Streptococcus dysgalactiae subsp. equisimilis (SDSE) has virulence factors similar to those of Streptococcus pyogenes. Therefore, it causes pharyngitis and severe infections indistinguishable from those caused by the classic pathogen. The objectives of this study were: to know the prevalence of SDSE invasive infections in Argentina, to study the genetic diversity, to determine the presence of virulence genes, to study antibiotic susceptibility and to detect antibiotic resistance genes. Conventional methods of identification were used. Antibiotic susceptibility was determined by the disk diffusion and the agar dilution methods and the E-test. Twenty eight centers from 16 Argentinean cities participated in the study. Twenty three isolates (16 group G and 7 group C) were obtained between July 1 2011 and June 30 2012. Two adult patients died (8.7%). Most of the isolates were recovered from blood (60.9%). All isolates carried speJ and ssa genes. stG62647, stG653 and stG840 were the most frequent emm types. Nineteen different PFGE patterns were detected. All isolates were susceptible to penicillin and levofloxacin, 6 (26.1%) showed resistance or reduced susceptibility to erythromycin [1 mef(A), 3 erm(TR), 1 mef(A)+erm(TR) and 1 erm(TR)+erm(B)] and 7 (30.4%) were resistant or exhibited reduced susceptibility to tetracycline [2 tet(M), 5 tet(M)+tet(O)]. The prevalence in Argentina was of at least 23 invasive infections by SDSE. A wide genetic diversity was observed. All isolates carried speJ and ssa genes. Similarly to other studies, macrolide resistance (26.1%) was mainly associated to the MLSB phenotype.
Streptococcus dysgalactiae subsp. equisimilis (SDSE) posee factores de virulencia similares a Streptococcus pyogenes y, en consecuencia, produce faringitis e infecciones graves indistinguibles de las generadas por este patógeno clásico. Los objetivos del estudio fueron conocer la prevalencia de SDSE en infecciones invasivas en Argentina, estudiar su diversidad genética, determinar la presencia de genes de virulencia, ensayar su sensibilidad a los antibióticos y conocer los genes de resistencia. Se emplearon métodos convencionales de identificación. La sensibilidad se determinó por difusión, Etest y dilución en agar. Participaron 28 centros de 16 ciudades argentinas. Se obtuvieron 23 aislamientos (16 del grupo G y 7 del grupo C) desde el 1-7-2011 hasta el 30-6-2012. Se registraron 2 muertes en adultos (8,7%). La mayoría de los aislamientos fueron obtenidos de sangre (60,9%). Todos eran portadores de los genes speJ y ssa. Los genotipos más frecuentes fueron stG62647, stG653 y stG840. Se detectaron 19 pulsotipos distintos. Todos los aislamientos fueron sensibles a penicilina y levofloxacina, 6 (26,1%) presentaron resistencia o sensibilidad disminuida a eritromicina (1 mef[A], 3 erm[TR], 1 mef[A]+erm[TR] y 1 erm[TR]+erm[B]) y 7 (30,4%) fueron resistentes o tuvieron sensibilidad disminuida a tetraciclina (2 tet[M], 5 tet[M]+tet[O]). La prevalencia anual en la Argentina fue de al menos 23 infecciones invasivas por SDSE y se observó una amplia diversidad genética. Todos los aislamientos presentaron los genes ssa y speJ. Como en otros estudios, la resistencia a macrólidos (26,1%) estuvo asociada, principalmente, al fenotipo MLSB.
β-Hemolytic streptococci are common pathogens that usually cause community-acquired infections.
In general, β-hemolytic streptococci isolated from human beings belong to Lancefield groups A, B, C, G, F, or more rarely, to L18. Until recently, group A streptococci (GAS, Streptococcus pyogenes)63 and group B streptococci (GBS, Streptococcus agalactiae)45,60 were considered the most important pathogens of this group of microorganisms in clinical settings.
GAS is responsible for a broad spectrum of human infections that result in significant morbidity and mortality, including pharyngitis, scarlet fever, skin and soft tissue infection (SSTI), streptococcal toxic shock syndrome (STSS), septicemia, pneumonia and rarely meningitis13. It is estimated that severe GAS diseases lead to more than 500,000 deaths each year via infections such as acute rheumatic fever, rheumatic heart disease, post streptococcal glomerulonephritis and invasive diseases8.
In contrast, group C and G Streptococcus dysgalactiae subsp. equisimilis (GCS and GGS) were long considered to be commensal organisms that only rarely cause invasive infections as opportunistic pathogens.
Since the mid-1980s there has been a marked increase in reported invasive group A infections, including cases of STSS in Europe and North America15,33,57.
These fulminant infections were characterized by acute hypotension, shock, multiorgan impairment and death. However, the factors underlying the worldwide resurgence of this pathogen remain unknown16.
In 1996, Vandamme et al62. proposed that a novel subspecies, S. dysgalactiae subsp. equisimilis (SDSE), was a clinically significant pathogen. This microorganism possesses group C or G antigens (rarely A or L), and exhibits strong β-hemolysis.
Beginning around the year 2000, invasive infections such as bacteremia caused by SDSE as well as those caused by GAS and GBS6,12,38,59 have been increasingly reported worldwide.
M proteins of GAS, which are encoded by emm genes and form elongated structures on the bacterial surface, play an important role in the pathogenesis of this microorganism. M proteins are known to be critical antiphagocytic constituents of GAS due to their role in resistance to opsonization4,14. GCS and GGS have shown a similar pathogenic pattern to S. pyogenes21, coincidently expressing homologs of the M virulence proteins of S. pyogenes7. Moreover, some strains contain superantigen genes firstly characterized in S. pyogenes25,29. As with the emm genes of S. pyogenes, GCS and GGS homologs have been used for sequence-based typing3,17,25.
We conducted a prospective study to assess the relative prevalence of SDSE versus GAS in invasive infections in Argentina during a twelve-month period. The objective of this study was to determine their epidemiological features, to explore the molecular characteristics of the infecting strains, to study their genetic diversity, virulence genes, antibiotic susceptibility and antibiotic resistance genes.
Materials and methodsAll S. pyogenes and SDSE isolated from invasive infections in 28 centers of 16 Argentinean cities from July 1, 2011 to June 30, 2012 were studied.
A microbiologist from each hospital was required to complete a data sheet of the isolates. During the study period, each laboratory collected one isolate per patient in accordance with the case definition. Isolates were appropriately sent to the reference center where they were preserved in 300μl of sheep blood at −80°C.
