Emerging evidence has demonstrated that long noncoding RNAs (lncRNAs) may be closely associated with Hepatitis C virus (HCV) infection and the development of hepatocellular carcinoma (HCC). In this study, we investigated the expression and functions of a lncRNA, LINC01189, in HCV-associated HCC.
Patients or Materials and MethodsLINC01189 expression was measured in HCC tumors, HCV-infected HCC tumors and HCV-infected HCC cells. LINC01189 was overexpressed in HCV-infected HepG2 cells to measure its function on HCV-correlated cancer proliferation. In HCC cell lines of Huh7 and Hep3B, LINC01189 was upregulated to investigate its effects on cancer cell proliferation and 5-FU chemoresistance. The competing endogenous RNA (ceRNA) target of LINC01189, human microRNA-155-5p (hsa-miR-155-5p) was probed by dual-luciferase assay and qRT-PCR. Hsa-miR-155-5p was upregulated in LINC01189-overexpessed Huh7 and Hep3B cells to investigate their epigenetic correlation on HCC development regulation.
ResultsLINC01189 is downregulated in HCV-infected HCC tumors and cell lines. LINC01189 overexpression inhibited HCC cancer cell proliferation and 5-FU chemoresistance. Hsa-miR-155-5p was confirmed to be a ceRNA target of LINC01189 in HCC. Upregulating hsa-miR-155-5p reversed the LINC01189-mediated inhibition on HCC proliferation and 5-FU chemoresistance.
ConclusionsLINC01189 downregulation is associated with HCV infection in HCC, and it has tumor-suppressing effects on HCC development through hsa-miR-155-5p.
Hepatocellular carcinoma (HCC) is one of the most commonly diagnosed and most malignant types of liver cancers worldwide. In USA, there are approximately 42,000 newly diagnosed cases of liver cancers and more than 30,000 associated deaths every year [1]. In China, liver cancer is the fourth-most diagnosed cancer (>390,000 cases / year) and third-most cause of cancer-related mortality (>360,000 cases / year) in both male and female patients [2,3]. While various risk factors may be associated with HCC or other liver cancer subtypes, the most common conditions for patients to eventually develop into HCC are hepatitis B virus (HBV) or hepatitis C virus (HCV) infections [4,5].
Long noncoding RNAs (lncRNAs) are families of long (>200 nucleotides) and non-protein-coding transcripts that had been recently shown to exert broad ranges of regulatory roles in various types of human disease [6,7]. Specifically, lncRNAs have been demonstrated to be aberrantly expressed, and exert promoting or suppressing functions in many types of human cancers [8,9]. Recently, a genomic study conducted by Yang and colleagues showed that there might be more than 1,000 lncRNAs that are recurrently deregulated in HCC [10]. In addition, prognostic and functional roles of lncRNAs had been revealed in HCC [11,12]. Particularly, it has been noticed that lncRNAs may be closely associated with HCV progression and the pathological development of HCC among patients with chronic liver diseases [13,14].
Similar to lncRNAs, microRNAs (miRNAs) are other families of non-protein-coding, but short (∼20 nucleotides) transcripts that are also shown to be aberrantly expressed and playing critical roles in human cancer development [15,16]. Moreover, new lines of evidence have demonstrated that lncRNAs could act as competing endogenous RNAs (ceRNAs) of miRNAs, so that the transcriptional regulations of lncRNAs-miRNAs axes could have even more significant impacts on regulating cancer cell oncogenesis, progression, apoptosis and metastasis [17,18].
In the current work, we examined the expression of a newly identified lncRNA, LINC01189 in HCC and its correlation with HCV-infected HCC. We also ectopically overexpressed LINC01189 in HCC cancer cells to study its functions on HCV-associated HCC proliferation and 5-FU chemoresistance. In addition, we studied the possible ceRNA candidate of LINC01189, hsa-miR-145-5p in HCC. The goal of this work is to further our understanding on the epigenetic regulation of lncRNA in HCC.
2Materials and methods2.1HCC human samplesA total of 67-paired hepatocellular carcinoma (HCC) tissues and surrounding non-tumorous tissues were obtained from the participating hospital. Among the HCC tissues, 21 were diagnosed with hepatitis C virus (HCV) infection. Human samples were immediately snap-frozen with liquid nitrogen and then stored at −75°C, until further processing.
2.2HCC cell lines and HCV-infectionHCC cell lines of HeG2, HuH7 and Hep3B were purchased from the American Type Culture Collection (ATCC, USA), and incubated in GlutaMAX-RPMI 1640 Medium (Thermo Fisher Scientific, USA) supplemented with 10% fetal bovine serum (FBS, Beyotime, China) and 1X Penicillin-Streptomycin (10,000 U/ml, Thermo Fisher Scientific, USA) in a humidified environment at 37°C with 5% CO2.
