinical infection of dairy cows in Córdoba province, Argentina. The aim of the present study was to determine the prevalence and antibiotic resistance profiles of CNS species isolated from bovine subclinical mastitis in 51 dairy herds in the central region of Argentina.
Materials and methodsSpecies identification of CNS isolates by PCR-RFLP analysis of a groEL gene sequenceIn this study, we included 219 (49.6%) CNS randomly selected isolates from a total of 441 isolates in pure cultures from composite milk samples with a SCC ≥200000cells/ml in a previous study8. The isolates were obtained from a previous cross-sectional study conducted on a sample randomly selected from 51 dairy farms located in Córdoba province, Argentina, with a herd size of around 100 and 250 cows37. The intramammary therapy of clinical cases was administered in 96.1% of the herds. Antibiotics used for dry cow therapy were applied to all cows in 58.8% of herds for at least one year. Among the milking hygienic practices, udder cleaning before milking was implemented in 94.1% of herds, whereas other practices such as using individual paper towels and, pre- and post-test dipping were not or were less frequently applied. On average, 45 lactating cows in each herd were sampled by the same investigator. In total, 2296 composite milk samples were collected. A volume of 0.01ml of milk was streaked on trypticase soy agar plates (Sigma–Aldrich, USA) containing 5% sheep blood and incubated at 37°C; plates were examined for growth at 24 and 48h. A mammary gland was considered infected by CNS when growth of ≥500CFU/ml of a particular organism and only one colony type on the plate were isolated8. Genomic DNA was extracted from all isolates and all type strains according to Aires-de-Sousa et al.1. PCR-RFLP analysis of a partial groEL gene sequence was performed as described by Santos et al.25. The CNS isolates were genotyped at species level as follows: (1) the presence of a DNA fragment (approximately 550bp) obtained by PCR of the partial groEL gene sequence was identified, (2) the banding patterns obtained after digestion by AluI restriction enzyme of the groEL gene of type strains were determined, (3) these patterns were compared with the known sequences from the NCBI GenBank database, using the NEB cutter program (version 2.0)36, (4) those CNS isolates with the same pattern of reference or type-T strains were assigned to a specific species. To differentiate S. chromogenes from S. hyicus and S. capitis, PCR products amplified from reference and type-T strains and/or IMI isolates were digested with HindIII and PvuII25. The following reference and type-T strains were included in this study: S. aureus ATCC 29740, S. capitis subsp. capitis ATCC 35661, S. caprae ATCC 35538T, S. chromogenes ATCC 43764T, S. epidermidis ATCC 12228, S. haemolyticus ATCC 29970T, S. hyicus ATCC 11249T, S. saprophyticus ATCC 49453, S. sciuri subsp. sciuri ATCC 29060, S. sciuri subsp. carnaticus ATCC700058T, S. simulans ATCC 11631, S. warneri ATCC 49454, S. xylosus ATCC 29971T.
Antibiotic susceptibility testingThe broth microdilution method was used to carry out in vitro antimicrobial susceptibility testing according to Clinical Laboratory Standards Institute guidelines (CLSI)6 document M31-A3. Custom-made microtiter plate panels were used (Trek Diagnostic Systems, Magellan Biosciences, UK-USA) (Sensititre-TREK). The antimicrobial agents and dilution ranges tested for each agent were as follows: penicillin (0.06–8μg/ml), oxacillin (0.25–4μg/ml), erythromycin (0.25–4μg/ml), tetracycline (2–16μg/ml) and clindamycin (0.5–2μg/ml). S. aureus strain ATCC 29213 was included in each assay as a control. The minimum inhibitory concentrations (MICs) results were evaluated based on veterinary interpretive criteria of the CLSI6, with resistance breakpoints of 0.25μg/ml for penicillin, 0.5μg/ml for oxacillin, 1μg/ml for erythromycin and 8μg/ml for tetracycline. Clindamycin is not licensed for the treatment of bovine mastitis; therefore, no approved breakpoints are available for the classification of bovine staphylococcal isolates from mastitis cases as susceptible or resistant. The MIC value required to inhibit 90% of the isolates tested was defined as MIC90. Isolates were categorized as susceptible or resistant, with intermediates classified as resistant. Isolates with a MIC for oxacillin >0.5μg/ml were additionally examined for the presence of the mecA gene by PCR as proposed by Mo and Wang14.
