Invasive Streptococcus pyogenes diseases represent the most severe form of infection produced by this microorganism. Early diagnosis and treatment are important, due to its potential severity. Etiological confirmation of invasive infection is performed by culture, which takes between 18 and 48h. We tested a rapid immunochromatographic assay directly from clinical samples from normally sterile sites and positive blood culture bottles when positive cocci chains were observed by Gram staining. Eighty samples were analyzed. The rapid test was positive in 35 samples: in 34 of them S. pyogenes was confirmed by culture. The immunochromatographic method showed 97.1% sensitivity and 97.8% specificity. The strept A® immunochromatographic rapid test allows to obtain reliable results in less than 10min and is accessible to any microbiology laboratory. This study demonstrates the potential use of a rapid immunochromatographic method directly from clinical samples and positive blood cultures.
La enfermedad invasiva por Streptococcus pyogenes representa la forma más grave de infección producida por este microorganismo y requiere un rápido diagnóstico, a fin de instaurar un tratamiento adecuado. La confirmación etiológica de esta infección se realiza por cultivo, lo que puede llevar entre 18 y 48h. En este estudio ensayamos una prueba inmunocromatográfica rápida directamente de muestras clínicas de sitios normalmente estériles y de botellas de hemocultivos positivos cuando la coloración de Gram evidenció cocos gram positivos en cadena. Se analizaron 80 muestras. La prueba rápida fue positiva en 35 muestras: en 34 de ellas se confirmó la presencia de S. pyogenes por cultivo. La sensibilidad y la especificidad de la prueba fueron del 97,1 y el 97,8%, respectivamente. La prueba inmunocromatográfica rápida monteBIO Strep A® permite obtener resultados confiables en menos de 10 min y es accesible para cualquier laboratorio de microbiología. Este estudio demuestra la utilidad de dicha prueba para ser practicada directamente en muestras clínicas y botellas de hemocultivos positivos.
Streptococcus pyogenes or group A Streptococcus is a gram positive coccus growing in chains of varying length, displaying white or gray β-hemolytic colonies larger than 0.5mm on 5% sheep blood agar after 24h of incubation. Its numerous virulence factors (M protein, pyrogenic exotoxins, hyaluronic acid capsule, hemolysins and other factors) allow it to cause a wide array of infections including pharyngitis, impetigo and other more serious infections such as pneumonia, cellulitis, and necrotizing fasciitis, and less frequently meningitis, endocarditis, sepsis and arthritis. Infections with toxin-producing strains can result in scarlet fever or more serious toxic shock-like symptoms. It is a strictly human pathogen, associated with 500000 annual deaths worldwide3,11,12. In recent decades there has been an overall increase in invasive infections in Europe and the United States, but the cause has not been determined. In Europe, the incidence is 2.79 cases/100000 inhabitants/year5.
Although up to now in Argentina, there has been no active surveillance of invasive S. pyogenes infections; several multicenter studies have been conducted. One of them carried out between 1998 and 1999 and including 40 centers in 16 cities, described 68 invasive S. pyogenes infections in six months8. Another one-year multicenter study, conducted between 2011 and 2012 and including 28 centers in 17 cities, identified 50 invasive S. pyogenes infections in pediatric patients7.
Invasive diseases due to S. pyogenes represent the most severe form of presentation. They have variable and often nonspecific clinical presentation. Early diagnosis and treatment are important, due to its potential severity, which sometimes requires intensive support and assistance4.
A validated rapid test for the etiological diagnosis of streptococcal pharyngitis from throat swab specimens is now available (monteBIO Strep A® immunochromatography kit). It is presented as a qualitative immunoassay, yielding results in less than an hour. However, in case of invasive infections, the etiological confirmation is performed by the isolation and subsequent identification of the pathogen, which takes between 18 and 48h. In the present study, this rapid immunochromatographic assay was tested directly from clinical samples from normally sterile sites and positive blood culture bottles.
This study was carried out in the clinical microbiology department of an Argentinean third level pediatric hospital between January 2016 and January 2019. S. pyogenes antigen detection was performed using the monteBIO Strep A® immunochromatographic kit (made in: Abon Biopharm, Hangzhou. P.R. China, imported and distributed by MONTEBIO S. R. L. Buenos Aires, Argentina) directly in 11 samples from normally sterile sites and 69 positive blood cultures. In all cases, the rapid test results were read blindly, without knowing the final result that would later be confirmed by culture. The interpretation was classified as: positive, when 2 lines appeared in the immunochromatographic strip, one in the region of the control and another in the test region, colored with the same intensity or when the intensity of the test line was greater than that of the control; weak positive, when the intensity of the line of the test region was very weak; and negative, when only the line in the control region was colored. Samples from pediatric patients with a clinical diagnosis of infection in skin and soft tissues, pneumonia with pleural effusion, pericarditis or septic arthritis were included when positive cocci chains were observed by direct examination of the specimen by Gram staining. Samples from patients with abdominal focus, post-surgical samples, and those in which polymicrobial microbiota was observed in the direct examination were excluded.
