Knowledge regarding the enzymatic machinery of fungi is decisive to understand their ecological role. The species of the genus Geastrum are known to grow extremely slowly in pure culture, which makes it difficult to evaluate physiological parameters such as enzyme activity. Qualitative assays were performed on isolates of four species of this genus, showing evidence of laccase, cellulase, pectinase, amylase and lipase activity and suggesting that a wide range of carbon sources can be exploited by these species. For the first time in this genus, quantitative assays verified manganese peroxidase activity (up to 0.6mU/g) in 30-day old cultures, as well as laccase, β-glycosidase and β-xylosidase activities.
El conocimiento de la maquinaria enzimática de un hongo es decisivo para entender su rol ecológico. Las especies del género Geastrum son conocidas por su crecimiento extremadamente lento en cultivos puros, lo que hace difícil la evaluación de parámetros fisiológicos como las actividades enzimáticas. Se realizaron ensayos cualitativos sobre aislamientos de 4 especies de este género, mostrando evidencias de actividades lacasa, celulasa, pectinasa, amilasa y lipasa, mostrando el amplio rango de fuentes de carbono que pueden ser explotadas por estas especies. Ensayos cuantitativos verificaron por primera vez en este género la actividad manganeso peroxidasa (hasta 0,6 mU/g) en cultivos de 30 días, así como también β-glucosidasa y β-xilosidasa.
Fungal extracellular enzymes have been a subject of study not only because of their intrinsic interest, but also due to their biotechnological applications. For this reason, most studies have been based on fast growing organisms that produce high enzyme activities, and on model species whose genetics and physiology are well known.
The finding of lignin-modifying enzymes in litter or soil fungi5 redirected the attention of many scientists to the adaptive role they may play in this environment. Recent studies based on genomic data provide invaluable information about the enzyme machinery of many fungal taxa in relation to their ecology11 and phylogenetic placement1; however, to date, no studies of this kind have included members of the family Geastraceae.
Since degradative fungal enzymes may have an important role in bioremediation scenarios, the study of enzyme activities is also relevant for soil and litter fungal species regardless of their possible industrial application. One of the problems in this field is the slow growth in culture of many species (e.g. ectomycorrhizal fungi), which is usually the main obstacle to their enzyme research. An example of this fact is the genus Geastrum, which, though entirely saprotrophic, shows an extremely slow growth rate. Many of its species are known to form extensive mycelial networks and cushions in all kinds of forests, tolerating inhibitory substrates such as Eucalyptus or Cupressaceae litter10.
This genus has been the subject of exhaustive taxonomic and floristic studies. Sunhede10 made a systematic revision and took thorough notes on its morphology, biology and ecology, however, at present just a few works make reference to its ecophysiology. Sunhede10 describes the growth of the isolates as “very slow, after six weeks varying from 3 to 22mm.” Ilko et al.4 indicate that the colony of Geastrum pouzarii reached 3–4mm after six weeks and Zamora et al.,14 report isolates of G. argentinum achieving up to 4mm after a week. This characteristic makes these species hard to isolate and to maintain in pure cultures, and this is the main reason why there are no reports of their exoenzyme activities. Sunhede10 reports oxidase enzymatic activity in some species by observing metachromatic staining of mycelia with some laccase and tyrosinase substrates. Zamora et al.13 carefully studied similar reactions in fresh basidiomes; however, no attempts to study in vitro enzymatic activity have been performed until now.
Four Geastrum species were isolated and studied, namely G. argentinum, G. papinuttii, G. morganii, and G. plicatum. Specimens were collected in the province of Buenos Aires and deposited in BAFC (Universidad de Buenos Aires), LPS (Universidad Nacional de La Plata) and MA-fungi (Real Jardín Botánico de Madrid) herbaria (Index Herbariorum, http://sweetgum.nybg.org/science/ih/)12. Isolates were obtained from the pseudoparenchymatous layer of the exoperidium in malt agar medium, and deposited in BAFC Culture Collection under the following numbers (herbarium collection numbers in parentheses): G. argentinum BAFC 3282 (LPS 48446), G. plicatum BAFC 3283 (MA-Fungi 83774), G. morganii BAFC 3284, G. papinuttii BAFC 3285 (MA-Fungi 83764). Three of the four species (G. argentinum, G. plicatum, and G. papinuttii) were molecularly characterized by Zamora et al.15,16, and GenBank accession numbers are available there for ITS (Internal Transcribed Spacer) nrDNA (nuclear ribosomal DNA), LSU (Large Subunit) nrDNA, rpb1 (polymerase II largest subunit gene) and atp6 (ATP synthase subunit 6 gene).
