Covalently closed circular (ccc)DNA acts as a viral reservoir in the liver of patients with a chronic hepatitis B (CHB) infection and can only be quantified in liver biopsies. Hepatitis B core-related antigen (HBcrAg) levels in plasma/serum have been proposed to reflect intrahepatic cccDNA-levels and may therefore monitor treatment efficacy. This study aimed to validate the relationship between HBcrAg and other intrahepatic and circulating viral markers in CHB patients with high viral load, before and after combination treatment.
Materials and methodsPlasma/serum levels of HBcrAg, HBsAg, HBV-DNA, and HBV pregenomic RNA (HBV-pgRNA), and intrahepatic cccDNA and HBV-DNA levels and fibrosis scores were measured in 89 CHB patients with HBV-DNA levels of >100,000 copies/mL (17,182 IU/mL). Measurements were done before and after a 48-week treatment with pegylated interferon alfa-2a and adefovir in a prospective study (ISRCTN77073364).
ResultsBaseline HBcrAg-values correlated strongly with intrahepatic cccDNA (ρ 0.77, p < 0.001), intrahepatic HBV-DNA (ρ 0.73, p < 0.001) and plasma/serum HBV-DNA (ρ 0.80, p < 0.001), HBV-pgRNA (ρ 0.80, p < 0.001), and to lesser extend HBsAg (ρ 0.56, p < 0.001). Baseline HBcrAg-levels could not predict functional cure (FC) but HBcrAg-levels declined more strongly in patients who developed FC or HBeAg-loss. Furthermore, most correlations persisted at the end of treatment and follow-up.
ConclusionsHBcrAg reflects cccDNA transcription activity more accurately than HBsAg and may replace HBV-DNA as a marker during future treatment regimens, especially when cccDNA transcription is targeted or nucleot(s)ide analogues are included in the treatment regime.
List of AbbreviationsCHB
Chronic Hepatitis B
HCCHepatocellular Carcinoma
cccDNACovalently Closed Circular DNA
NANucleot(s)ide Analogue
PEG-IFNPegylated Interferon
pgRNAPregenomic RNA
CCComplete Cure
FCFunctional Cure
HBsAgHepatitis B surface antigen
HBcrAgHepatitis B core-related antigen
HBeAgHepatitis B e antigen
p22cr22 kDa precore protein
AMCAcademic Medical Centre
EOTEnd Of Treatment
EOFUEnd Of Follow-up
LoDLower limit of Detection
PCRPolymerase Chain Reaction
LoQLower limit of Quantification
ALTAlanine transferase
iHBV-DNAintrahepatic Hepatitis B Virus DNA
PCAPrincipal Component Analysis
ROCReceiver Operating Characteristic
Lay summary
The standard viral markers for monitoring viral activity in the plasma/serum of patients with a chronic hepatitis B infection (CHB), may not be of use in future combination therapies where these markers are supressed. In this study, we assessed the use of the novel viral markers: Hepatitis B core-related antigen (HBcrAg) in patients treated with a combination of a viral suppressor and immune modulator. We found that HBcrAg levels before treatment could not predict treatment response. However, HBcrAg levels gave a good reflection of the viral activity and the viral reservoir in the liver, before and after treatment. Therefore, HBcrAg can replace the currently used markers for monitoring CHB. This of special interest for future therapies under which standard markers lose their value.
Chronic hepatitis B (CHB) patients are at increased risk of developing liver cirrhosis, liver failure and Hepatocellular carcinoma (HCC). Worldwide, more than 250 million people are chronically infected with the hepatitis B virus (HBV), leading to a yearly mortality rate of 887,000 [1].
In the infected hepatocytes, the cell nucleus contains covalently closed circular (ccc)DNA that acts as the template for HBV replication [2]. This viral reservoir of cccDNA poses the biggest challenge in developing a curative treatment for CHB [3]. Current treatment options are: viral suppression using nucleot(s)ide analogues (NAs) and in some cases immunomodulatory treatment with pegylated interferon (PEG-IFN). Treatment with NAs prevents the formation of new viral particles by blocking reverse transcription of pregenomic RNA (pgRNA) into DNA, but it does not interfere with cccDNA transcription and the formation of viral proteins. PEG-IFN is less preferable due to its many side effects and moderate effectiveness. Elimination of the virus including cccDNA is considered ideal but currently unfeasible. A more achievable goal is a functional cure (FC), a state in which cccDNA transcription is silenced and is defined by the most recent guidelines as undetectable serum Hepatitis B surface antigen (HBsAg) [4-6]. Patients without liver cirrhosis, who develop FC are not at risk of developing HBV-related cirrhosis, low risk of HCC and do not need medical surveillance. Currently, there are several new compounds under development that aim to achieve FC, including immune modulators, capsid assembly modulators, RNA-interference, antisense molecules, entry inhibitors and HBsAg-release inhibitors [7]. Combining therapies is likely essential to achieve a high rate of functional cure after treatment. These therapies, most presumably, will include synergic direct acting antivirals and an immune modulator [4].
Quantifying cccDNA and its transcriptional activity would be an ideal marker for monitoring treatment outcome [8,9]. However, liver biopsies are needed for cccDNA measurement. This procedure should be avoided since it is invasive and unsuitable for frequent repeated measurements over time. Therefore, viral activity and treatment response in CHB patients is currently monitored using plasma/serum HBV-DNA and HBsAg-levels [5]. In NA-treated patients however, HBV-DNA is supressed and loses its correlation with cccDNA-activity. Quantitative HBsAg is a less reliable surrogate marker for cccDNA [10,11]. Nevertheless, HBsAg has been used as marker for treatment efficacy in various new antiviral therapies currently under development [7].