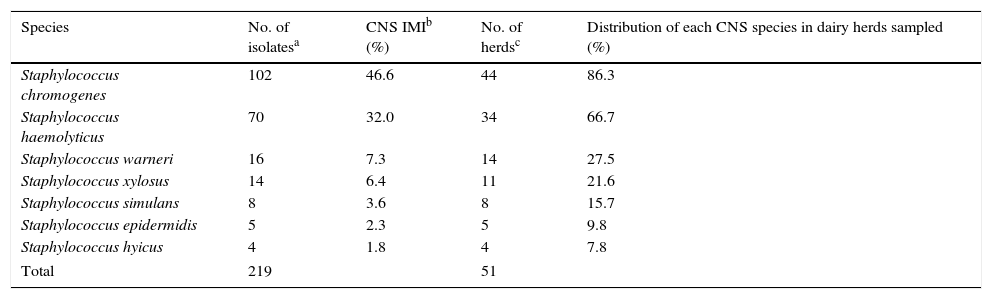
ResultsDistribution of CNS speciesTwo hundred and nineteen CNS isolates from cows with SCM were identified to the species level by PCR-RFLP of the groEL gene. The distribution of different species among identified CNS is shown in Table 1, where S. chromogenes and S. haemolyticus were the predominant species. A minimum of three different CNS species were present in 41.2% of the herds. S. chromogenes was isolated in 86.3% of all farms, whereas S. haemolyticus was isolated in 66.7% of them (Table 1).
Species distribution of coagulase-negative staphylococci (CNS) isolates from bovine subclinical mastitis in 51 Argentinean dairy herds
| Species | No. of isolatesa | CNS IMIb (%) | No. of herdsc | Distribution of each CNS species in dairy herds sampled (%) |
|---|---|---|---|---|
| Staphylococcus chromogenes | 102 | 46.6 | 44 | 86.3 |
| Staphylococcus haemolyticus | 70 | 32.0 | 34 | 66.7 |
| Staphylococcus warneri | 16 | 7.3 | 14 | 27.5 |
| Staphylococcus xylosus | 14 | 6.4 | 11 | 21.6 |
| Staphylococcus simulans | 8 | 3.6 | 8 | 15.7 |
| Staphylococcus epidermidis | 5 | 2.3 | 5 | 9.8 |
| Staphylococcus hyicus | 4 | 1.8 | 4 | 7.8 |
| Total | 219 | 51 | ||
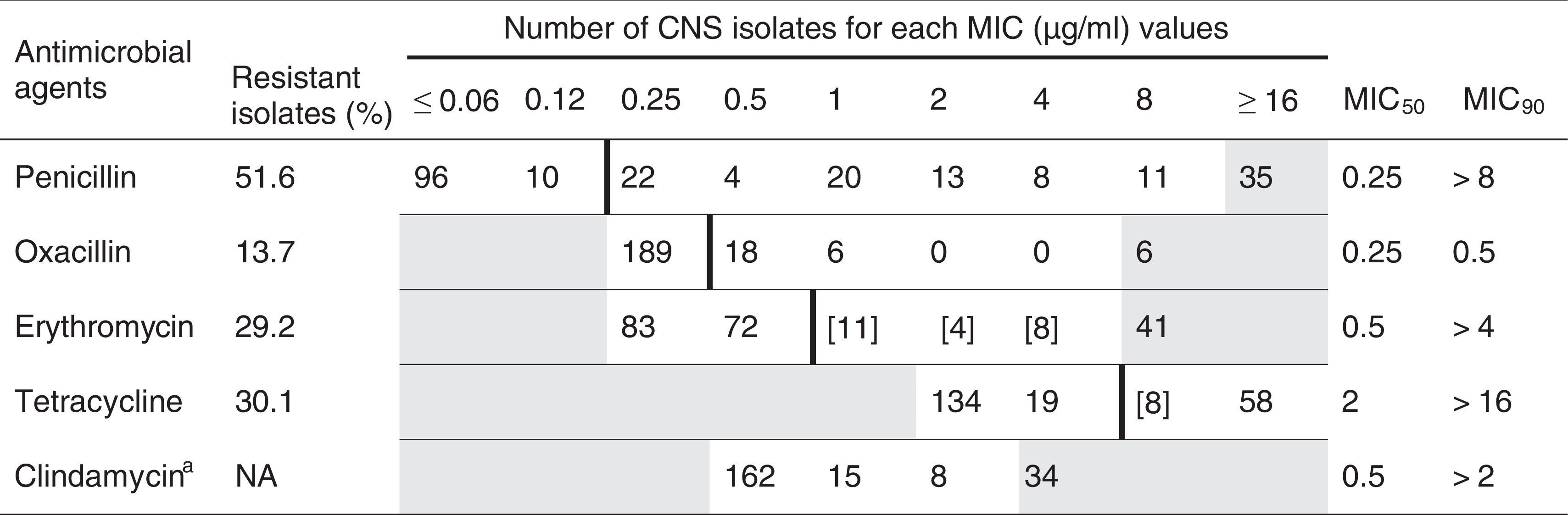
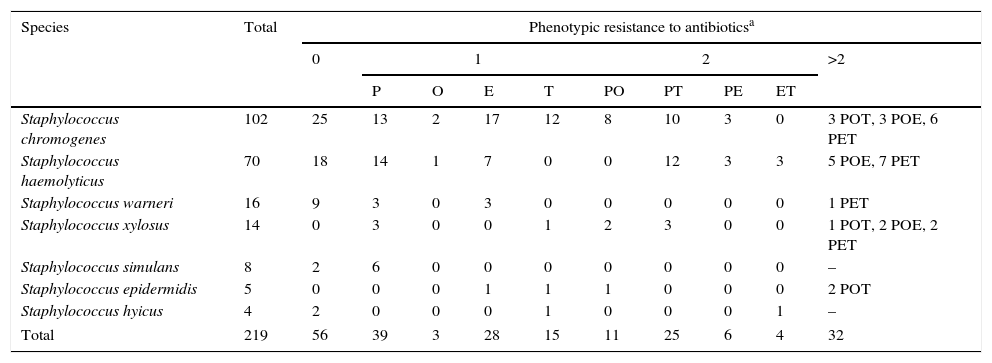
In the present study 219 CNS isolated from cows with SCM and identified by genotyping were evaluated for susceptibility to five antimicrobials (Table 2). Penicillin was used as the representative of penicillinase-labile penicillins and is the most frequently used in Argentina for bovine mastitis treatment. One hundred and thirteen strains (51.6%) of the 219 strains were resistant to penicillin; The MIC90 value for this antibiotic was higher than 8μg/ml, well above the recommended breakpoint. Thirty (13.7%) of all CNS isolates tested were resistant to oxacillin; the MIC90 value