The method of infecting HCC cell line with HCV was described in a previous publication with slight modifications [19]. Briefly, HepG2 cells were cultured in FBS-free medium and inoculated with human serum collected from HCV-infected patients. After 2hours, HepG2 cells were replenished with FBS-containing medium, supplemented with 4% polyethylene glycol (Beyotime, China) and cultured for another 24hours. QRT-PCR was then performed to examine the infection efficacy of HCV viral RNA.
2.3RNA extraction and quantitative real time-PCRTotal RNA was extracted and purified using a PureLink™ Pro 96 total RNA Purification Kit (Thermo Fisher Scientific, USA) according to the manufacturer’s recommendation. The quality of RNA were verified using a Vairoskan Lux uDrop kit (Thermo Fisher Scientific, USA), and reverse transcription was performed using a SuperScript™ VILO™ cDNA Synthesis Kit (Invitrogen, USA) according to the manufacturer’s recommendations. An ABI PRISM® 7000 Sequence Detection System (Applied Biosystems, USA) was used for quantitative real time-PCR (qRT-PCR). To quantitatively compare HCV viral RNA, the method was described in the previous publication [19]. To compare lncRNA LINC01189 expression level, a customized TaqMan™ noncoding RNA Assay (Invitrogen, USA) was used. To measure hsa-miR-155-5p expression level, a customized TaqMan™ microRNA Assay (Invitrogen, USA) was used. Finally, the relative expression levels were calculated by the 2(−ΔΔCt) method.
2.4LINC01189 upregulation and downregulation assayThe whole sequence of human LINC01189 was amplified and sub-cloned into the pcDNA/3.4 plasmid (Addgene, USA) to generated a LINC01189 overexpressing vector, pc_1189. An empty pcDNA/3.4 plasmid (Addgene, USA) was used as the non-specific overexpression vector, pc_NS in this study. Alternatively, a siRNA specifically targeting human LINC01189, si_1189 and a non-specific lncRNA siRNA, si_NS, were designed and manufactured by GenePharma (Shanghai GenePharma, Shanghai, China). In Hu7, Hep3B, and HCV-infected HepG2 (HepG2-HCV) cells, they were transfected with pc_1189, pc_NS, si_1189 and si_NS for 48hours using the Lipofectamine 3000 transfection reagent (Thermo Fisher Scientific, USA). After that, qRT-PCR was performed to examine the transfection efficacies.
2.5Proliferation assayHCC cell proliferation was measured using a CCK-8 Assay (Dojindo, Japan) according to the manufacturer’s recommendation. With the application of a highly water-soluble tetrazolium salt, WST-8, this assay produces a water-soluble formazan dye upon reduction in the presence of an electron mediator, thus providing quantitative method to assess proliferating cells in the culture. To do so, HCC cells were cultured in a 96-well plate, with a starting density of 1500 cells /well, for 5 days. Every 24h, CCK-8 reagent (10μL) was mixed into tested wells for 2hours at 37 C. Then, proliferating rates were directly estimated by measuring the optical absorbance at 570nm using a Varioskan Lux microplate reader (Thermo Fisher Scientific, USA) according to the manufacturer’s recommendation.
2.65-FU chemoresistance assayThe method of examining HCC fluorouracil (5-FU) chemoresistance was described in a previous publication with slight modification [20]. Briefly, HCC cells were cultured in a 96-well plate, with a starting density of 5,000 cells /well. 5-FU was added into cell culture at concentration (μM) of 0, 2, 5, 10, 20 for 48h. The relative cell viability was then measured using the CCK-8 assay.
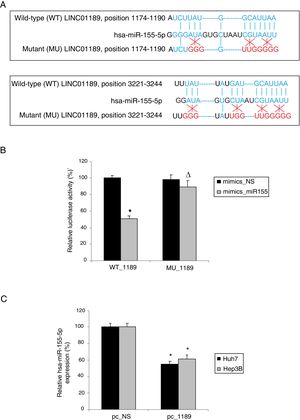
2.7Dual-luciferase reporter assayUsing online bioinformatics database of StarBase 2.0 [21], we identified two DNA sequences on LINC01189 3’-UTR that might be putative binding sites for human microRNA-155-5p (hsa-miR-155-5p). We thus constructed two pmiR-RB-REPORT vectors (Shanghai GenePharma, Shanghai, China), one containing the wild-type (WT) LINC01189 3’-UTR (WT_1189), and the other containing a mutant LINC01189 3’-UTR (MU_1189) to deactivate hsa-miR-155-5p binding. In addition, a synthetic hsa-miR-155-5p mimics (mimics_miR155) and a non-specific human miRNA mimics (mimics-NS) were commercially bought from GenePharma (Shanghai GenePharma, Shanghai, China). Then, a dual-luciferase reporter assay was conducted using the method described in our previous publication [22].
2.8Hsa-miR-155-5p overexpression assayIn Huh7 and Hep3B cells which were pre-transfected with pc_1189, they were secondary transfected with mimics_miR155 or mimics-NS using the Lipofectamine 3000 reagent for 48h. QRT-PCR was then performed to examine the overexpression efficacy.