DefinitionsInfections in deep tissues, blood, cerebrospinal fluid, or other liquids obtained by puncture, in which causative organisms were isolated from otherwise sterile samples, were defined as invasive infections.
STSS was defined as an invasive infection due to the presence of a β-hemolytic streptococcus and causing hypotension and two or more of the following conditions: renal impairment, coagulopathy, liver abnormalities, acute respiratory distress syndrome, extensive tissue necrosis and erythematous rash55.
IdentificationS. pyogenes and SDSE were identified according to the differentiating characteristics described by Ruoff et al49. Hemolysis was detected on 5% sheep blood Columbia agar and the observation of chains was performed in Gram smears prepared with drops of overnight cultures in thioglycolate broth. Conventional identification included the following tests: pyrrolidonyl-β-naphthylamide hydrolysis (PYR), bacitracin susceptibility, cell morphology and Gram staining characteristics, Voges-Proskauer, arginine dihydrolase, esculin, starch, sorbitol and trehalose fermentation,α-galactosidase and β-glucuronidase. PYR and bacitracin tests were performed by using Britania® disks (Buenos Aires, Argentina). Sorbitol, trehalose, α-galactosidase and β-glucuronidase were tested by using DIATABS® commercial tablets (Rosco, Taastrup, Denmark) according to the manufacturer's instructions. Voges-Proskauer, arginine dihydrolase, esculin, and starch tests were performed by standard methods. Moreover, identification was performed using API 20 Strep® (Biomérieux Argentina). Identification and grouping were completed by using the latex agglutination method (Slidex Strepto kit®, Biomérieux Argentina).
emm typingGAS emm DNA fragment preparation was conducted according to the protocol of the Centers for Disease Control and Prevention (CDC)9.
As specified by the CDC protocol, primers 1 and 2 were used for amplifying the N-terminal region of the emm gene [primer 1 was 5′-TATT(C/G) GCTTAGAAAATTAA-3′, and primer 2 was 5′-GCAAGTTCTTCAGCTTGTTT-3′]. The DNA amplicons were sent for DNA sequencing. The first 160 bases of the 5 end of the emm gene were compared with those in the CDC emm sequence database (http://www.cdc.gov/ncidod/biotech/strep/strepblast.htm). An emm type showing more than 98% identity with a CDC reference strain was identified as that particular emm type.
PFGE analysisAll isolates were digested by SmaI (Promega, Madison, WI, USA). Pulsed field gel electrophoresis (PFGE) was carried out according to the previously described protocol10.
The digested fragments of DNA were separated by agarose gel 1.2% (TBE 0.5X) in the CHEF-DR III System (BioRad, Hercules, CA, EE.UU.) for 22h (0.5–40s pulses at 6V/cm, 120°C).
The genetic relationship between the bacterial strains was evaluated based on the levels of similarity among the SmaI PFGE patterns. A dendrogram was constructed using the unweighted-pair group method with arithmetic mean (UPGMA) algorithm using BioNumerics software version 6.0 (Applied Maths, Kortrijk, Belgium).
Superantigen (SAg) gene detectionThe isolates were tested for the presence of SAg genes, including speA, speB, speC, speF, speG, speH, speI, speJ, ssa, and smeZ25.
Antimicrobial susceptibility testsDisk diffusion tests were performed by the Bauer and Kirby method according to CLSI guidelines with 5% sheep blood Mueller–Hinton agar11.
Testing disks containing penicillin (PEN 10U), erythromycin (ERY 15μg), clindamycin (CLI 2μg), tetracycline (TET 30μg), ofloxacin (5μg), pefloxacin (5μg), norfloxacin (10μg) and levofloxacin (5μg) were from Britania® Laboratories (Buenos Aires, Argentina). Incubation was performed at 35°C for 24h in ambient air. Streptococcus pneumoniae ATCC® 4961, Staphylococcus aureus ATCC® 25923, Escherichia coli ATCC® 25922 and Pseudomonas aeruginosa ATCC® 27853 were used as reference strains.
The agar dilution method was used for susceptibility testing of five antibiotics (PEN, ERY, CLI, TET and LEV) with 5% sheep blood Mueller-Hinton agar plates according to CLSI guidelines11. Antibiotics were gently provided by the Servicio de Antimicrobianos, INEI-ANLIS “Dr. Carlos G. Malbrán”, Buenos Aires. Staphylococcus aureus ATCC® 29213 and Enterococcus faecalis ATCC® 29212 were used as reference strains.
Phenotypic expression of MLS resistanceWhen performing the disk diffusion tests, blunting of the CLI inhibition zone near an ERY disk placed at 12mm from the edge of the CLI disk, indicated an inducible type of resistance to macrolides, lincosamides, and streptogramin B (MLSB), while no blunting indicated the probability of the efflux-mediated M resistance phenotype (resistance only to 14- and 15-membered macrolides). Resistance to both CLI and ERY indicated a constitutive type of MLSB resistance50.
Detection of different macrolide and tetracycline resistance genesGenotypic characterization of antimicrobial resistance genes was carried out by multiplex PCR [erm(B), erm(TR), mef(A), tet(M) and tet(O) genes]. Methods used to detect antibiotic resistance genes have been previously described27,41.
ResultsEpidemiological, clinical and demographic characteristics of the patients with SDSE invasive diseaseOne hundred and eleven invasive infections due to groups A, C, or G β-hemolytic streptococci were recorded. Eighty eight of them (79.3%) were identified as S. pyogenes, 16(14.4%) as GGS and 7(6.3%) as GCS. The annual prevalence in Argentina (2011–2012) was of at least 23 SDSE invasive infections.
Most cases of SDSE invasive infections (65.2%) were diagnosed in Buenos Aires City and its surroundings (n: 9 cases in Ciudad Autónoma de Buenos Aires, n: 4 in San Martín, and n: 2 in Lanús). The rest of the cases were detected in Mar del Plata (2), Bahía Blanca (1), General Pico (La Pampa) (2), Santa Fe (2) and Concordia (Entre Ríos) (1). STSS was recorded in 11 cases (9 GAS, 1 SDSE group C and 1 SDSE group G) and mortality was 6.3% (4 adults and 1 child died as a consequence of a S. pyogenes infection and 2 adults died of GGS SDSE infections).