Blood samples were inoculated in PF Plus® bottles and FN plus® and incubated in the BacT/Alert 3D® automated system for 5 and 7 days respectively. The immunochromatographic test was performed on positive blood culture bottles that met the inclusion criteria. Briefly, 4 drops of reagent A, 4 drops of reagent B (provided in the kit) and 2 drops of the positive blood sample were added to the extraction tube. Then, it was incubated for 1min at room temperature and finally the test strip was introduced. After 5min the result was recorded following the manufacturer's recommendations. To perform the immunochromatographic assay in purulent samples, these were diluted in equal parts with sterile physiological solution and the same procedure as in the positive blood culture broths was followed.
To ensure that test reagents were working properly and the test was correctly performed, an external positive control was carried out. Two hundred microliters of a 103S. pyogenes (ATCC 19615) inoculum plus 2ml of sterile sheep blood were inoculated into a PF Plus® bottle, which was incubated in a BacT/Alert 3D® automated system until positivity was detected. Then the immunochromatographic test was performed as indicated above. In this way, the entire assay was monitored.
Broths, samples of normally sterile liquids such as pleural fluid, joint fluid, material from skin puncture and soft tissue were further cultured on chocolate agar and 5% sheep blood agar and incubated for 48h at 37°C in a 5% CO2 atmosphere. Gram positive cocci in chains were identified from the isolated colonies by the MALDI-TOF MS method (Vitek MS® system).
Sensitivity, specificity, positive predictive value and negative predictive value of the technique were calculated. Data were loaded in an Excel spreadsheet, and Stata 10 software was used for the statistical analysis.
The control bottle containing S. pyogenes (ATCC 19615) was positive in the BacT/Alert 3D® automated system at 0.51 days and the rapid immunochromatographic test was positive.
During the study period, 80 samples were analyzed, including 69 blood cultures, five samples of cellulitis, two of pleural fluids, three of joint fluids and one of pericardial fluid. S. pyogenes was isolated from 35 samples. Different species of other positive cocci were isolated from 43 samples. A non-spored gram positive rod was isolated in one sample. In one case the culture was negative.
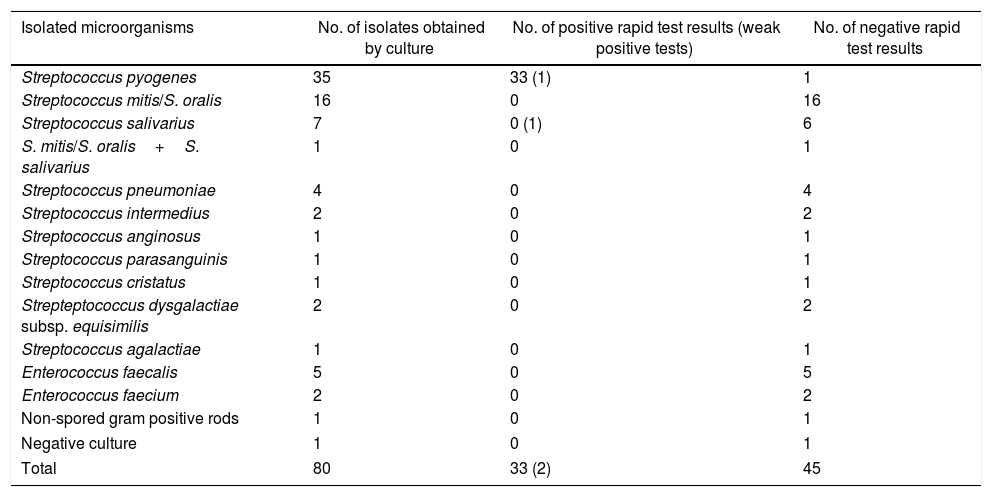
The rapid test was positive for 35 samples: S. pyogenes was detected by culture in 34 of them, and Streptococcus salivarius was identified in one case. Negative tests were recorded in 45 cases: S. pyogenes was detected in one of them by culture (false negative), other organisms grew in 43 samples, and in only one case the culture was negative (Table 1).
Rapid immunochromatographic assay results.
| Isolated microorganisms | No. of isolates obtained by culture | No. of positive rapid test results (weak positive tests) | No. of negative rapid test results |
|---|---|---|---|
| Streptococcus pyogenes | 35 | 33 (1) | 1 |
| Streptococcus mitis/S. oralis | 16 | 0 | 16 |
| Streptococcus salivarius | 7 | 0 (1) | 6 |
| S. mitis/S. oralis+S. salivarius | 1 | 0 | 1 |
| Streptococcus pneumoniae | 4 | 0 | 4 |
| Streptococcus intermedius | 2 | 0 | 2 |
| Streptococcus anginosus | 1 | 0 | 1 |
| Streptococcus parasanguinis | 1 | 0 | 1 |
| Streptococcus cristatus | 1 | 0 | 1 |
| Strepteptococcus dysgalactiae subsp. equisimilis | 2 | 0 | 2 |
| Streptococcus agalactiae | 1 | 0 | 1 |
| Enterococcus faecalis | 5 | 0 | 5 |
| Enterococcus faecium | 2 | 0 | 2 |
| Non-spored gram positive rods | 1 | 0 | 1 |
| Negative culture | 1 | 0 | 1 |
| Total | 80 | 33 (2) | 45 |
Tested directly in samples from normally sterile sites or positive blood cultures in which positive cocci in chains were observed on direct examination.