2.9Statistical analysisAll experiments were performed in at least three independent repeats. The averaged data were presented as mean±standard error of mean (mean±S.E.M.). One-way analysis of variance (ANOVA) followed by Tukey’s multiple-comparison tests in a SPSS software (SPSS, version 16.0, USA) was used for all analysis. Significance was set at 0.05 for all statistical tests.
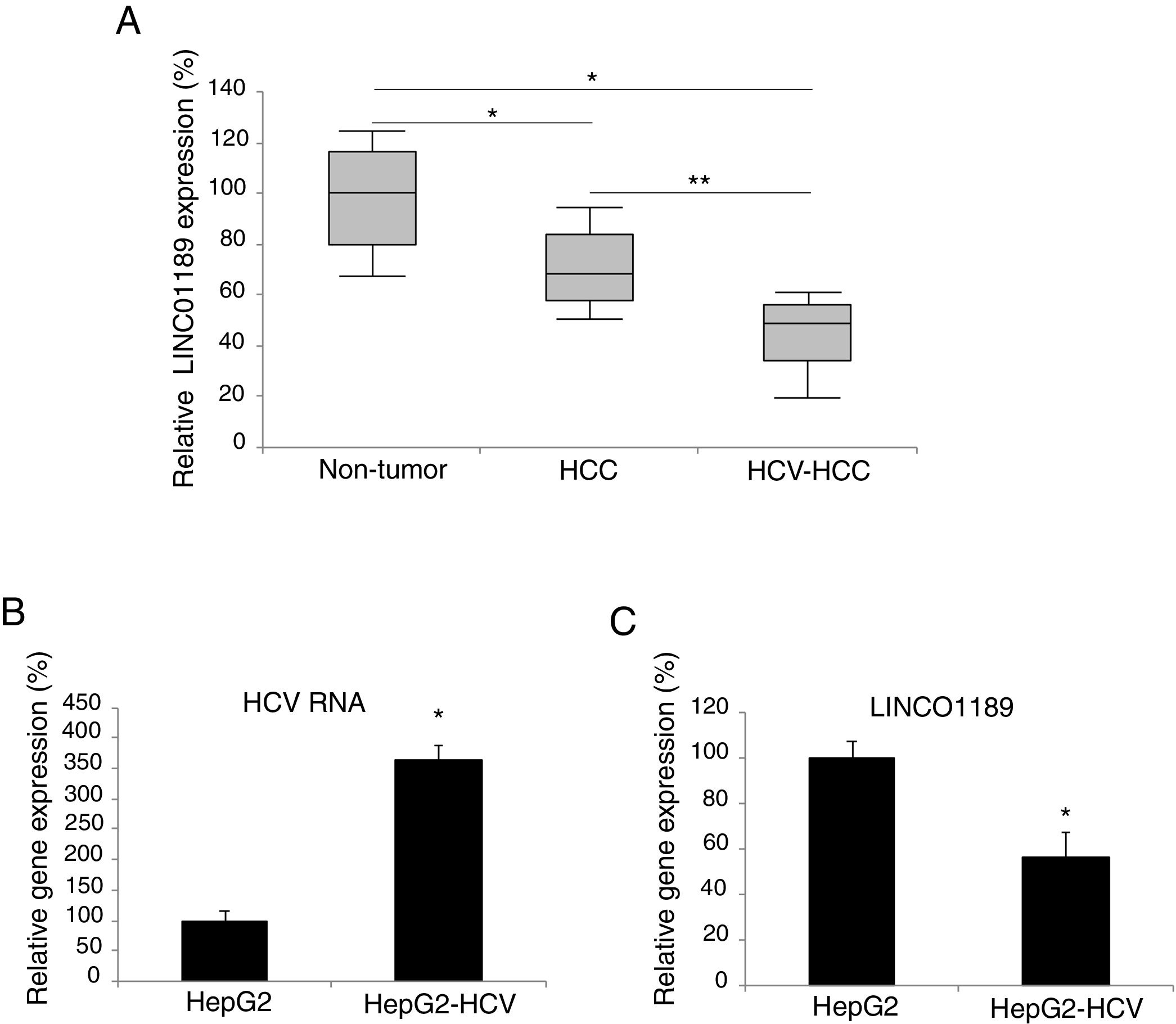
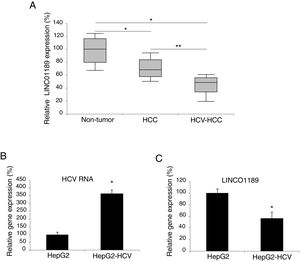
3Results3.1LINC01189 is lowly expressed in HCV-infected HCC tumors and cell linesWe used qRT-PCR to detect the expression pattern of LINC01189 in HCC tumors and cell lines. It was demonstrated that LINC01189 expression is markedly lower in HCC tumors than in adjacent tissues (Fig. 1A, * P<0.05). In addition, LINC01189 expression was even lower in HCC tumors with HCV-infected (Fig. 1A, ** P<0.05).