Most of the SDSE isolates were obtained from blood (60.9%). Other foci of infection were osteoarticular (21.7%) and soft tissue (17.4%). Only GAS was isolated from pleural effusion (n: 5) and ascites (n: 3).
The total number of patients was 111. Fifty-four of them were adults (≥16 years old) and fifty-seven were children (<16 years old). With regard to invasive infections by SDSE, nineteen patients were adults and four were children. The median age of the patients was 54 years old (20 months–90 years old) and 69.6% of the patients were male.
Almost 70% (69.6%) of the infected patients presented at least one predisposing condition and 17.4% more than one. The most frequent predisposing conditions for invasive disease were diabetes (n: 4), renal disease (n: 2), arterial hypertension (n: 2) and HIV infection (n: 2).
More than 80% (82.6%) of the patients had been treated with antibiotics. Clindamycin was only administered to 5 patients whereas immunoglobulins were not administered at all. None of the patients who died were treated with clindamycin.
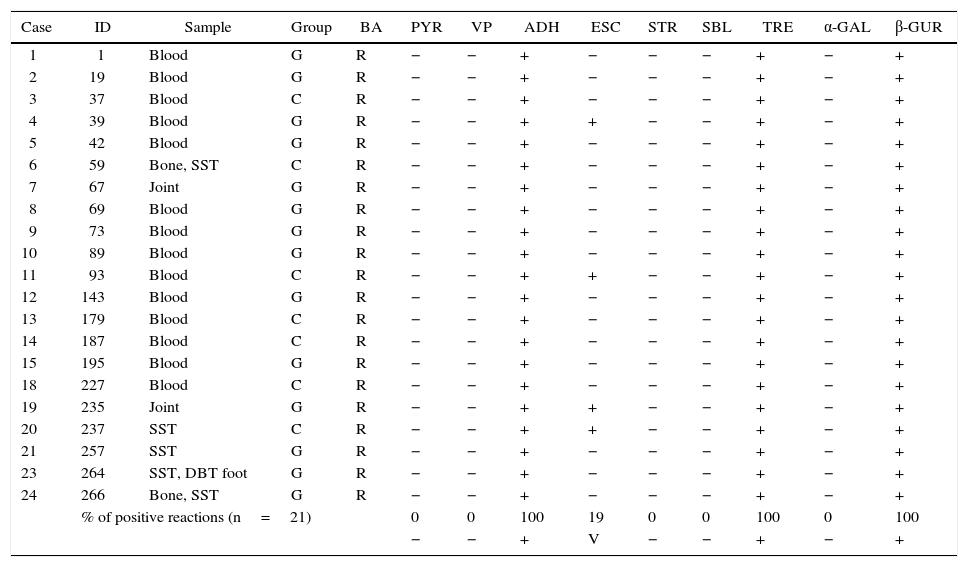
Phenotypic identificationAll GCS and GGS isolates obtained from invasive infections (n: 21) were identified by conventional biochemical tests and API20 Strep. Fourteen of them were isolated from blood, 3 from soft tissue, 2 from bone and 2 from joint-bone fluid.
Table 1 shows the results of the manual biochemical tests and Table 2 summarizes API 20 Strep results with the percentage of positive values obtained from the bibliography. The percentage of positive tests is summarized at the end of both tables.
Characteristics of Streptococcus dysgalactiae subsp. equisimilis isolated from invasive infections in Argentina (2011–2012). The percentage of positive tests is summarize at the end of the table
| Case | ID | Sample | Group | BA | PYR | VP | ADH | ESC | STR | SBL | TRE | α-GAL | β-GUR |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | 1 | Blood | G | R | − | − | + | − | − | − | + | − | + |
| 2 | 19 | Blood | G | R | − | − | + | − | − | − | + | − | + |
| 3 | 37 | Blood | C | R | − | − | + | − | − | − | + | − | + |
| 4 | 39 | Blood | G | R | − | − | + | + | − | − | + | − | + |
| 5 | 42 | Blood | G | R | − | − | + | − | − | − | + | − | + |
| 6 | 59 | Bone, SST | C | R | − | − | + | − | − | − | + | − | + |
| 7 | 67 | Joint | G | R | − | − | + | − | − | − | + | − | + |
| 8 | 69 | Blood | G | R | − | − | + | − | − | − | + | − | + |
| 9 | 73 | Blood | G | R | − | − | + | − | − | − | + | − | + |
| 10 | 89 | Blood | G | R | − | − | + | − | − | − | + | − | + |
| 11 | 93 | Blood | C | R | − | − | + | + | − | − | + | − | + |
| 12 | 143 | Blood | G | R | − | − | + | − | − | − | + | − | + |
| 13 | 179 | Blood | C | R | − | − | + | − | − | − | + | − | + |
| 14 | 187 | Blood | C | R | − | − | + | − | − | − | + | − | + |
| 15 | 195 | Blood | G | R | − | − | + | − | − | − | + | − | + |
| 18 | 227 | Blood | C | R | − | − | + | − | − | − | + | − | + |
| 19 | 235 | Joint | G | R | − | − | + | + | − | − | + | − | + |
| 20 | 237 | SST | C | R | − | − | + | + | − | − | + | − | + |
| 21 | 257 | SST | G | R | − | − | + | − | − | − | + | − | + |
| 23 | 264 | SST, DBT foot | G | R | − | − | + | − | − | − | + | − | + |
| 24 | 266 | Bone, SST | G | R | − | − | + | − | − | − | + | − | + |
| % of positive reactions (n=21) | 0 | 0 | 100 | 19 | 0 | 0 | 100 | 0 | 100 | ||||
| − | − | + | V | − | − | + | − | + | |||||
BA: bacitracin, PYR: pirrolidonil-arilamidase, VP: Voges–Proskauer, ADH: arginine dihydrolase, ESC: esculin, STR: starch, SBL: sorbitol, TRE: trehalose, α-GAL: α-galactosidase, β-GUR: β-d-glucuronidase, +: positive, −: negative, V: variable, SST: skin and soft tissue, and DBT: diabetic.