Two of the 35 rapid test positive samples were considered to be weak positive: one was a joint fluid sample with a positive culture for S. pyogenes and the other was a blood culture sample with S. salivarius (false positive).
The immunochromatographic method showed a sensitivity of 97.1% (95% CI: 85.5–99.5%), a specificity of 97.8% (95% CI: 84.5–99.6%), with a positive predictive value of 97.1% and a negative predictive value of 97.8%.
A delay in the initiation of effective antimicrobial therapy for streptococcal invasive diseases has a high impact on mortality. Therefore, the early recognition of the disease is a matter of real concern6.
The rapid Strep A immunochromatographic strip test is an easy-to-perform method that was only validated for diagnosing streptococcal pharyngitis from throat swab samples. The clinical sensitivity and specificity of this method in throat swab samples are 97% (95% CI: 91–99%) and 95% (95% CI: 92–97%) respectively according to the manufacturer's correlation studies.
With this method performed on samples from normally sterile sites with gram positive cocci chains observed by Gram staining we obtained a false positive result in a blood culture sample in which S. salivarius was isolated.
The kit manufacturer specifies in the package insert that: “the color intensity of the line in the region of the test line may vary depending on the concentration of the Strep A antigen present in the sample. Therefore, any color tonality in the region of the test line should be considered positive”. However, the manufacturer did not include S. salivarius in the validation test to evaluate cross-reactivity. For this reason, when a weak color line is observed in the region of the test line in blood culture samples, we consider that it would be prudent not to interpret it as “positive”.
Although the immunochromatographic rapid test showed high sensitivity and specificity, we recommend confirming this result by culture and subsequent identification.
On the other hand, in only one case the rapid Strep A immunochromatographic strip was negative and S. pyogenes was identified by culture. This false negative result could be explained by the purulent nature of the sample, by an inoculum level below the limit of detection of the test or by the prozone effect.
There are other methodologies for detecting S. pyogenes directly from clinical samples, such as: qPCR, which is a quantitative polymerase chain reaction assay that detects and amplifies bacterial DNA and sequences the genes that encode the 16S ribosomal RNA region2,9. Although this is a sensitive, reliable and faster method than conventional cultures, it requires equipment, supplies and an adequate infrastructure for molecular biology. This technique can take from 2 to 6h approximately. Filmarray® (Biomerieux) is another molecular technique that can be performed from positive blood cultures. It is a multiplex PCR that integrates sample preparation, amplification, detection and analysis in a closed platform. It may detect 24 etiological agents of sepsis, including S. pyogenes in 1h. Its sensitivity exceeds 96% for all microorganisms included in the panel10. An advantage of this technique is that a specific area of molecular biology in laboratories is not required. However, equipment and supplies may be very expensive.
Mass spectrometry, MALDI-TOF MS, is a rapid and highly specific technique that can be performed from positive blood culture samples. However, it requires preprocessing of the sample by washing and centrifugation to eliminate red blood cells and interfering culture medium. This step can take approximately 30–40min. Although reagents are not expensive, the limitation of this methodology is the high cost of the equipment and its maintenance1.
In comparison, the Strep A immunochromatographic rapid test allows to obtain reliable results in less than 10min and is accessible to any microbiology laboratory since special equipment or infrastructures are not needed.
This study demonstrates the potential use of a rapid immunochromatographic method directly from clinical samples and positive blood cultures, despite the manufacturer's warning note as to its limitations indicating that blood may interfere and could be responsible for false positive results.
To our knowledge this is the first study to evaluate the Strep A immunochromatographic rapid test in samples from normally sterile sites. The main strength of this study is the type and number of samples where the test was evaluated.
Ethical considerationsThe samples analyzed in this research were processed without the patients’ intervention and without altering the standardized microbiological diagnostic operating procedures for our hospital. Patient details were anonymized before data analysis, ensuring the absolute confidentiality of the information obtained and for the unique purpose of the present investigation. The procedures followed were in accordance with the Personal Data Protection Law No. 25.326, art 11, subsection 3, point d; in line with the Declaration of Helsinki. For this reason, the Ethics Committee of our Hospital did not consider it necessary to obtain informed consent from individual patients.
Funding sourcesThis research did not receive any specific grant from funding agencies in the public, commercial, or not-for-profit sectors.
Conflicts of interestNone declared.
The authors thank Prof. Dr. Horacio Lopardo for his continued support in our researches.