LINC01189 may be closely associated with HCV infection in HCC. (A) Expression of LINC01189 was probed, by qRT-PCR in paired HCC tumor tissues and adjacent non-tumor tissues (n=67) (* P<0.05). In addition, LINC01189 was also probed in a sub-set of HCV-infected HCC tumors (n=21) (** P<0.05). (B) A HCC cell line, HepG2 was infected with HCV virus. After that, HCV RNA expression levels were compared, by qRT-PCR, between infected (HepG2-HCV) and un-infected cells (* P<0.05). (C) Relative expression of LINC01189 in infected (HepG2-HCV) and un-infected HepG2 cells was probed by qRT-PCR (** P<0.05).
Next, HCC cell line of HepG2 was infected with HCV virus. QRT-PCR confirmed that HCV RNA expression was significantly upregulated in HCV-infected HepG2 cells (Fig. 1B, * P< 0.05; Note: expression level of 100% was based on arbitrary value). On the other hand, LINC01189 expressed at a much lower level in HCV-infected HepG2 cells (Fig. 1C, * P<0.05).
Thus, these data indicated that LINC01189 downregulation may be closely associated with HCV infection in HCC.
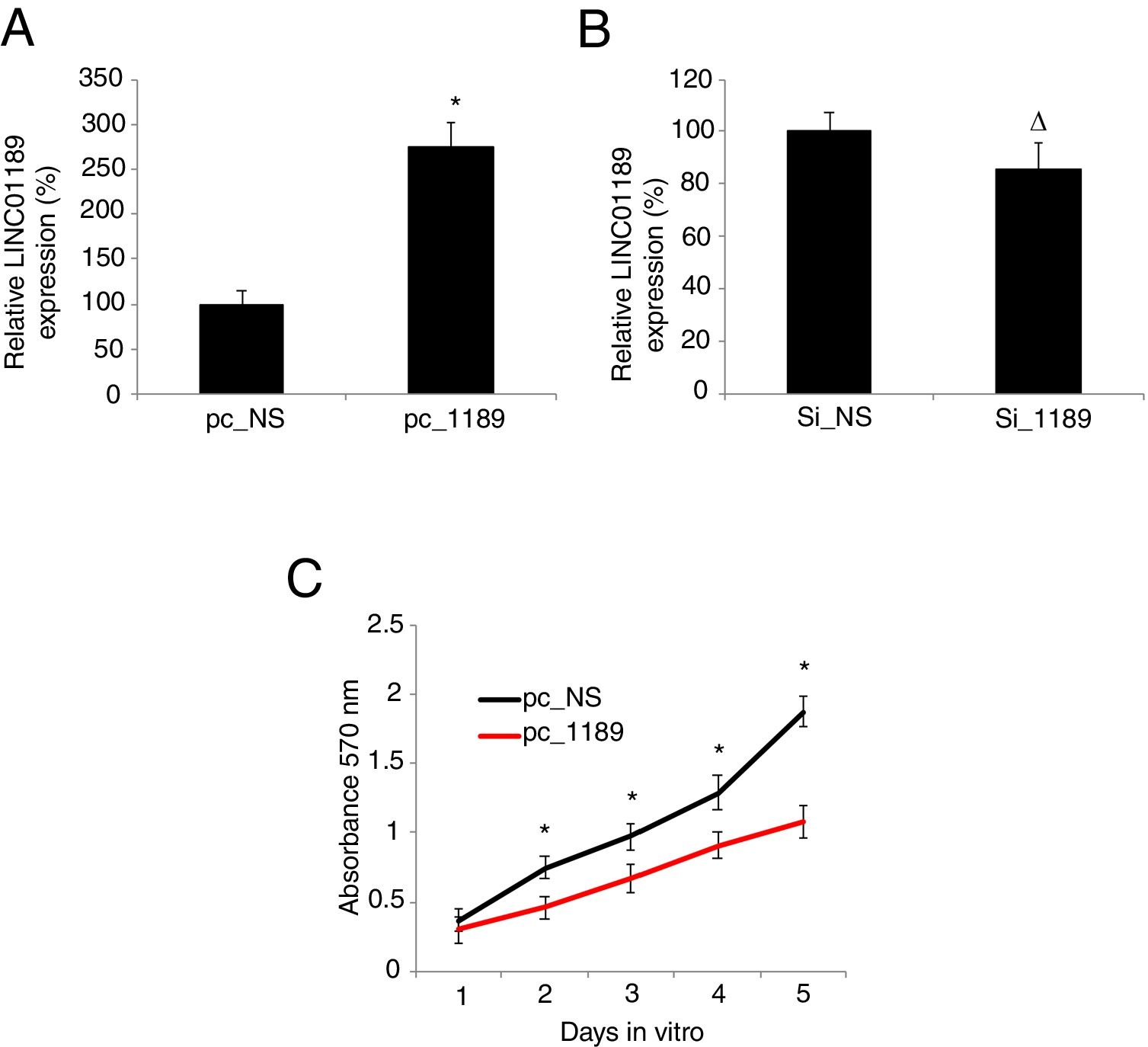
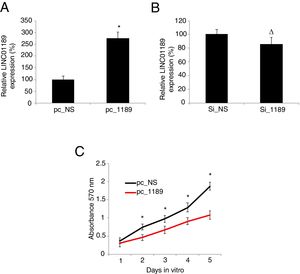
3.2LINC01189 overexpression inhibited proliferation in HCV-infected HCC cell linesWe either overexpressed or downregulated LINC01189 in HCV-infected HepG2 cells. The result of qRT-PCR showed that, in cells transfected with pc_1189, endogenous LINC01189 expression was much higher than in cells transfected with pc_NS, confirming the overexpression strategy (Fig. 2A, * P<0.05). However, LINC01189 expression levels were not different between cells transfected Si_1189 and Si_NS, indicating that LINC01189 was unable to be downregulated in HCV-infected HepG2 cells (Fig. 2B, Δ P>0.05). Then, a CKK-8 assay was conducted for 5 consecutive days to compare proliferation between pc_1189- and pc_NS- transfected HCV-HepG2 cells. It showed that LINC01189 overexpression significantly inhibited proliferation in HCV-infected HCC cells. (Fig. 2C, * P<0.05).