An agarized medium containing glucose and glutamic acid (GG medium)3 was utilized for the screening. Laccase (EC:1.10.3.2) activity was quantified according to Paszczynski et al.8 and MnP (manganese peroxidase; EC:1.11.1.13) following Paszczynski et al.9 with and without MnCl2 0.1mM as inducer. In all cases, aliquots of supernatant were replaced by agar plugs (5mm diameter) provided by 60-day malt agar cultures. β-xylosidase (EC:3.2.1.37) and β-glucosidase (EC:3.2.1.21) enzyme activities were determined by measuring the release of p-nitrophenol from p-nitrophenyl-β-d-xylopyranoside (pNPX) and p-nitrophenyl-β-d-glucopyranoside (pNPG). Agar plugs were incubated in 900μl of 1 mM substrate at 50°C for 30min. The reaction was stopped by adding 2ml of Clark and Lubs buffer pH 9.8. The release of p-nitrophenol was measured using a UV-vis spectrophotometer at λ 405nm and 1 unit of β-xylosidase activity was defined as the amount of the enzyme releasing 1μmol p-nitrophenol equivalent per minute under the assayed conditions. Hydrolytic enzyme activities were qualitatively evaluated according to Pardo and Forchiassin7, replacing glucose in each assay by starch 0.1% (for amylase EC:3.2.1.1 activity), pectin 0.1% (for pectinase EC:3.2.1.15 activity), carboxymethyl-cellulose (CMC) 0.1% (for endoglucanase EC:3.2.1.4 activity) and xylan 0.1% (for xylanase EC:3.2.1.8 activity). Lipase (EC:3.1.1.3) qualitative assay was as indicated by Nikoleit et al.6 using Tween 80 as a carbon source but replacing the medium by basal GG medium3 without a carbon source. Laccase and LiP (lignin peroxidase) activity were also qualitatively evaluated in a GG medium by adding dimethoxyphenol 0.1mM and Azure B. All data presented are the means of the results of triplicates and error bars indicate SE.