for this antibiotic was 0.5μg/ml. Resistance to oxacillin was attributed to the presence of the mecA gene in 2 of 12 (16.7%) of the oxacillin-resistant isolates with a MIC >0.5μg/ml. Moderate resistance to erythromycin and tetracycline was detected among CNS isolates, 29.2% and 30.1%, respectively. In our study, the MIC90 values of erythromycin and tetracycline for CNS were more than one or two dilution steps higher than those of the recommended breakpoint, respectively. Table 3 shows phenotypic resistance profiles of CNS obtained from bovine SCM. Of 219 CNS isolates, 56 (25.6%) were phenotypically susceptible to all antimicrobial agents tested. Eighty-five (38.8%) isolates expressed resistance to a single compound, and 11 isolates (5%) expressed resistance to 2 related compounds (β-lactams antimicrobial agents) and 67 (30.6%) isolates expressed resistance to compounds belonging to different antimicrobial classes. In S. chromogenes and S. haemolyticus, the most prevalent isolated CNS species, a large proportion of the strains investigated were resistant to penicillin, 45.1% and 58.6%, respectively. Although resistance to oxacillin was lower than that found in other 2 resistant species to the same antibiotic, the number of isolates of these species was too low to draw conclusions. For S. warneri, the third most common CNS species, the proportion of resistant isolates to penicillin was less common than in other species and no resistance to oxacillin was observed. Finally, in S. xylosus, penicillin resistance was the most common among the species tested. For other species, S. xylosus, S. epidermidis and S. hyicus, the number of resistant isolates was too low to be analyzed. The percentage of resistant S. haemolyticus strains to two or more non-related antimicrobials was higher than that observed in S. chromogenes, 42.8% and 24.5%, respectively.