Upregulating LINC01189 reduced proliferation in HCV-infected HepG2 cells. (A) HCV-infected HepG2 cells were transfected with a LINC01189 overexpressing vector, pc_1189 or a non-specific overexpression vector, pc_NS. Relative LINC01189 expressions were detected by qRT-PCR (* P<0.05). (B) HCV-infected HepG2 cells were transfected with a siRNA specifically targeting human LINC01189, si_1189 or a non-specific lncRNA siRNA, si_NS. After that, relative LINC01189 expressions were detected by qRT-PCR (Δ P>0.05). (C) CCK-8 assay was conducted to compare proliferation in HCV-infected HepG2 cells transfected with pc_1189 and those transfected with pc_NS (* P<0.05).
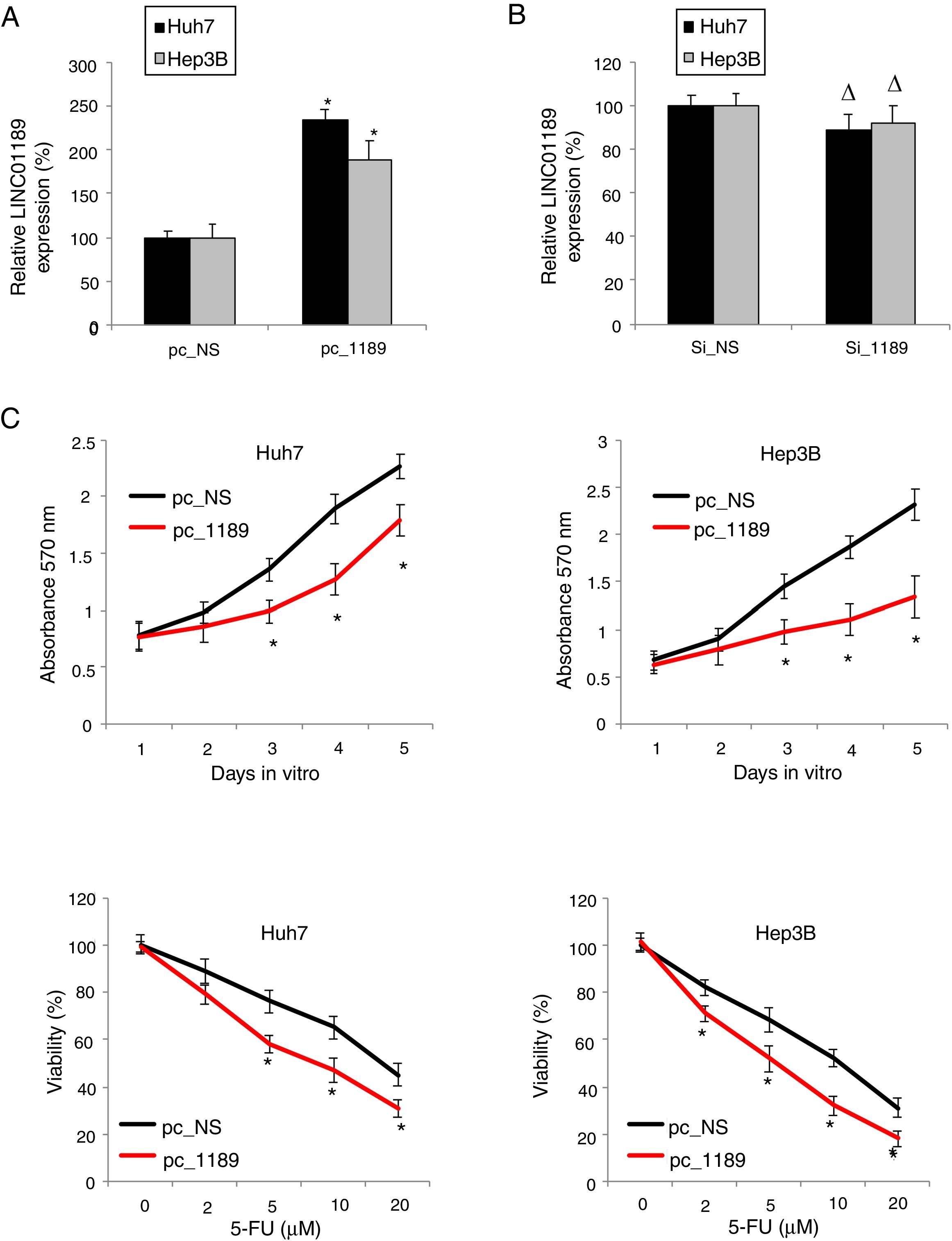
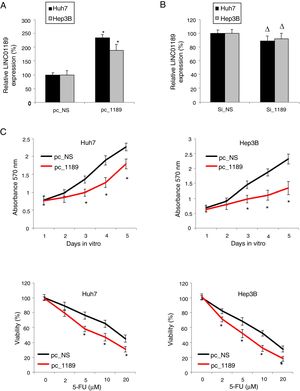
To further investigate the effects of LINC01189 on HCC development, we either overexpressed or downregulated LINC01189 in two HCC cell lines, Huh7 and Hep3B. In one set of experiments, HCC cells were transfected with either pc_1189 or p_NS. QRT-PCR showed that, in HCC cells transfected with pc_1189, endogenous LINC01189 expression was much higher than in those transfected with pc_NS (Fig. 3A, * P< 0.05). In the other set of experiments, Huh7 and Hep3B were transfect with either si_1189 or si_NS. This time, qRT-PCR did not show significantly difference in endogenous LINC01189 expressions (Fig. 3B, Δ P>0.05), suggesting that genetic modification was unable to knock down LINC01189 in Huh7 or Hep3B cells.