New, non-invasive markers for the intrahepatic viral reservoir activity are needed. Hepatitis B core-related antigen (HBcrAg) and HBV-pgRNA in plasma, are suggested to serve this purpose. The HBcrAg assay measures the shared epitope of HBcrAg, Hepatitis B e antigen (HBeAg) and the 22 kDa precore protein (p22cr). These proteins are products of transcription of the (pre)core gene of cccDNA [12]. HBV-pgRNA is a direct product from cccDNA transcription and the precursor of HBV-DNA. Both HBcrAg and HBV-pgRNA have been reported to correlate with HBV-DNA-levels and intrahepatic cccDNA-activity, especially in Asian cohorts [13,14]. In contrast to HBV-pgRNA, HBcrAg is measured using a well-standardized, commercially available test, which is of particular interest to monitor the efficacy of new antiviral therapies targeting cccDNA. The value of HBcrAg as surrogate marker for cccDNA during and before CHB treatment is however largely unknown.
The aim was to validate the clinical value of HBcrAg measurements in CHB patients with a high viral load, before and after peg-IFN and NA combination treatment by comparing HBcrAg levels to both standard (HBV-DNA, HBsAg) and new serum/plasma markers (HBV-pgRNA) as well as intrahepatic markers (cccDNA and HBV-DNA) at different time points. In addition, we aimed to investigate if HBcrAg-levels at baseline were predictive for FC after treatment.
2Materials and Methods2.1PatientsCHB patients (n=92) participated in a prospective investigator-initiated study in the Academic Medical Centre (AMC) and the Erasmus University Medical Centre (EMC) Rotterdam, from 2009 to 2011 as described previously [2],[15],[16]. Patients with HBV-DNA-levels of >100,000 copies/mL (17,182 IU/mL) were included and treated with 180 μg peg-IFN-alfa-2a (Pegasys; Hoffman La Roche, Basel, Switzerland) once weekly and 10 mg of adefovir dipivoxil (Hepsera; Gilead Sciences, Foster City, CA, USA) daily, for 48 weeks (end-of-treatment (EOT)). Patients were followed-up for a total of 144 weeks after start of study (end of follow-up, EOFU). Serum and plasma samples were collected and stored (-80°C) at various time-points including week 42, week 48 (EOT), week 56 and week 72 from baseline. Liver biopsies were performed at baseline and at EOT. Treatment outcomes were FC, defined as HBsAg-loss; and HBeAg-loss in HBeAg-positive patients. The study was conducted according to the guidelines of the Declaration of Helsinki and the principles of good clinical practice, and was approved by local ethics committees (ISRCTN 77073364). All patients gave written informed consent.
2.2HBcrAg measurementHBcrAg-testing was performed on serum/plasma samples at baseline, EOT and week 56 according to the manufacturing protocol (Fujirebio Europe, Gent, Belgium). Samples were treated with a pre-treatment solution and denatured at 60℃ for 30 minutes. HBcrAg-quantification was done using Lumipulse G HBcrAg assay on the Lumipulse G1200 analyzer (Fujirebio Europe, Gent, Belgium). Measurement range was 3-7 log10 U/mL. Samples that reached upper limit of detection were diluted with Specimen Diluent 1 for Lumipulse assays and retested.
2.3Virological and biochemical markersHBV-DNA level in plasma was determined by the Cobas TaqMan 48 assay (F. Hoffmann-La Roche, Ltd), with detection range 20-1.70×108 IU/mL. Samples under the lower limit of detection (LoD) were set at 10 IU/mL (58 copies/mL or 1.76 log10 copies/mL) for statistical analysis. For the detection of HBV-pgRNA, an in-house developed sensitive polymerase chain reaction (PCR) assay was used [2] (lower limit of quantification (LoQ) 2.85 log10 copies/mL, LoD 1.85 log10 copies/mL). Samples under LoQ or LoD were set at 2.55 log10 copies/mL and 1.55 log10 copies/mL respectively. Quantitative HBsAg-levels were measured using the Architect assay (Abbott, Abbott Park, Illinois), (LoD <0.05 IU/mL). HBV-genotype was determined by sequencing a part of the polymerase gene with dideoxynucleotide technology or using the INNO-LiPA assay (Fujirebio, Europe, Gent, Belgium). Alanine transaminase (ALT) levels were analysed in the local clinical chemistry lab with normal values of 0-34 U/L for female and 0-45 U/L for male patients.
2.4Liver biopsy testsLiver biopsies were cryo-preserved in liquid nitrogen. Intrahepatic cccDNA and intrahepatic HBV-DNA (iHBV-DNA) levels were determined using in-house developed assays [2,17]. Total DNA was extracted and purified from liver tissue. Before cccDNA quantification, relaxed DNA was digested using plasmid safe adenosine triphosphate (ATP) dependent DNase (Epicentre Biotechnologies, Madison, WI, USA). A real-time PCR was used for the detection and quantification of cccDNA (LoQ, 150 copies/biopsy) using selective primers which target the gap region between the two Direct Repeat regions (DR1 and DR2) only present in cccDNA. Amounts of iHBV-DNA were quantified using a real-time PCR with genotype independent primers. CccDNA and iHBV-DNA-levels were corrected for the number of hepatocytes per biopsy. Fibrosis in liver biopsies was histologically assessed using the Ishak scoring system (0-6) [18].