Phenotypic characteristics determined by the API 20 Strep system for Streptococcus dysgalactiae subsp. equisimilis isolated from invasive infections in Argentina (2011–2012). The percentage of positive tests is summarize at the end of the table and results with the percentage of positive values obtained from bibliography (identification table API 20 strep V 7.0)
| Case | ID | Sample | Group | VP | HIP | ESC | PYR | α-GAL | β-GUR | β-GAL | PAL | LAP | ADH | RIB | ARA | MAN | SBL | LAC | TRE | INU | RAF | AMD | GLYG | HEM | Profile |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | 1 | Blood | G | − | − | − | − | − | + | − | + | + | + | − | − | − | − | − | + | − | − | + | − | + | 0 4 6 1 0 1 5 |
| 2 | 19 | Blood | G | − | − | − | − | − | + | − | + | + | + | + | − | − | − | − | + | − | − | + | − | + | 0 4 6 3 0 1 5 |
| 3 | 37 | Blood | C | − | − | − | − | − | + | − | + | + | + | − | − | − | − | − | + | − | − | + | − | + | 0 4 6 1 0 1 5 |
| 4 | 39 | Blood | G | − | − | + | − | − | + | − | + | + | + | + | − | − | − | − | + | − | − | + | − | + | 4 4 6 3 0 1 5 |
| 5 | 42 | Blood | G | − | − | − | − | − | + | − | + | + | + | + | − | − | − | − | + | − | − | + | + | + | 0 4 6 3 0 1 7 |
| 6 | 59 | Bone, SST | C | − | − | − | − | − | + | − | + | + | + | + | − | − | − | + | + | − | − | + | − | + | 0 4 6 3 4 1 5 |
| 7 | 67 | Joint | G | − | − | − | − | − | + | − | + | + | + | + | − | − | − | − | + | − | − | + | − | + | 0 4 6 3 0 1 5 |
| 8 | 69 | Blood | G | − | − | − | − | − | + | − | + | + | + | + | − | − | − | + | + | − | − | + | − | + | 0 4 6 3 4 1 5 |
| 9 | 73 | Blood | G | − | − | − | − | − | + | − | + | + | + | + | − | − | − | − | + | − | − | + | − | + | 0 4 6 3 0 1 5 |
| 10 | 89 | Blood | G | − | − | − | − | − | + | − | + | + | + | − | − | − | − | − | + | − | − | + | − | + | 0 4 6 1 0 1 5 |
| 11 | 93 | Blood | C | − | − | + | − | − | + | − | + | + | + | + | − | − | − | − | + | − | − | + | − | + | 4 4 6 3 0 1 5 |
| 12 | 143 | Blood | G | − | − | − | − | − | + | − | + | + | + | + | − | − | − | − | + | − | − | + | − | + | 0 4 6 3 0 1 5 |
| 13 | 179 | Blood | C | − | − | − | − | − | + | − | + | + | + | + | − | − | − | − | + | − | − | + | − | + | 0 4 6 3 4 1 5 |
| 14 | 187 | Blood | C | − | − | − | − | − | + | − | + | + | + | + | − | − | − | + | + | − | − | + | − | + | 0 4 6 3 4 1 5 |
| 15 | 195 | Blood | G | − | − | − | − | − | + | − | + | + | + | + | − | − | − | − | + | − | − | + | − | + | 4 4 6 3 0 1 5 |
| 18 | 227 | Blood | C | − | − | − | − | − | + | − | + | + | + | + | − | − | − | − | + | − | − | + | − | + | 4 4 6 3 0 1 5 |
| 19 | 235 | Joint | G | − | − | + | − | − | + | − | + | + | + | + | − | − | − | − | + | − | − | + | − | + | 4 4 6 3 0 1 5 |
| 20 | 237 | SST | C | − | − | + | − | − | + | − | + | + | + | + | − | − | − | − | + | − | − | + | − | + | 4 4 6 3 0 1 5 |
| 21 | 257 | SST | G | − | − | − | − | − | + | − | + | + | + | + | − | − | − | − | + | − | − | + | − | + | 0 4 6 3 0 1 5 |
| 23 | 264 | SST, DBT foot | G | − | − | − | − | − | + | − | + | + | + | + | − | − | − | − | + | − | − | + | − | + | 0 4 6 3 0 1 5 |
| 24 | 266 | Bone, SST | G | − | − | − | − | − | + | − | + | + | + | + | − | − | − | + | + | − | − | + | − | + | 0 4 6 3 4 1 5 |
| % of positive reactions (n=21) | 0 | 0 | 19 | 0 | 0 | 100 | 0 | 100 | 100 | 100 | 86 | 0 | 0 | 0 | 19 | 100 | 0 | 0 | 100 | 5 | 100 | ||||
| S. dysgalactiae subsp. equisimilis | 0 | 1 | 25 | 1 | 1 | 99 | 1 | 99 | 100 | 97 | 97 | 1 | 1 | 1 | 45 | 99 | 0 | 1 | 98 | 40 | 94 | ||||
VP: Voges–Proskauer, HIP: hippurate, ESC: esculin, PYR: pyrrolidonyl-β-naphthylamide hydrolysis, α-GAL: α-galactosidase, β-GUR: β-glucuronidase, β-GAL: β-galactosidase, PAL: alkaline phosphatase, LAP: leucinaminopeptidase, ADH: arginine dihydrolase, RIB: ribose, ARA: arabinose, MAN: manitol, SBL: sorbitol, LAC: lactose, TRE: trehalose, INU: inulin, RAF: raffinose, AMD: amigdaline, GLYG: glycogen, HEM: hemolysis, +: positive, −: negative, SST: skin and soft tissue, and DBT: diabetic.
Six distinct biochemical profiles were obtained using the API 20 Strep identification system: 19% of the strains fermented lactose and the same percentage hydrolyzed esculin, 86% fermented ribose and only 5% fermented glycogen.