Proportion of resistant isolates and minimum inhibitory concentration (MIC) distribution for 219 isolated coagulase-negative staphylococci (CNS) from bovine subclinical mastitis
White fields denote the range of dilutions tested for each substance. MICs above the range are given as the concentration closest to the range. MICs equal to or lower than the lowest concentration tested are given as the lowest tested concentration. Bold vertical lines indicate breakpoints based on veterinary interpretive criteria (CLSI 2008). Numbers in brackets indicate the number of isolates with a MIC in the intermediate resistant range.
a Clindamycin is not licensed for the treatment of bovine mastitis. As a consequence, no approved breakpoints are available for the classification of bovine staphylococcal isolates from mastitis cases as susceptible or resistant; NA, not applicable.
Phenotypic resistance profiles of coagulase-negative staphylococci (CNS) obtained from bovine subclinical mastitis
| Species | Total | Phenotypic resistance to antibioticsa | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| 0 | 1 | 2 | >2 | ||||||||
| P | O | E | T | PO | PT | PE | ET | ||||
| Staphylococcus chromogenes | 102 | 25 | 13 | 2 | 17 | 12 | 8 | 10 | 3 | 0 | 3 POT, 3 POE, 6 PET |
| Staphylococcus haemolyticus | 70 | 18 | 14 | 1 | 7 | 0 | 0 | 12 | 3 | 3 | 5 POE, 7 PET |
| Staphylococcus warneri | 16 | 9 | 3 | 0 | 3 | 0 | 0 | 0 | 0 | 0 | 1 PET |
| Staphylococcus xylosus | 14 | 0 | 3 | 0 | 0 | 1 | 2 | 3 | 0 | 0 | 1 POT, 2 POE, 2 PET |
| Staphylococcus simulans | 8 | 2 | 6 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | – |
| Staphylococcus epidermidis | 5 | 0 | 0 | 0 | 1 | 1 | 1 | 0 | 0 | 0 | 2 POT |
| Staphylococcus hyicus | 4 | 2 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 1 | – |
| Total | 219 | 56 | 39 | 3 | 28 | 15 | 11 | 25 | 6 | 4 | 32 |
Genotypic methods have shown to have higher specificity and sensitivity39 than other methods for discriminating among species, resulting in a better alternative for the identification of CNS isolates. A number of PCR amplicon sequencing-based methods for the identification of CNS have been reported5,26,30. Amplified fragment length polymorphism (AFLP)20,31 and PCR-restriction fragment length polymorphism (RFLP) applied to the groEL or gap genes have been used to identify CNS isolated from bovine milk15,25. The discrepancy observed in the distribution of CNS species identified by genotyping causing SCM in different countries, might be due to differences in experimental design10,20,30,31,35,38 however, S. chromogenes was one of the most prevalent species both in this work and in previous studies carried out in other countries such as Finland, Sweden, Belgium, Canada and Brazil, ranging between 23.3% and 78.8% of CNS isolates. To our knowledge, only Piessens et al.20 revealed that S. haemolyticus was the second most prevalent CNS species as in our study causing 27.6% of cases. Special attention should be given to both species, considering that the udder was found to be their main reservoir20.
In our study, a minimum of three different species of CNS were present in 41.2% of the herds. S. chromogenes and S. haemolyticus were isolated in most of the farms. This is in line with other studies, where these species have been isolated from all analyzed herds20,30. Although the reasons for the increased prevalence of some CNS species are not known, these might be attributed to the ability of each species to adapt to the mammary gland10,20 or to the different management patterns of mastitis control schemes30.