2.5Statistical analysisData were analysed using the statistical program for Social Sciences (SPSS 25.0.0.1, Chicago, Illinois, USA). Missing data points were not replaced or included in analysis. Groups were compared using student's t-test, Mann-Whitney-U test, Kruskal-Wallis or ANOVA where appropriate. Multivariable logistic regression was used to assess HBcrAg as independent predictor of therapy response. Correlations between variables were analysed by the Spearman-rank correlation test and compared using a Fisher r-to-z transformation. Data reduction was done by a principal component analysis (PCA) followed by k-means unsupervised clustering. Diagnostic performance of HBcrAg-levels as marker for HBeAg-seropositivity was evaluated using receiver operating characteristic (ROC) curves. Statistical significance was set at a p-value of <0.05. All authors had access to the study data and reviewed and approved the final manuscript.
3Results3.1Patient characteristicsOf the 92 patients included in the initial study, plasma/serum of 89 patients was available for HBcrAg testing and those patients were included in the analysis (Supplementary Figure 1). In total, 88/89 patients had a baseline HBcrAg level >LoQ. Liver biopsies of 54 patients were available at baseline and of 46 patients at EOT, 40 patients underwent a biopsy at both time points. Three baseline biopsies had cccDNA levels below LoQ and levels were set at 50 copies/biopsy before normalising values to copies/haptocyte. Characteristics of patients with and without an available liver biopsy at baseline were similar (supplementary table 1). Patient characteristics are shown in Table 1. Included patients had an average age of 39.3 years, were more often male and various ethnic backgrounds. Genotype A and D were most common. HBeAg-positive patients (n=44) had higher (p < 0.001) levels of: HBcrAg, HBsAg, HBV-DNA, and HBV-pgRNA in plasma/serum as well as intrahepatic cccDNA and cccDNA transcriptional activity (ratio intrahepatic HBV DNA/ cccDNA) [10] compared to HBeAg-negative patients (n=45) (Table 1).
Patient characteristics at baseline.
| Characteristics and markers | Total group (n= 89) | HBeAg-positive (n = 44) | HBeAg-negative (n = 45) | p-value* |
|---|---|---|---|---|
| Mean age, years (SD) | 39.3 (10.3) | 35.8 (9.5) | 42.7 (10.1) | 0.0011 |
| Female sex (%) | 22 (24.7) | 9 (20.5) | 13 (30.0) | n.s.2 |
| Ethnicity (%)CaucasianAsianAfrican | 27 (30.3)33 (37.1)29 (32.6) | 16 (36.4)20 (45.4)8 (18.2) | 11 (24.4)13 (28.9)21 (46.7) | 0.0172 |
| Median ALT U/L (IQR) | 81 (49.5-138.5) | 97.0 (50.8 – 211.3) | 69.0 (46.0 – 120.5) | 0.0393 |
| HBV genotype*ABCDE | 27 (30.3)15 (16.9)11 (12.4)27 (30.3)9 (10.1) | 18 (40.9)8 (18.2)7 (15.9)9 (20.5)2 (4.5) | 9 (20.0)7 (15.6)4 (8.9)18 (40.07 (15.6) | 0.0472 |
| Mean HBsAg, log10 IU/mL (SD) | 3.8 (0.9) | 4.3 (0.7) | 3.3 (0.8) | <0.0011 |
| Mean HBV-DNA, log10 IU/mL (SD) | 6.8 (1.7) | 8.0 (1.2) | 5.6 (1.1) | <0.0011 |
| Mean HBV-pgRNA, log10 C/mL (SD) | 5.6 (1.8) | 6.9 (1.5) | 4.3 (1.2) | <0.0011 |
| Median HBcrAg, log10 U/mL (IQR) | 5.8 (4.5 – 8.0) | 8.0 (6.6 – 8.4) | 4.5 (3.9 – 5.6) | <0.0013 |
| Intrahepatic virology markers | Total group(n= 54) | HBeAg-positive(n = 24) | HBeAg-negative(n = 30) | p-value† |
| Median cccDNA, C/hep (IQR) | 1.5 (0.3 – 7.0) | 7.7 (3.0 – 9.5) | 0.6 (0.1 – 1.3) | <0.0013 |
| Mean iHBV-DNA, log10 C/hep (SD) | 2.1 (1.3) | 3.1 (0.7) | 1.3 (1.0) | <0.0011 |
| Mean iHBV DNA/cccDNA, ratio (SD) | 2.1 (0.9) | 2.4 (0.4) | 1.9 (1.1) | 0.0161 |
Lower baseline HBcrAg-levels were seen in patients aged ≥40 years compared to <40 (5.5 log10 U/mL versus 6.7 log10 U/mL, p=0.002). Patients with genotype D (median 5.2 log10 U/mL) had lower (p=0.04) HBcrAg-levels and patients with genotype A (median 7.7 log10 U/mL) had higher (p=0.032) HBcrAg-levels, compared to other genotypes combined. Furthermore, patients from African origin (median 4.8 log10 U/mL) had lower HBcrAg-levels compared to Asian (median 6.1 log10 U/mL, p=0.05) or Caucasian patients (median 7.0 log10 U/mL, p=0.05) (Supplementary Table 2).
3.3Treatment outcome and HBcrAg as baseline predictorHBsAg-loss (FC) was observed in 8 of the 89 patients (9%) at 72 weeks after start of the study and in 13 patients (15%) at EOFU. Of the 44 HBeAg-positive patients, 15 (34%) had HBeAg-loss at EOT, 16 (36%) at week 72 and 20 (45%) at EOFU. HBcrAg baseline levels did not vary between patients who achieved FC or HBeAg-loss (among HBeAg-positive patients) at EOFU, and who did not (Table 2). In an univariate logistic regression analysis, no relationship was found between FC at EOFU and baseline HBcrAg-levels (Supplementary Table 3). Only baseline HBsAg-levels were related to FC (OR 0.14 [CI, 0.03-0.58], p=0.007) in HBeAg-negative patients.