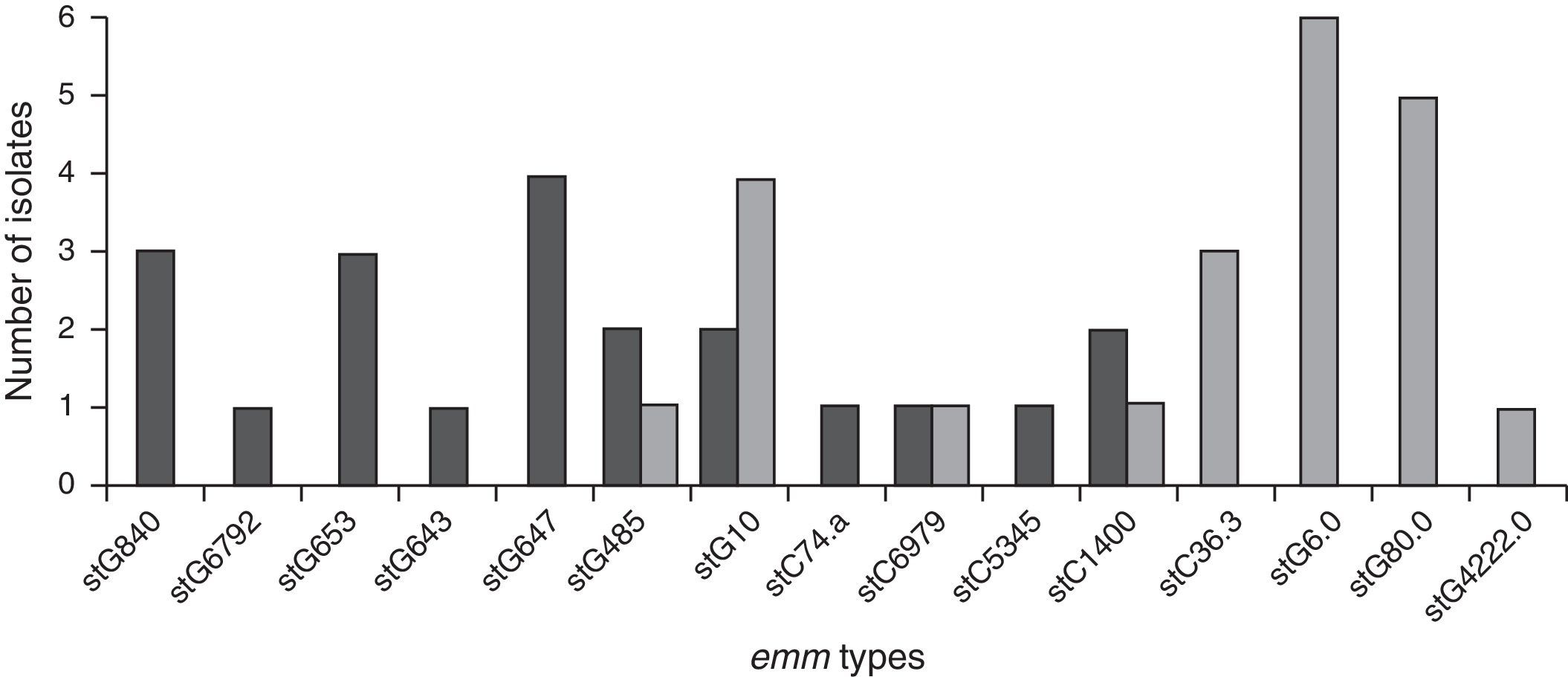
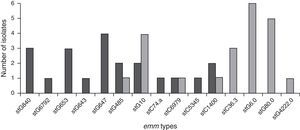
Genetic diversityA total of 23 isolates, 16 GGS and 7 GCS were isolated from invasive infections and 11 different emm genes were identified. Three types, stG62647 (n: 4), stG840 (n: 3) and stG653 (n: 3) predominated among the 21 emm invasive strains typed.
Figure 1 shows the distribution of invasive types. The comparison with the types found in the 1998–1999 Argentinean multicenter study is also shown. All the sequences were compared with the CDC emm-sequence-database.
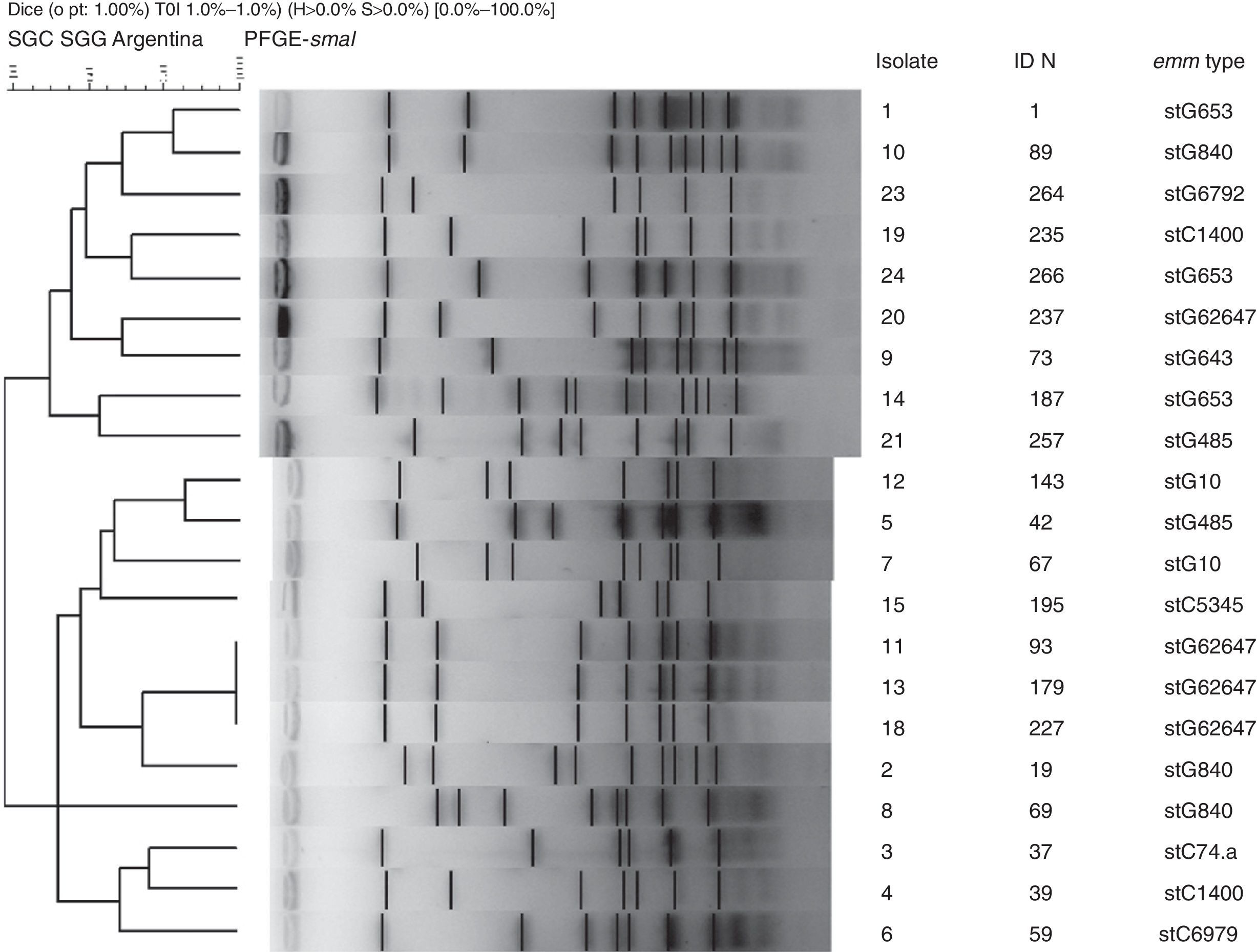
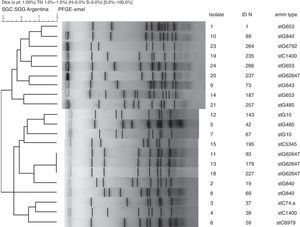
Nineteen distinct PFGE patterns were detected, with no significant relationship among them. The PFGE profiles of invasive strains digested with the restriction enzyme SmaI are shown in Figure 2. Strains 11, 13 and 18 exhibited the same pattern and all were stG62647 type.