All isolates showed active growth in each assayed carbon source (xylan, cellulose, starch and pectin). The use of lipid esters as a carbon source was also demonstrated in G. plicatum through the active growth and degradation of Tween 80.
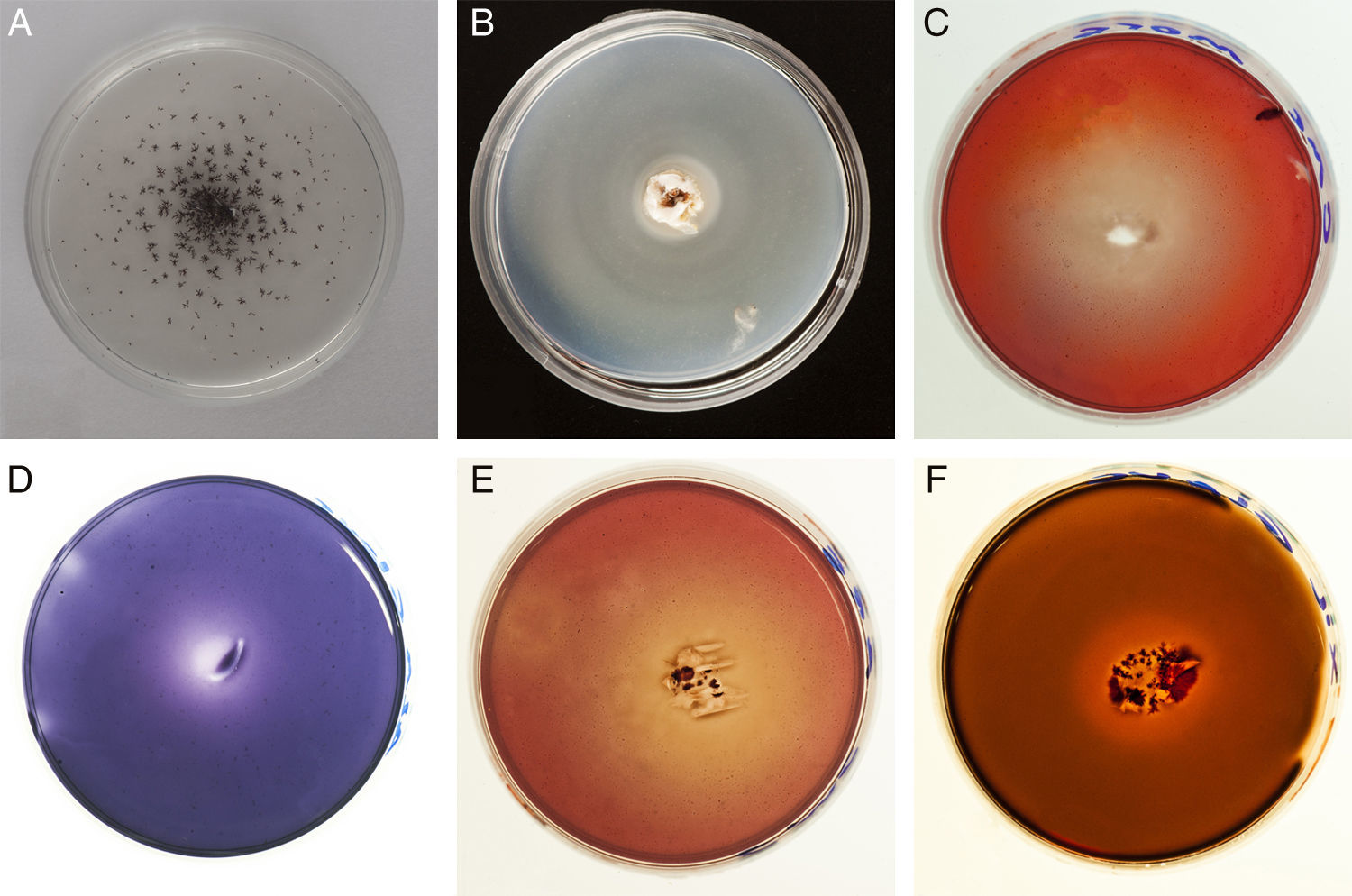
Enzyme activities evidenced by qualitative assays are shown in Fig. 1. Activity halos reaching higher diameter than the colony evidence the activity of exoenzymes. The degradation of carbon sources is represented in Table 1 by the diameters of degradation and growth halos corresponding to day 20. A weak decolorization halo in the cultures containing Azure B suggests LiP activity in G. papinuttii, while laccase activity was evidenced 24h after inoculation in every isolate.
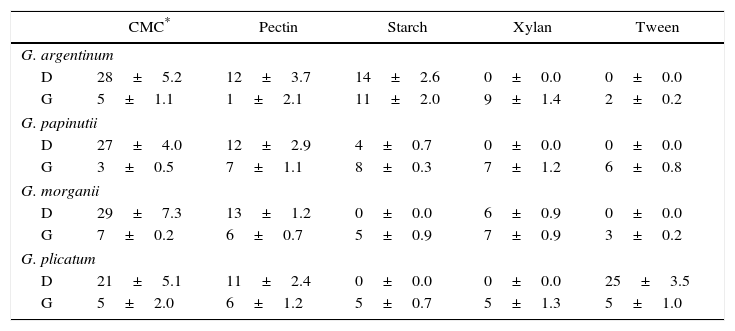
Diameter of degradation (D) and growth (G) halos (mm) on different carbon sources after 30 days of culture by the four isolates
| CMC* | Pectin | Starch | Xylan | Tween | |
|---|---|---|---|---|---|
| G. argentinum | |||||
| D | 28±5.2 | 12±3.7 | 14±2.6 | 0±0.0 | 0±0.0 |
| G | 5±1.1 | 1±2.1 | 11±2.0 | 9±1.4 | 2±0.2 |
| G. papinutii | |||||
| D | 27±4.0 | 12±2.9 | 4±0.7 | 0±0.0 | 0±0.0 |
| G | 3±0.5 | 7±1.1 | 8±0.3 | 7±1.2 | 6±0.8 |
| G. morganii | |||||
| D | 29±7.3 | 13±1.2 | 0±0.0 | 6±0.9 | 0±0.0 |
| G | 7±0.2 | 6±0.7 | 5±0.9 | 7±0.9 | 3±0.2 |
| G. plicatum | |||||
| D | 21±5.1 | 11±2.4 | 0±0.0 | 0±0.0 | 25±3.5 |
| G | 5±2.0 | 6±1.2 | 5±0.7 | 5±1.3 | 5±1.0 |
Each value represents the means of three independent cultures±standard error.