Baseline HBcrAg-levels in patients selected by treatment outcome.
| Median baseline HBcrAg (IQR) | HBeAg positive (n = 44) | HBeAg negative (n = 45) | Total group (n= 89) | p-value |
|---|---|---|---|---|
| HBsAg loss (FC) week 144Yes, n= 13No, n= 76 | 8.3 (6.3-8.4)8.0 (6.3-8.4) | 4.2 (3.6-5.9)4.9 (4.1-5.6) | 6.1 (3.9-8.1)5.6 (4.7-8.0) | 0.769 |
| HBeAg loss week 144Yes, n= 20No, n= 24 | 8.0 (6.7-8.5)8.0 (6.7-8.4) | n.a.n.a. | 8.0 (6.7-8.5)8.0 (6.7-8.4) | 0.620 |
Groups compared using a Mann-Whitney U test. HBcrAg, hepatitis B core-related antigen; HBeAg, hepatitis B e antigen; HBsAg, hepatitis B surface antigen; n.a., not applicable.
HBsAg-levels at baseline were previously described as an independent predictor for FC in HBeAg-negative patients in this cohort [15]. When adding HBcrAg-levels at baseline to this model, HBsAg-levels remained predictive of FC and HBcrAg was no additional predictor of FC (Supplementary Table 4).
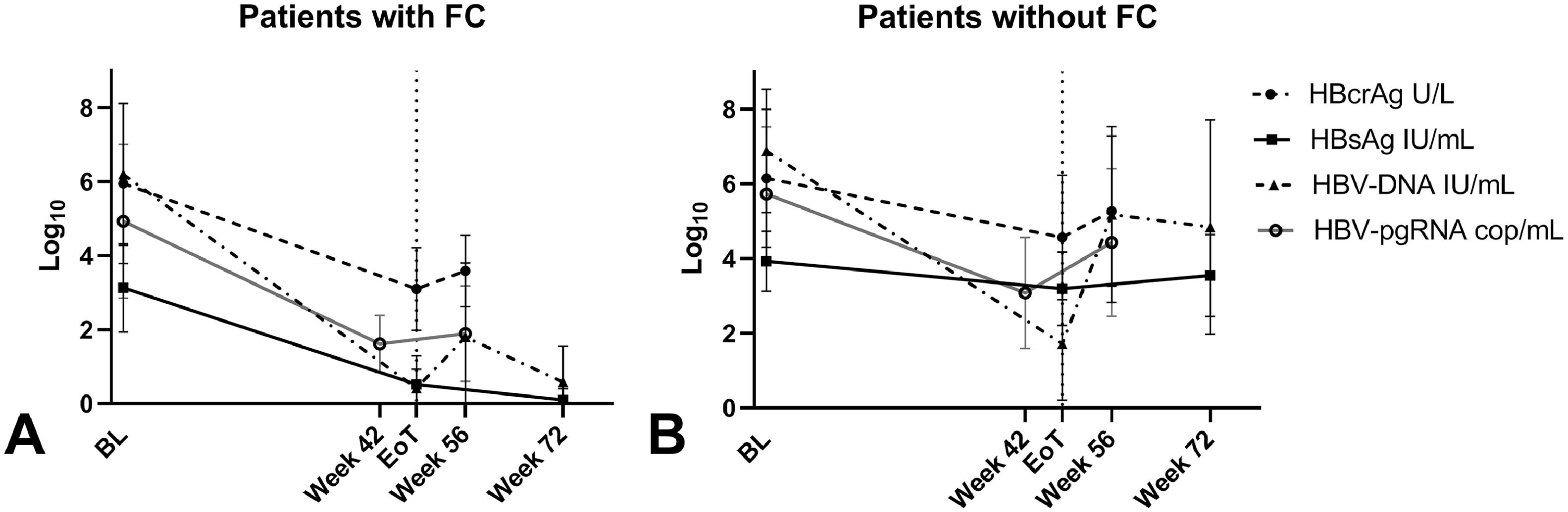
3.4HBcrAg dynamicsDuring treatment, overall HBcrAg-levels declined significantly (p < 0.001) from baseline (median 5.8 log10 U/mL) to EOT (median 4.3 log10 U/mL, IQR 2.9-5.8) and increased at week 56 to a median level of 4.6 log10 U/mL (IQR 3.5-6.2) but remained below the baseline levels (p < 0.001). Variations in HBsAg, HBV-DNA, HBV-pgRNA and HBcrAg-levels during treatment are shown in Supplementary Figure 2. The decrease in median HBcrAg-levels that was seen in patients that achieve FC (from 6.1 log10 U/mL at baseline to 2.7 log10 U/mL at EOT, p < 0.001) was significantly steeper (p=0.026) compared to the decline that was seen in patients who did not achieve FC (from baseline 5.8 log10 U/mL to EOT 4.5 log10 U/mL, p < 0.001) (Fig. 1). HBcrAg-levels were still detectable at EOT in most (6/8) patients with FC after treatment. HBeAg-positive patients with HBeAg-loss at EOFU, showed a more pronounced (p < 0.001) decline of median HBcrAg during treatment (8.0 log10 U/mL to 4.8 log10 U/mL) compared to patients without HBeAg-loss (8.0 log10 U/mL to 6.2 log10 U/mL).