Virulence factorsToxigenic genes speA, speB, speC, speF, speG, speH, speJ, smeZ and ssa were analyzed. All the strains only carried speJ and ssa genes simultaneously.
Antibiotic susceptibility and macrolide and tetracycline resistance gene studyAll SDSE isolates showed in vitro susceptibility to PEN (MIC90 0.016μg/ml) and to LEV (MIC90 0.5μg/ml).
The percentage of isolates not susceptible to CLI was 19.0% (MIC90 0.25μg/ml), to ERY 26.1% (MIC90 4μg/ml) and to TET 30.4% (MIC90 32μg/ml).
Only one isolate was found to be less susceptible to NOR, PEF and OFL by the disk diffusion method (13mm, 6mm and 19mm, respectively). Breakpoints were only available for ofloxacin11 and 19mm was inside the category of susceptibility.
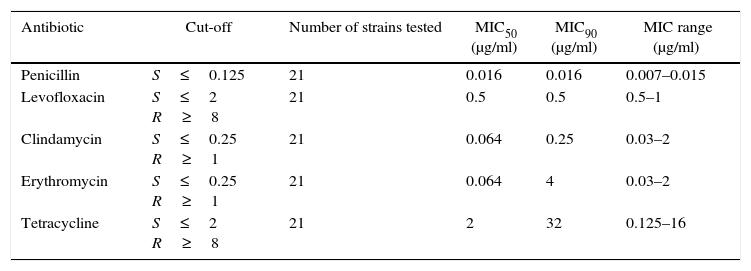
Table 3 summarizes the minimum inhibitory concentration (MIC) ranges, MIC50 and MIC90 values of antimicrobial agents for SDSE. Table 4 summarizes the macrolide and TET resistance gene study.
Minimum inhibitory concentration (MIC) ranges, MIC50 and MIC90 values of antimicrobial agents for Streptococcus dysgalactiae subsp. equisimilis
| Antibiotic | Cut-off | Number of strains tested | MIC50 (μg/ml) | MIC90 (μg/ml) | MIC range (μg/ml) |
|---|---|---|---|---|---|
| Penicillin | S≤0.125 | 21 | 0.016 | 0.016 | 0.007–0.015 |
| Levofloxacin | S≤2 R≥8 | 21 | 0.5 | 0.5 | 0.5–1 |
| Clindamycin | S≤0.25 R≥1 | 21 | 0.064 | 0.25 | 0.03–2 |
| Erythromycin | S≤0.25 R≥1 | 21 | 0.064 | 4 | 0.03–2 |
| Tetracycline | S≤2 R≥8 | 21 | 2 | 32 | 0.125–16 |
MIC: minimum inhibitory concentration.
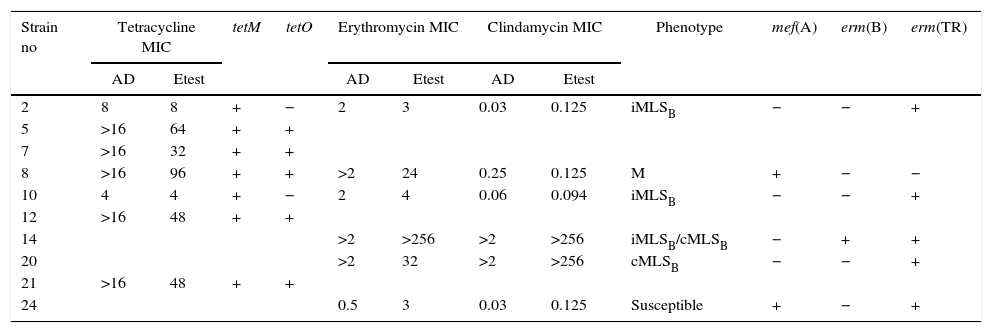
Characteristics of macrolide and tetracycline resistance among Streptococcus disgalactiae subsp. equisimilis from Argentina
| Strain no | Tetracycline MIC | tetM | tetO | Erythromycin MIC | Clindamycin MIC | Phenotype | mef(A) | erm(B) | erm(TR) | |||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| AD | Etest | AD | Etest | AD | Etest | |||||||
| 2 | 8 | 8 | + | − | 2 | 3 | 0.03 | 0.125 | iMLSB | − | − | + |
| 5 | >16 | 64 | + | + | ||||||||
| 7 | >16 | 32 | + | + | ||||||||
| 8 | >16 | 96 | + | + | >2 | 24 | 0.25 | 0.125 | M | + | − | − |
| 10 | 4 | 4 | + | − | 2 | 4 | 0.06 | 0.094 | iMLSB | − | − | + |
| 12 | >16 | 48 | + | + | ||||||||
| 14 | >2 | >256 | >2 | >256 | iMLSB/cMLSB | − | + | + | ||||
| 20 | >2 | 32 | >2 | >256 | cMLSB | − | − | + | ||||
| 21 | >16 | 48 | + | + | ||||||||
| 24 | 0.5 | 3 | 0.03 | 0.125 | Susceptible | + | − | + | ||||
MIC: minimum inhibitory concentration; AD: agar dilution.