Upregulating LINC01189 suppressed HCC proliferation and chemoresistance. (A) HCC cell lines, Huh7 and Hep3B cells, were transfected with a LINC01189 overexpressing vector, pc_1189 or a non-specific overexpression vector, pc_NS. After that, relative LINC01189 expressions were detected by qRT-PCR (* P<0.05). (B) Huh7 and Hep3B cells were transfected with a siRNA specifically targeting human LINC01189, si_1189 or a non-specific lncRNA siRNA, si_NS. After that, relative LINC01189 expressions were detected by qRT-PCR (Δ P>0.05). (C) CCK-8 assay was conducted to compare proliferation in HCC cells transfected with pc_1189 and those transfected with pc_NS (* P< 0.05). (D) For Huh7 and Hep3B cells transfected with either pc_1189 or pc_NS, they were incubated with 5-FU at 0, 2, 5, 10 and 20μM for 24h. After that, chemoresistance was compared using a viability assay (* P<0.05).
Then, pc_1189- or pc_NS- transfected Huh7 and Hep3B were examined by the CKK-8 assay for 5 days. It showed that LINC01189 overexpression markedly reduced proliferation in HCC cells (Fig. 3C, * P<0.05).
In addition, pc_1189- or pc_NS- transfected Huh7 and Hep3B were treated by low-to-high concentrations of 5-FU to compare their chemoresistance. The results showed that LINC01189 overexpression also significantly reduced 5-FU chemoresistance in HCC cells (Fig. 3D, * P<0.05).
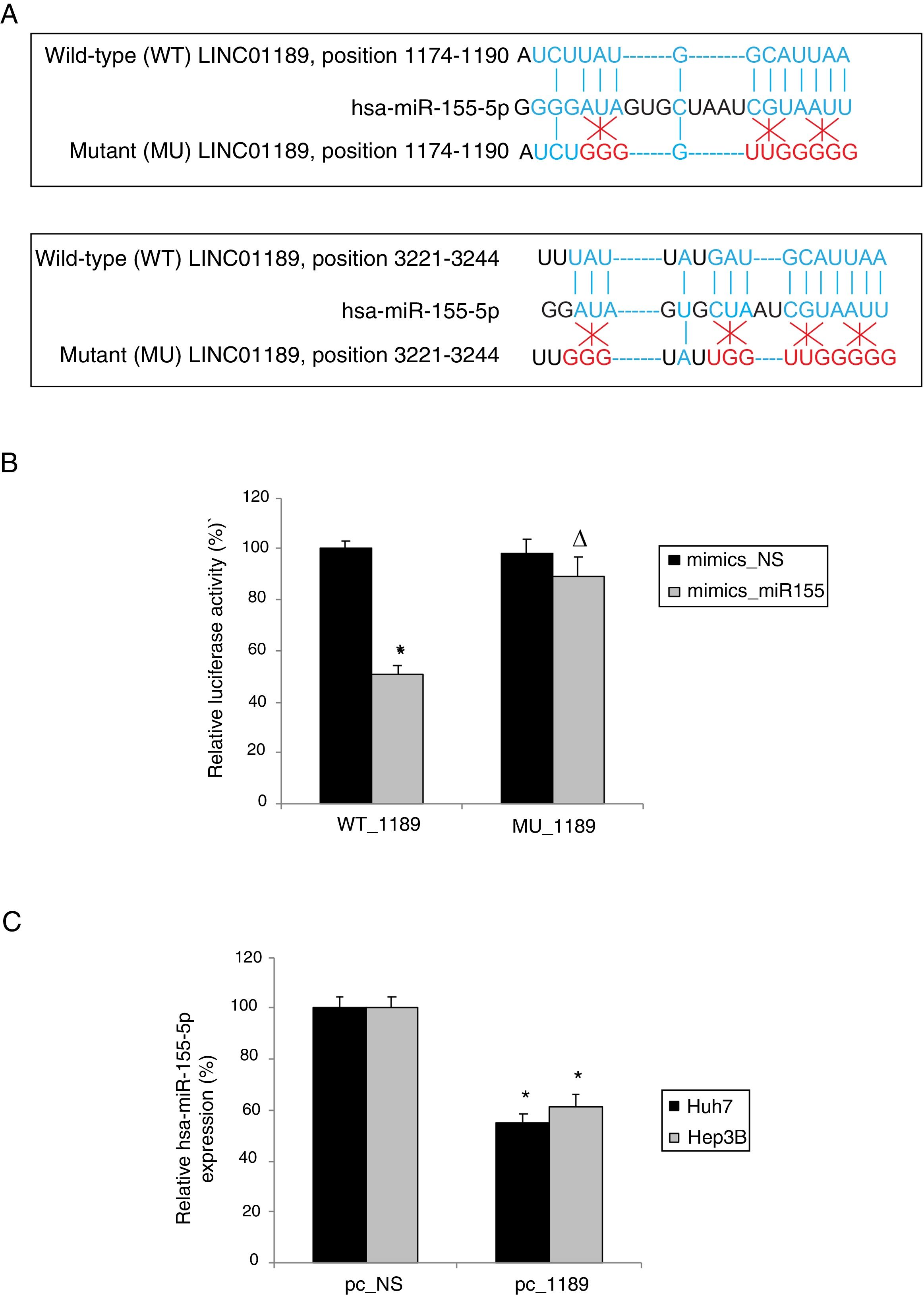