3.5Correlation between HBcrAg and alternative plasma/serum/intrahepatic markersAt baseline, HBcrAg-levels correlated positively with plasma/serum: HBV-DNA (ρ 0.80, p < 0.001), HBV-pgRNA (ρ 0.80, p < 0.001), HBsAg (ρ 0.56, p < 0.001) and ALT (ρ 0.29, p=0.006) in the total cohort. HBcrAg also correlated strongly to intrahepatic cccDNA (ρ 0.77, p < 0.001 and iHBV-DNA (ρ 0.73, p < 0.001). When separating patients by HBeAg status (Supplementary Figure 3), HBcrAg was not correlated to HBsAg in the HBeAg-negative group. In addition, in the HBeAg positive group, HBcrAg was not correlated to ALT or iHBV-DNA.
At EOT, the positive correlation between HBcrAg-levels and HBV-DNA and HBsAg persisted in the total cohort while the correlations between HBcrAg and cccDNA or iHBV-DNA were only found in the HBeAg-positive patients separately. Later, at week 56, a positive correlation was observed between HBcrAg and HBV-DNA or HBV-pgRNA in the total cohort. HBcrAg-levels in HBeAg-negative patients separately, did not correlate with HBV-pgRNA-levels (Table 3).
HBcrAg correlations.
| Baseline | End of treatment (Wk 48) | Follow up (Wk 56) | ||||
|---|---|---|---|---|---|---|
| Laboratory marker | ρ | p-value | ρ | p-value | ρ | p-value |
| HBeAg-positive | ||||||
| HBsAg†, log10 IU/mL | 0.51 | <0.001 | 0.54 | <0.001 | n.a. | |
| HBV-DNA†, log10 C/mL | 0.59 | <0.001 | 0.70 | <0.001 | 0.93 | <0.001 |
| HBV-pgRNA, log10 C/mL | 0.65 | <0.001 | n.a. | 0.91 | <0.001 | |
| ALT, IU/L | -0.04 | 0.820 | -0.20 | 0.220 | 0.19 | 0.915 |
| cccDNA‡, log10 C/hep | 0.50 | 0.014 | 0.55 | 0.012 | n.a. | |
| iHBV-DNA‡, log10 C/hep | 0.31 | 0.116 | 0.67 | 0.001 | n.a. | |
| HBeAg negative | ||||||
| HBsAg†, log10 IU/mL | 0.02 | 0.886 | 0.39 | 0.028 | n.a. | |
| HBV-DNA†, log10 C/mL | 0.75 | <0.001 | 0.48 | 0.002 | 0.59 | <0.001 |
| HBV-pgRNA, log10 C/mL | 0.73 | <0.001 | n.a. | 0.43 | 0.235 | |
| ALT, IU/L | 0.45 | 0.002 | 0.19 | 0.466 | 0.23 | 0.137 |
| cccDNA‡, log10 C/hep | 0.57 | 0.001 | -0.20 | 0.524 | n.a. | |
| iHBV-DNA‡, log10 C/hep | 0.51 | 0.004 | 0.11 | 0.751 | n.a. | |
Correlation between HBcrAg and alternative circulating and intrahepatic viral markers. ALT, alanine transaminase; cccDNA, covalently closed circular DNA; C/hep, copies/hepatocyte; HBsAg, hepatitis B surface antigen; IQR, interquartile range; p, correlation coefficient; wk, week. †at end of treatment, 58 patients had HBV-DNA-levels
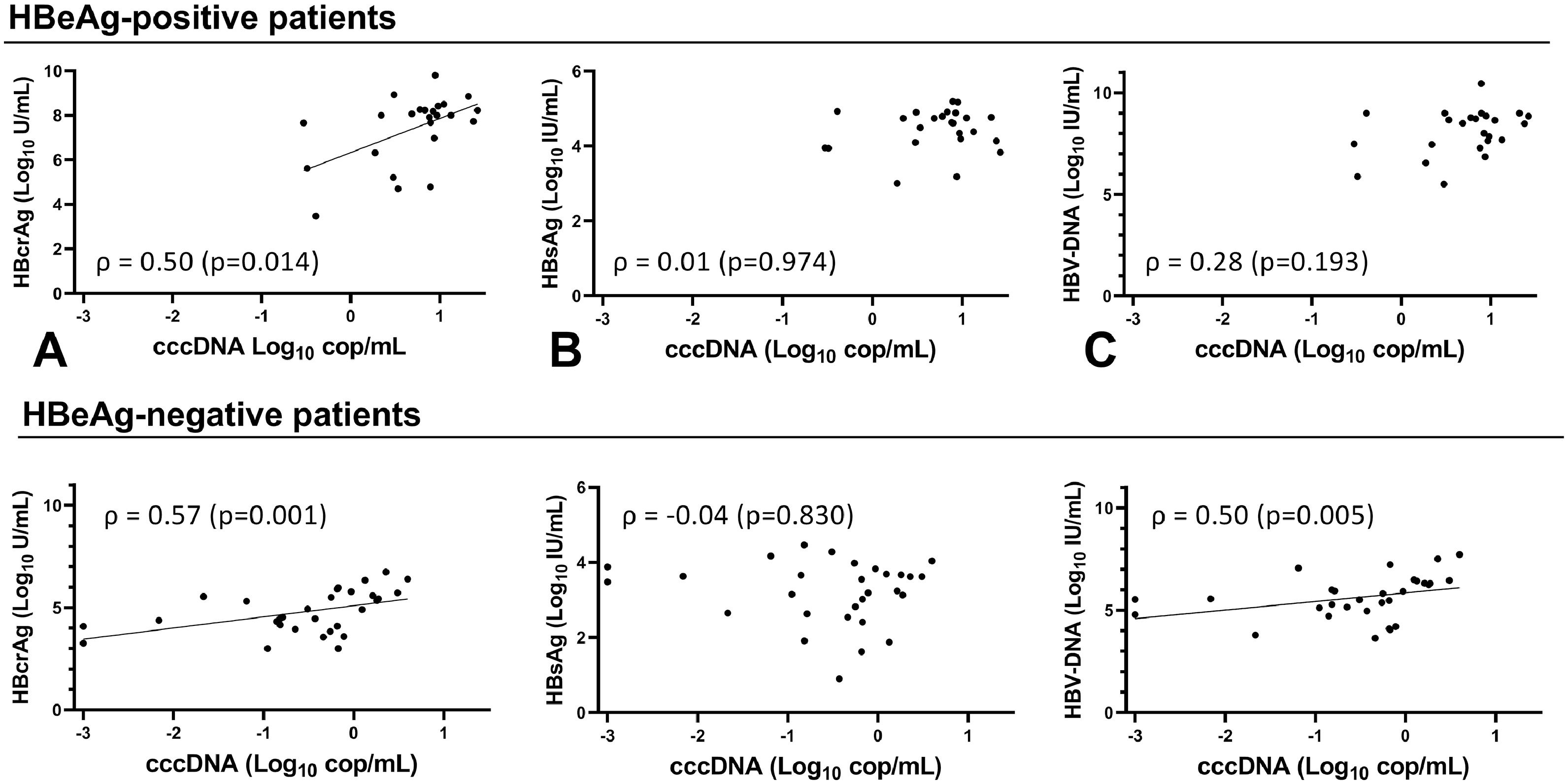
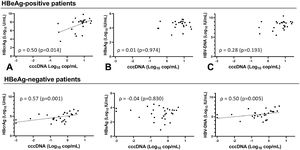