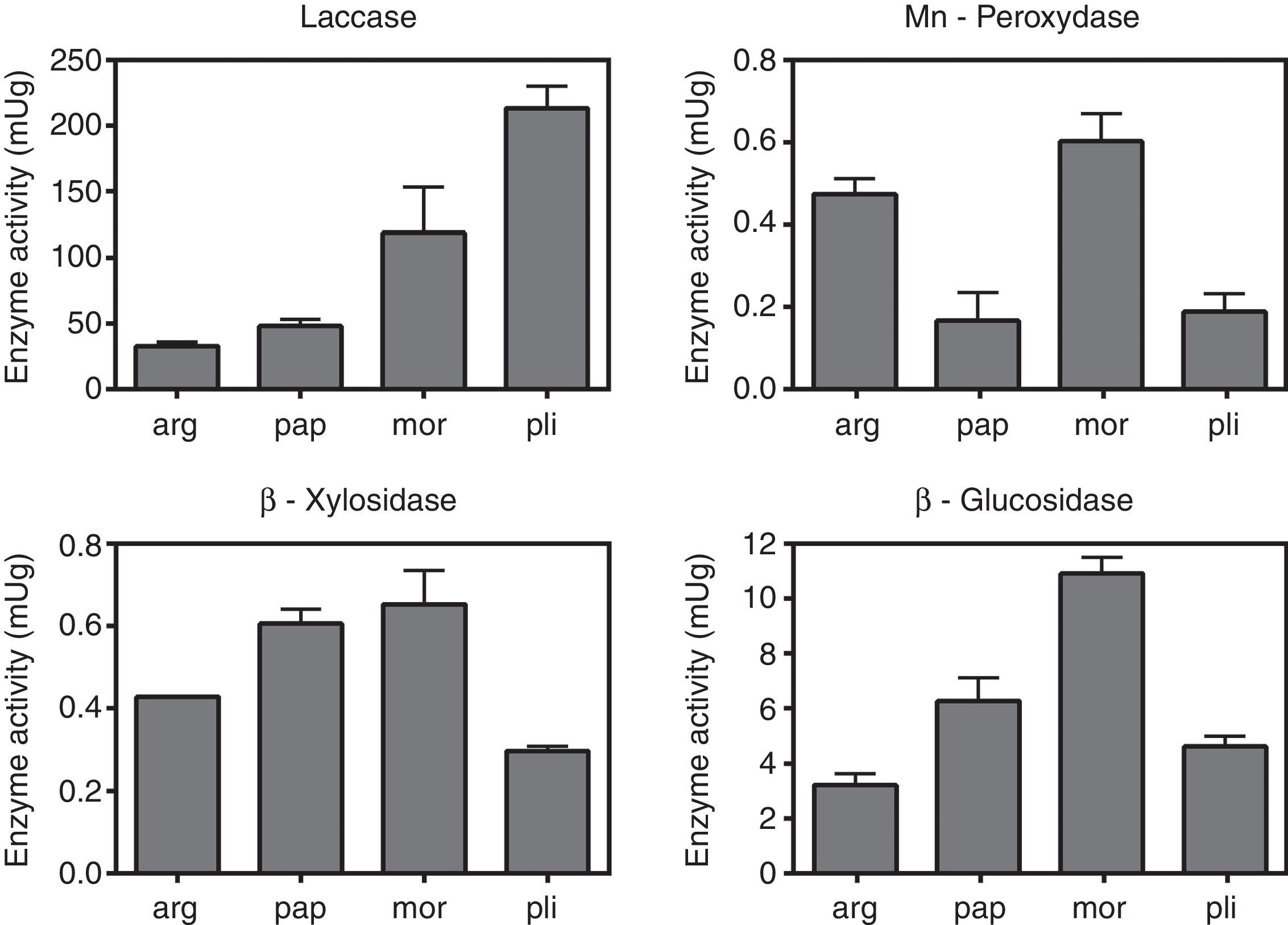
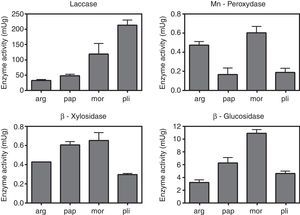
The values of laccase, MnP, β-glucosidase (EC:3.2.1.21) and β-xylosidase (EC:3.2.1.37) activities are shown in Fig. 2. Uninduced cultures did not show MnP activity. The results corresponding to this enzyme were measured in cultures induced with MnCl2 0.1mM. The absence of degradation halos in some qualitative assays (Table 1) may reflect that the carbon source was not actively degraded by the strain, and the positive growth in those cases could be explained by the use of the amino acid, instead. Manganese peroxidase activity was only detectable after induction with MnCl2. Laccase, as well as β-glucosidase and β-xylosidase showed intrinsic activity in the absence of inducers.
Quantification of enzimatic activities in agar plugs. arg, G. argentinum; pap, G. papinuttii; mor, G. morganii; pli, G. plicatum. Bars represent the mean enzyme activity of laccase, MnP, β-glucosidase and β-xylosidase per gram of agarized medium. MnP quantification corresponds to cultures induced with MnCl2. Uninduced cultures did not show MnP activity. Values represent means±SE of three independent cultures.
Since the whole Geastraceae family forms dense mycelia, matting together organic debris10, the enzymatic machinery should be adapted to decay this substrate. In fact, Fuji et al.2, suggest that laccase and MnP (which showed to be active in our isolates) are involved in the early stages of litter decomposition. G. argentinum is often associated with fallen trees13; however, the advanced decay of the logs suggests that the lignocellulolytic activities found in our work are not correlated with an active degradation of wood matter. Sunhede10 also reports that basidiomes of Geastrum species are often found on moldering stumps. β-Xylosidase and β-glucosidase activities, together