3.4Hsa-miR-155-5p is a downstream ceRNA candidate of LINC01189 in HCCWe then explored the downstream ceRNA candidates of LINC01189. Through bioinformatics research (StarBase 2.0) [21], it was noticed that human microRNA-155-5p (hsa-miR-155-5p) may be attached to two complementary DNA sequences in the 3′-UTR of LINC01189 (Fig. 4A). Based on this information, two luciferase reporters were constructed. One included the wild type LINC01189 3’-UTR, WT_1189. The other included a mutant LINC01189 3’-UTR, MU_1189, with hsa-miR-155-5p binding sequences point-mutated. Then, in a dual-luciferase activity assay, it was confirmed that hsa-miR-155-5p is a downstream ceRNA candidate of LINC01189 (Fig. 4B, * P<0.05; Δ P>0.05). Moreover, in Huh7 and Hep3B cells transfected with pc_1189, qRT-PCR demonstrated that their endogenous hsa-miR-155-5p expression levels were significantly lower than in HCC cells transfected with pc_NS (Fig. 4C, * P<0.05).
LINC01189 attaches to hsa-miR-155-5p. (A) Two DNA sequences on wild-type LINC01189 3’-UTR were predicted to attach to hsa-miR-155-5p. The binding sites were then point-mutated. (B) The wild type and mutant LINC01189 3’-UTRs were used to generate two luciferase reporter vectors, WT_1189 and MU_1189. Then, hsa-miR-155-5p mimics (mimics_miR155) or a non-specific miRNA mimics (mimics_NS) were co-transfected with WT_1189 or MU_1189 in human HEK293T cells. And a dual-luciferase activity assay was conducted 48h later. (C) In pc_1189- or pc_NS- transfected Huh7 and Hep3B cells, qRT-PCR was performed to compare hsa-miR-155-5p expressions levels (* P<0.05).