Overall, cccDNA correlation with HBcrAg was similar to its correlation with circulating HBV-DNA (ρ 0.74, p < 0.001) and HBV-pgRNA (ρ 0.70, p < 0.001) when comparing correlation coefficients. However, cccDNA correlated more strongly (p=0.001) to HBcrAg than HBsAg (ρ 0.51, p < 0.001). The strongest correlation was seen between cccDNA and iHBV-DNA (ρ 0.86, p < 0.001). cccDNA at baseline correlated only with HBcrAg (ρ 0.50, p=0.014) and iHBV-DNA (ρ 0.88, p < 0.001) in HBeAg-positive patients (Fig. 2A-C, Supplementary Figure 4). At EOT, cccDNA correlated with HBcrAg (ρ 0.55, p=0.012), HBV-DNA (ρ 0.56, p=0.010) and iHBV-DNA (ρ 0.67 p=0.001) but not with HBsAg. cccDNA correlated with HBcrAg (ρ 0.57, p= 0.001), HBV-DNA (ρ 0.50, p= 0.005) and iHBV-DNA (ρ 0.58, p= 0.001) at baseline, but not with HBsAg in HBeAg-negative patients (Fig. 2D-F, Supplementary Figure 5). At EOT, cccDNA-levels were low or undetectable in most HBeAg-negative patients so no correlations could be calculated.
3.7Identifying patient clusters based on HBV markersThe relation between all HBV markers was further explored in 54 patients with available liver biopsy markers at baseline. Included were: serum/plasma levels of HBcrAg, HBV-DNA, HBV-pgRNA, HBsAg, ALT and intrahepatic cccDNA and iHBV-DNA-levels. Ishak score for fibrosis showed a very low correlation to all markers (data not shown) and was excluded from analysis. After excluding 3 outliers (patients with cccDNA levelsFig. 3A,C).
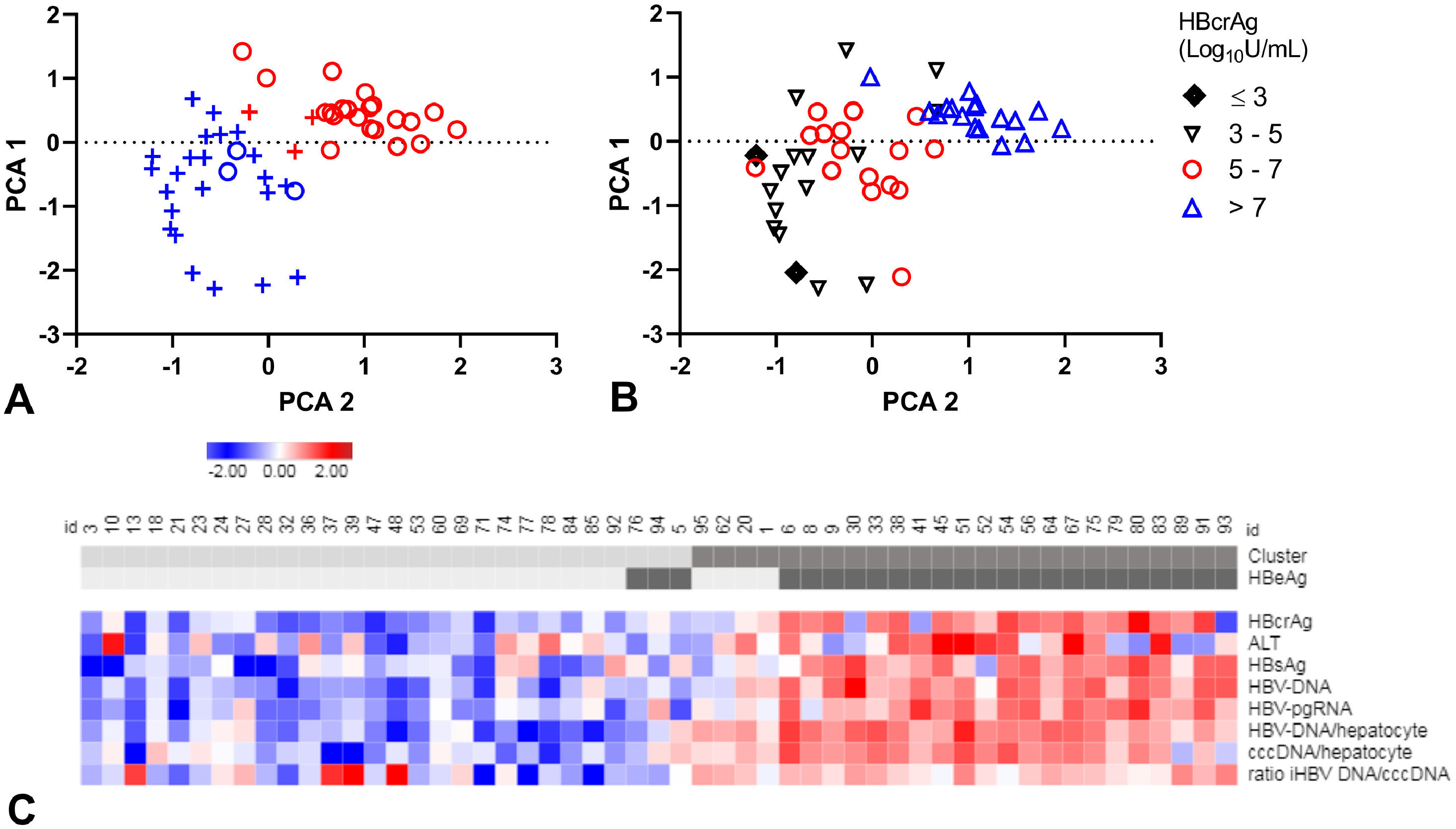
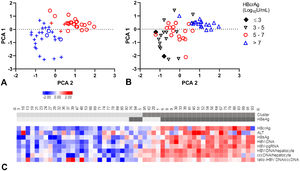
A-C. Principal component analysis (PCA) and clustering.
PCA plots, each patient resembling a dot (A-B). Clusters distinguished in colour, clusters 1 (blue), 2 (red) and HBeAg-negative (+) and HBeAg-positive (o) patients in symbols (A). Patients divided by HBcrAg values (B). Heat map (C) showing all included markers: red represents values above 2SD and blue below -2SD from the mean for each value. HBeAg-seropositivity and clusters of each patient are shown in the top bar, cluster 1 and HBeAg negative patients are in lighter grey. cccDNA; covalently closed circular DNA, HBcrAg; hepatitis B core-related antigen, HBeAg; hepatitis B e antigen, HBsAg; hepatitis B surface antigen, HBV-RNA; HBV pregenomic RNA.
Patients in cluster 1 were of older age (45 years, versus 37 years), with lower ALT levels (median: 73 U/L, versus 128 U/L) and lower levels of all virology markers compared to cluster 2 (Supplementary Table 5). Median HBcrAg-levels were 4.9 log10 U/mL (IQR, 4.1 - 5.6) in cluster 1 and 8.0 log10 U/mL (IQR, 6.6 - 8.3) in cluster 2 (Fig. 3B). Remarkably, cluster 1 included almost all (88.5%) HBeAg-negative patients and cluster 2 almost all (84.0%) HBeAg-positive patients (PCA-plot,Fig. 3A and C). In a ROC curve, a HBcrAg cut-off of 6.0 log10 U/mL could distinguish between HBeAg-positive and -negative patients with a sensitivity of 0.84 and specificity of 0.91 (Supplementary figure 6).