The six isolates with resistance or diminished susceptibility to ERY presented the following phenotypes and genotypes: 1M [mef(A)], 2 iMLSB[erm(TR)], and 1 cMLSB[erm(TR)]. Isolate 14 presented two subpopulations: one resistant to ERY and CLI (CIM>256μg/ml; cMLSB) and another resistant to ERY (CIM>256μg/ml) but susceptible to CLI (iMLSB phenotype). Isolate 24 was susceptible to ERY by the disk diffusion method but showed low level resistance to ERY (MIC=3μg/ml) and carried resistance genes mef(A) and erm(TR).
Two TET resistant strains harbored only the tet(M) gene. The other five strains carried both the tet(M) and tet(O) genes.
DiscussionInvasive SDSE infections included sepsis with an unknown focus, cellulitis, septic arthritis, pneumonia, necrotizing fascitis, meningitis, infectious endocarditis, STSS, abscesses at sites other than skin, osteomyelitis, and others.
In our study most of the SDSE isolates were obtained from blood (n: 14). The rest of the isolates were obtained from bone, joint fluid or soft tissue (n: 7).
All our strains were correctly identified by both systems (manual and API 20 Strep) as SDSE. The correlation was excellent between the tests with ROSCO tablets (esculin, sorbitol, trehalose, α-galactosidase and β-glucuronidase) and the same biochemical tests in the API 20 Strep panel.
The frequency of invasive SDSE infection has increased in Asia5,38, in Europe19,34,36,37,48,53,54, and in America1,6,40 over the years. In particular, since 2003, the number of invasive SDSE infections, including STSS and severe soft tissue infection22,26,44, has gradually increased in Japan each year.
By the year 2011 the Argentine population was 40.73 million inhabitants, therefore, the annual prevalence of SDSE invasive infections (n: 23) was at least 0.06/100,000 in Argentina during the 2011–2012 period. This prevalence seems to be lower than that recorded in a previous multicenter study conducted in 1998–1999, in which 27 invasive SDSE infections were reported during six-months; however, more health institutions participated in that study40.
As for age distribution, a population-based surveillance carried out by Broyles et al. (n: 212)6 found that most patients suffering from invasive diseases due to SDSE (59.0%) were adults under 65 years old. Moreover, Takahashi et al. analyzed 286 SDSE infections from August 2006 to December 200960 and all patients with invasive infections were adults, often elderly (74.0%), and most of them with underlying diseases. Another investigation58 also indicated that invasive SDSE infection (n=42) mostly occurred in elderly adults (60–80 years old). Severe underlying conditions (i.e., diabetes mellitus, liver or renal dysfunction, and others) were associated with 85.7% of these invasive SDSE infections.
In our study, the age range was 21–86 years; 57.1% of the patients were adults older than 50 years old (12 out of 21 patients).
With regard to underlying diseases, in U.S. surveillance reports, 96.2% of patients with invasive infections possessed underlying medical conditions13.
Similarly to the study by Sunaoshi et al.58, 69.9% of SDSE-infected patients in Argentina presented at least one underlying disease and 17.4% had more than one, being diabetes the most frequent one.
Concerning SDSE disease outcomes, the mortality rate in a Japanese study (12.7%) was similar to that previously described in Hong Kong and the United States (12 and 15%)6,60,64. In our series, the mortality rate due to SDSE invasive infections was 8.7% (2 cases) and both were adults. The same percentage corresponded to STSS cases. Three STSS cases and 3 deaths were recorded in a previous six-month Argentinean study40.
Neither fatalities nor SDSE-mediated STSS were detected in children. SDSE-mediated STSS was a consequence of infections due to types stG840 and stG62647 and fatal infections were associated with stC74.a and stG62647.
The dominant emm types in Japan (stG6792 and stG485) were different from those in the United States (stG6, stG245, stG2078, and stG643). The geographical factor may not be the only cause accounting for such difference since both study periods differed in these studies (2002–2004 vs. 2006–2009)6,60. The emm type stG6792 was the most frequently found in SDSE Japanese isolates (n: 65; 22.7%) and was more strongly related to poor outcome of SDSE diseases than other SDSE emm types60. Furthermore, those isolates displayed similar DNA profiles with PFGE suggesting the clonal expansion of a specific subpopulation of strains rather than the spread of distinct strains.
In another study58, 3 emm types, stG6792, stG485, and stG2078, predominated among the 42 invasive strains; strains having the same emm type showed uniform DNA profiles by PFGE. Moreover, previous reports described emm types and the DNA profiles by PFGE as variable among SDSE strains22,26. In two other SDSE bacteremia studies carried out in the U.S. and Israel, stG485, stG6, stG245 and stG2078 types were the most frequently found6,12.
In our study, 19 distinct DNA profiles by PFGE were detected, not having significant relationship among them. These results were expected because strains were not isolated in the context of an epidemic outbreak and were obtained from different geographic places. Predominant emm-types were stG62647 (n: 4), stG840 (n: 3) and stG653 (n: 3). stG6792 (n: 1), stG485 (n: 2) and stG2078 (n: 0) types were less represented in the present study, in contrast to the study conducted by Sunaoshi et al58. The types in this study were also different from those reported in the previous Argentinean multicenter study (1998), in which stG6.0 (n: 6), stG480 (n: 5) and stG10.0 (n: 4) types40 predominated.
Therefore, emm typing is an excellent comparative epidemiological tool for the analysis of SDSE strains belonging to different geographical regions or from the same region but isolated at different times.
The complete genomic sequence of SDSE GGS_124 (stG480.0) isolated from patients with STSS (GenBank accession No. AP010935) have been recently determined. The genome size was 2.1 Mbp, and sequence coverage with GAS genomes was 61–63% identity20. Interestingly, many genes encoding virulence factors in GAS were identified in SDSE.
SDSE possesses many virulence factors shared with GAS28, such as M protein, streptolysin O, streptolysin S, streptokinase, fibronectin binding protein, collagen binding protein and DNase. They also exhibit homology in streptococcal inhibitor of complement (SIC). However, they do not possess certain virulence factors, such as some superantigens, cysteine protease (SPE-B) and the ABC operon52. However, it was reported that some strains of SDSE may contain superantigen genes, which were firstly characterized in S. pyogenes25,29.
It has been suggested that these factors were transmitted from GAS to SDSE species30.
We analyzed speA, speB, speC, speF, speG, speH, speJ, smeZ and ssa genes. All the strains harbored only speJ and ssa genes simultaneously.
In the study by Ikebe et al. from Japan, all isolates were negative to speA, speB, speC, speH, speI, speJ, speL and speM, however, 12 out 16 isolates had the speG gene26.