with CMC and pectin halos (shown by all isolates), account for their capacity of exploiting cellulosic substrates. The differential behavior of other hydrolytic enzymes such as xylanases, amylases and lipases suggests different strategies of organic matter degradation. For instance, the positive reaction of G. plicatum in the lipase assay may be related to the degradation of persistent waxy cuticles of leaves; however, more isolates of each species are needed to confirm these patterns. The possible interactions of Geastrum species with other organisms enhancing the degradation of cellulosic materials as well as the role of some Geastrum species in late stages of wood debris degradation need to be further investigated. The detection of MnP and laccase activities in the tested isolates suggests that they might be a promising source of enzymes with the ability to transform xenobiotics, although their extremely slow growth entails the fact that no high scale biotechnological application of these enzymes is possible without cloning them in more efficient organisms.
Ethical disclosuresProtection of human and animal subjectsThe authors declare that no experiments were performed on humans or animals for this study.
Confidentiality of dataThe authors declare that no patient data appear in this article.
Right to privacy and informed consentThe authors declare that no patient data appear in this article.
Conflict of interestThe authors declare that they have no conflicts of interest.
This work was supported by Consejo Nacional de Investigaciones Científicas y Técnicas, Argentina (PIP 11220120100408CO), Universidad de Buenos Aires (UBACyT, 20020120200085), Ministerio de Ciencia and Tecnología e Innovación ProductivaPICT 3457/14. We also thank the Consejo Superior de Investigaciones Científicas (Spain) for supporting JCZ with the grant JAE-Pre.