The occurrence of HBeAg-loss or FC after treatment was not different between the clusters (Supplementary table 5 and figure 7A, B). All HBeAg-positive patients who were appointed to cluster 1 (n=3) became HBeAg-negative after treatment. The four HBeAg-negative patients in cluster 2 had the highest HBcrAg and HBV-DNA-levels of all HBeAg-negative patients.
4DiscussionIn this study, the value of HBcrAg quantification in CHB patients was analysed, before and after combination treatment. We found that HBcrAg-values at baseline correlated strongly to cccDNA, intrahepatic HBV-DNA, and circulating HBV-DNA, HBV-pgRNA but in lesser extent to HBsAg. Furthermore, baseline HBcrAg-levels could not predict FC after treatment with Peg-IFN and adefovir. HBcrAg levels were however more rapidly declined after end of treatment in patients who developed FC or HBeAg-loss. Almost all correlations persisted to the end of the 48-week treatment and during follow-up.
The HBcrAg-test used, is validated, standardized and widely available, making it suitable for clinical use. However, the high LoQ of 3.0 log10 U/mL complicates research in patients with low replicative activity. Especially HBeAg-negative patients since HBeAg proteins are one of three antigens measured in the HBcrAg-test [10]. We included patients with a high viral load (HBV-DNA >10,000 copies/mL). As a result, HBeAg-positive as well as HBeAg-negative patients had quantifiable HBcrAg-levels. This enabled the analysis of HBcrAg-levels before and after treatment but also limited the applicability of our findings in CHB patients with a low viral load.
Baseline HBcrAg-levels were found to be strongly correlated to HBV-DNA and HBV-pgRNA in plasma/serum and to intrahepatic cccDNA and iHBV-DNA levels but not to HBsAg. Furthermore, the correlation between HBcrAg and HBsAg was even less strong in the HBeAg-positive group and absent in the HBeAg-negative group. This is in line with earlier findings [10] and may possibly be explained by the attribution of HBsAg from integrated HBV-DNA rather than cccDNA. Especially since integrated HBV DNA is more prominent in HBeAg-negative patients [19]. This would in turn suggest that HBcrAg is a better marker for cccDNA transcription than HBsAg. Indeed, in our study, cccDNA correlated best to HBcrAg (ρ = 0.77) and significantly better to HBcrAg than to HBsAg (ρ = 0.51) when comparing circulating markers. A very recent meta-analysis by Caviglia and colleagues supports this finding and describes an average higher correlation between cccDNA and HBcrAg (ρ = 0.665) compared to HBsAg (ρ = 0.475) [33]. Based on this, HBcrAg may be considered as first choice in monitoring treatment effect of drugs that target cccDNA transcription.