One limitation to the present study is the fact that we cannot be sure that the CNS were isolated from the diseased quarter since other non-infectious factors could cause an inflammatory response. However, CNS in pure culture allow to ensure that they are the only etiological agents causing subclinical mastitis.
The direct comparison of studies on antimicrobial susceptibility of CNS is often difficult mainly because of the use of different methodologies and breakpoints for testing susceptibility. A great variability has been reported in the MIC90 values for penicillin in CNS, ranging from 0.12μg/ml to 32μg/ml2,11,12,19,22,24,34. The MIC90 value for penicillin found in our study was well above the recommended breakpoint (CLSI, 2008). Moreover, the penicillin resistance percentage was higher than that described by Gentilini et al.11 in Argentina and those reported in other countries such as Uruguay12, The Netherlands27, USA22 and Sweden2. In the last decade, Gentilini et al.11 reported methicillin resistance among CNS isolated from bovine mastitis in our country. Since continuous surveillance is needed for the early detection of this kind of resistance, oxacillin has been used in this study to test susceptibility to methicillin. The MIC90 value for oxacillin was similar to those reported by different authors11–13,24,28. The percentage of resistant strains found was higher than that reported by other authors2,11–13, and similar to that reported in studies from Finland, Germany, USA, Sweden and The Netherlands19,21,22,27,28. The variability in the MIC90 values of the antimicrobial activity of penicillin and oxacillin against CNS highlights the relevance of determining β-lactams susceptibility patterns for this bacterial group. Macrolides are frequently used in Argentina for bovine mastitis treatment, since high concentrations in milk are obtained following parenteral administration. Although erythromycin was evaluated as representative of this group, tylosin is used in the therapy. Tetracycline was included in this study for epidemiological purposes as representative of tetracyclines; however, they are not recommended for mastitis treatment in Argentina because of the risk of an extended residue presence in milk. The MIC90 value for erythromycin and tetracycline was higher than that observed by several authors in Argentina, Finland, Germany, Sweden and USA2,11,13,19,21,28. Although clindamycin is not used in the treatment of bovine mastitis, the determination of the MIC for this antibiotic was conducted in order to test susceptibility to lincomycin; however, specific interpretative breakpoints are defined for pirlimicyn6. Susceptibility to pirlimycin could not be inferred from clindamycin results because some mechanism (enzymatic modification) does not exhibit cross-resistance. The MIC90 value for clindamycin obtained in this study was similar to that previously described by Gentilini et al.11 in our country and higher than that reported by other investigators in different countries2,13,19,21. The proportion of CNS resistant to a single compound or two related compounds obtained in our study was found to be similar to that described by Sampimon et al.27S. haemolyticus and S. chromogenes were the most frequent resistant species; therefore, it would also be hypothesized that they should be more “at risk” of developing antibiotic resistance or acquiring resistance determinants. Conversely, the absence or lower percentages of resistant isolates to multiple drugs for both species were found by Sampimon et al.27 We conclude that CNS species from bovine subclinical mastitis differ in prevalence and antimicrobial resistance profiles, which may have implications for treatment and management decisions when CNS are the predominant bacterial group on dairy farms.
Ethical disclosuresProtection of human and animal subjectsThe authors declare that no experiments were performed on humans or animals for this study.
Confidentiality of dataThe authors declare that no patient data appear in this article.
Right to privacy and informed consentThe authors declare that no patient data appear in this article.
Conflict of interestThe authors declare that they have no conflicts of interest.
This research was supported by grants from the Secretaría de Ciencia y Técnica de la Universidad Nacional de Río Cuarto, Argentina (SECyT-UNRC) and FONCyT (PICTO 21-30368/05). C. Vissio, M. Pellegrino and E. Reinoso are Career Investigators from the Consejo Nacional de Investigaciones Científicas y Técnicas (CONICET) and S. Dieser is a recipient of a posdoctoral fellowship from CONICET.