As shown in the present study and in previous reports, SDSE secretes only a few previously known superantigen exotoxins23,35,46.
The clinical manifestations of STSS caused by SDSE are similar to EGA. speA, speB, and speC genes were not observed in our study although they are the most frequent detected exotoxins in S. pyogenes. Therefore, we suspect that not yet identified superantigen exotoxins or exoenzimes may exist, which could play an important role in the development of STSS caused by SDSE.
Although 60 years have passed since the introduction of PEN, β-hemolytic streptococci still continue being susceptible to this antibiotic, even though some S. agalactiae strains showed reduced susceptibility32.
Macrolide, CLI and TET resistance has been a matter of real concern because of the development of ERY-resistant S. pyogenes outbreaks in Japan, Australia and several Europeans countries since the 70s42,51,56. Macrolide and TET resistance has been observed both in GAS and SDSE31,43.
SDSE isolates (n=212) collected in a multicenter surveillance study by Broyles et al.6 in the United States showed resistance rates of 28.8% to ERY, 4.2% to CLI, and 0.9% to fluoroquinolones.
In Korea39, high frequency of the TET resistance-mediating tet(S) gene was demonstrated (68.8%), while ERY, CLI, and chloramphenicol resistance rates were low (9.4, 3.1 and 9.4%, respectively)61.
Of 231 SDSE isolates in the study by Takahashi et al.60, four harbored the mef(A) gene, and 13 and six isolates carried the erm(A) and erm(B) genes, respectively.
In our study, 19.0% were non-susceptible to CLI and 26.1% to ERY. Only one strain was resistant to ERY (iMLSB phenotype, ermTR positive) and no CLI constitutive resistance was detected in the previous multicenter study40.
In our study, TET resistance was 30.4%. Two tetracycline-resistant strains only carried the tet(M) gen while five others carried both the tet(M) and tet(O) genes.
TET resistance was also common in SDSE during the 1998–1999 period in Argentina (33.3% in group C SDSE and 40% in group G SDSE)40. In such study, TET resistance was 40.7% and all TET resistant strains carried only the tet(M) gen.
In Portugal, a high rate of fluoroquinolone resistance (12%) was detected between 1998 and 200547, although a previous study from Europe and the United States detected less than 1% resistance toward this group of antibiotics2.
All our isolates were in vitro susceptible to LEV. Only one isolate (strain 9) was found to have reduced susceptibility to NOR, PEF and OFL.
Fluoroquinolones act by inhibition of bacterial DNA gyrase and DNA polymerase. The alterations of these enzymes were the prevalent mechanism of resistance in the Streptococcus genus24. The mutation in the gyrA subunits (gyrase DNA) and parC (DNA topoisomerase) are found in the so-called quinolone resistance determining regions (QRDR). The mechanisms involved in the small diameters observed by the disk diffusion test with strain 9 deserve to be further studied.
In the future, we should continue to survey invasive SDSE infections to further clarify the prevalence of infection, the study of new virulence factors, the distribution of emm types and to monitor antibiotic susceptibility rates in the Argentinean population.
Ethical disclosuresProtection of human and animal subjectsThe authors declare that no experiments were performed on humans or animals for this study.
Confidentiality of dataThe authors declare that no patient data appear in this article.
Right to privacy and informed consentThe authors declare that no patient data appear in this article.
Conflict of interestThe authors declare that they have no conflicts of interest.
To Prof. Giselle Blanchiman for the careful proofreading of the manuscript.
The “National Collaborative Group for the Study of Streptococci and Related Bacteria” consists of twenty eight centers belonging to a collaborative group for the study of streptococci and related bacteria: 5 centers from Buenos Aires City (C. Hernández, L. Casimir, M. Litterio and M. del C. Ceinos, Hospital de Pediatría Prof. Dr. Juan P Garrahan; A. Famiglietti, C. Rodríguez and S. García, Hospital de Clínicas G. J. San Martín; C. Vay, Sanatorio Mater Dei; D. Ballester and F. Amalfa, Hospital Parmenio Piñero; R. Pereda, Hospital Pedro Elizalde), 4 from Rosario (N. Borda and R. Notario, Hospital Español; E. Sutich, J. Pérez, G. Cera, M. J. Spoletti, I. Demaría and D. Aguila, Hospital Provincial del Centenario; A. Ernst, A. Badano and A. Aletti, Hospital de Niños Víctor J. Vilela; A. Ponessa, R. Notario, T. Gambandé and L. All, Cátedra de Microbiología, Facultad de Ciencias Médicas Universidad Nacional de Rosario), 2 from Santa Fe (S. Virgolini, Ma. R. Barone and G. Ezcurra, Hospital de Niños Dr. O. Alassia; E. Méndez, Hospital Dr. José María Cullen) 2 from Córdoba (M. Bottiglieri, Clínica Privada Reina Fabiola; P. Montanaro, A. Oreccini and A. Garnero, Hospital de Niños de la Santísima Trinidad), 2 from San Juan (H. Castro, Hospital Marcial Quiroga; O. Navarro, M. López and M. Mengual, Hospital Guillermo Rawson) and 1 each from Mar del Plata (M. Vallejo, N. Rosales, V. Fanjul and M. Gordovil, Hospital Privado de la Comunidad), San Rafael (A. Acosta and C. Baldoni, Hospital Teodoro Schestakow), Mendoza (M. A. Di Stéfano and L. Contreras, Hospital Central de Mendoza), Lanús (A. Togneri, L. Podestá and M. Pérez, Hospital Evita), La Plata (A. Pacha, Hospital San Juan de Dios), Pilar (V. Vilches, Hospital Austral), Bahía Blanca (M. Rizzo, Ma. Luz Benvenutti and L. Giordano, Hospital General de Agudos Dr. José Penna), Neuquén (M. R. Núñez, Hospital Provincial Dr. Castro Rendón), Gral. Pico (S. Cirimele, A. Baroni and D. Ruderman, Establecimiento Asistencial Gobernador Centeno), Santa Rosa (G. Almada, Hospital Lucio Molas), Esquel (O. Daher, Hospital Zonal de Esquel), Posadas (M.E. von Specht, L. Leguizamón and Oscar López, Hospital Provincial de Pediatría Dr. F. Barreyro), and Concordia (Ma. Ofelia Moulins, L. Otaegui and L. Bernhardt, Hospital Masvernat).
The complete list of centers of the National Collaborative Group for the Study of Streptococci and Related Bacteria is included in Appendix 1.