Although adefovir and Peg-IFN treatment is currently no longer first- of choice in clinical care, the results of this study is relevant to both current and future therapies for several reasons. At present, adefovir is rarely used due to the risk of viral resistance development in the long-term (mean 20 months) [20]. However, adefovir is a potent antiviral with the same mode of action as NAs used in current practice. It is therefore suitable for evaluation of the effect of NAs on viral activity, especially in study context and in short-term use such as 48 weeks. Peg-IFN treatment on its turn has been losing ground as a treatment for CHB due to its unfavourable side effect profile. However, Peg-IFN is the only registered immune modulator for CHB and thus is expected to be of growing importance in future combination treatments that aim to achieve functional cure. As affirmed by the many phase 1 and 2 trials that combine Peg-IFN with developmental compounds such as: nucleic acid polymers, Myrcludex-B and therapeutic vaccinations [21-23].
We found that baseline HBcrAg-levels could not predict PEG-IFN/NA treatment outcome. Treatment with Peg-IFN leads to FC in around 3-5% of patients and 1-2% in NA-based therapies [24]. Due to these low FC-rates, investigating the predictive value of HBcrAg for FC can be challenging as seen in previous studies [25,26]. Here, this was not a limiting factor with an observed FC-rate of 15%. One other prospective study [27] investigated the prediction of FC after combination treatment with Peg-IFN plus tenofovir and described similar findings. A smaller cohort was however included in this study (n=32) and patients had lower HBcrAg-levels at baseline (3.87 log10 U/mL, SD 1.26) suggesting that some patients might have had HBcrAg-levels below LoQ.
The use of HBcrAg as marker of treatment response is attributed by the finding that, HBcrAg remained detectable in most patients and remained correlated to all virological markers and cccDNA after treatment. Furthermore, a more pronounced decline in HBcrAg-levels was seen between start and end of treatment in patients who developed FC or HBeAg-loss compared to non-responders. HBcrAg dynamics earlier in treatment have been described in Peg-IFN treatment with or without entecavir. A HBcrAg-decline of <0.5 log10 U/mL at 12 weeks had a negative predictive value of 94.6% for FC (sensitivity 63.6%, specificity 63.6%) [28]. HBcrAg-decline in the first 4-24 weeks of Peg-IFN monotherapy or NA-monotherapy has also been described to predict HBeAg-loss or non-response in several, mainly retrospective, studies [26,29]. So HBcrAg dynamics under treatment, rather than baseline levels, may be suitable to predict treatment response.
Under NA-treatment, plasma HBV-DNA reaches undetectable levels causing the need for alternative markers for viral replication [30]. HBV-pgRNA seems a less suitable alternative marker than HBcrAg for several reasons; HBcrAg correlated better to cccDNA than HBV-pgRNA in our study, there is currently no standardized, validated and commercially available test for HBV-pgRNA and finally, HBV-pgRNA-levels are known to decline under NA treatment and may reach an undetectable level sooner than HBcrAg [2]. This last reason is remarkable since the inhibition of reverse transcription should not affect HBV RNA nor HBV protein synthesis. This difference may thus be explained by the low sensitivity of the HBV-pgRNA plasma test and that the HBV-pgRNA is only present in virions in the plasma [2] which are released in much lower amounts than viral proteins. For example, the amount of virions in sera is exceeded by sub-viral particles consisting of mainly HBsAg proteins, by a factor between 102-105[34]. In addition, proteins are generally more stable then RNA in plasma and the HBcrAg combines 3 proteins which further enhances the stability of the test. Furthermore, HBcrAg may be detectable even after HBsAg-loss, [31] as confirmed in the majority of patients who achieved FC at EOT. This indicates that HBcrAg may reflect the persistence of low cccDNA transcription after FC that could account for the low but persisting risk of HCC after FC [32] and reactivation under immunosuppression.
In depth analysis allocated HBeAg-positive and -negative patients to two separate clusters while HBeAg-status was not incorporated in the cluster analysis. A recent study identified two HBeAg-negative subgroups where viral replication markers but not the cccDNA-levels differed between patients [10]. Since we only included patients with a high viral load, an additional HBeAg negative cluster could not be identified in our study. Analysing the role of HBcrAg in the clusters showed that HBcrAg-levels alone were sufficient to identify HBeAg-seropositivity with a cut-off value of 6.0 log10 U/mL. The few HBeAg positive-patients allocated to the ‘HBeAg negative’ cluster, developed HBeAg-seroconversion upon treatment. Suggesting that HBcrAg-levels may distinguish patients with an overall high and low HBV-replicative activity better than HBeAg-status.
In summary, we confirmed that HBcrAg reflects cccDNA transcription activity more accurately than HBsAg and may replace HBV-DNA as a marker during treatment, especially in future treatment regimens which target cccDNA transcription or include NA treatment.
5Author contributionsRE acts as guarantor of this article. Study concept and design was formed by HR, HZ and NK; funding obtained by HR, NK and RE; acquisition of data by RE, HZ, EB and BT; technical and material support by EB; drafting of the manuscript by RE; statistical analysis by RE; analysis and interpretation of data by RE and NK; critical revision of the manuscript for important intellectual content by HZ, SW, BT, HR and NK.