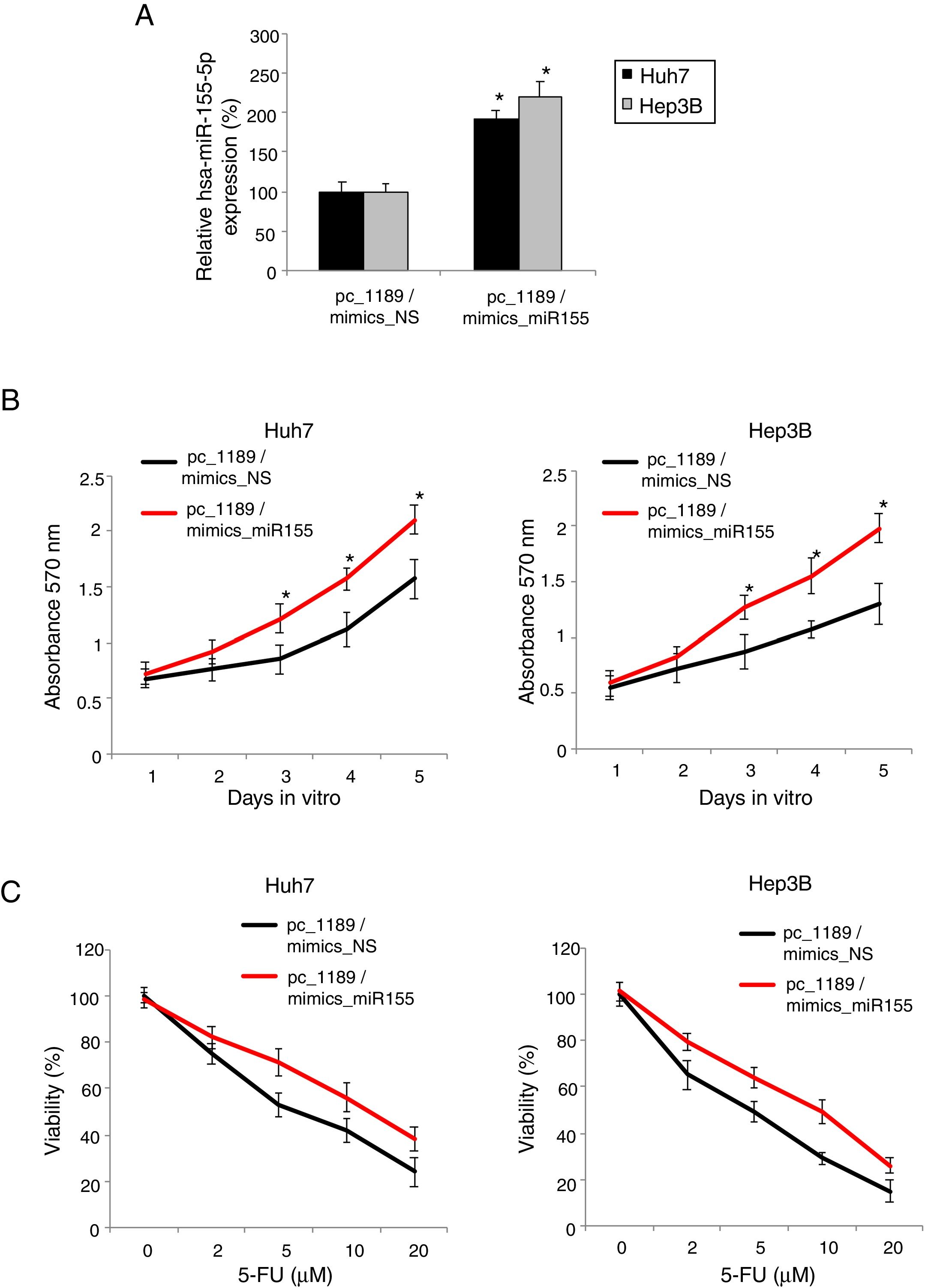
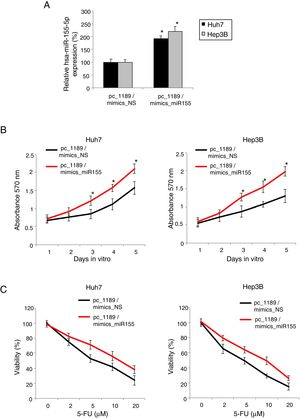
Finally, we investigated the functional relationship between hsa-miR-155-5p and LINC01189 in HCC. In Huh7 and Hep3B cells transfected with pc_1189, we double-transfected them with mimics_miR155 to upregulate hsa-miR-155-5p expression. QRT-PCR demonstrated that, as compared to mimics_NS transfection, mimics_miR155 transfection significantly drove up hsa-miR-155-5p expressions in pc_1189-overexpressed HCC cells (Fig. 5A, * P< 0.05).
LINC01189 -mediated HCC proliferation and chemoresistance was regulated by hsa-miR-155-5p. (A) Pc_1189-transfected Huh7 and Hep3B cells were double-transfected with a hsa-miR-155-5p mimics (mimics_miR155) or a non-specific miRNA mimics (mimics_NS). QRT-PCR was used to compare endogenous hsa-miR-155-5p expression levels (* P<0.05). (B) CCK-8 assay was conducted to compare proliferation in HCC cells transfected with pc_1189 / mimics_miR155 and those transfected with pc_1189 / mimics_NS (* P<0.05). (C) For double-transfected Huh7 and Hep3B cells, they were incubated with 5-FU at 0, 2, 5, 10 and 20μM for 24h. After that, chemoresistance was compared using a viability assay (* P< 0.05).
Then, double-transfected Huh7 and Hep3B were examined by the CKK-8 assay for 5 days. It showed that upregulating hsa-miR-155-5p markedly promoted proliferation in LINC01189-overexpessed HCC cells (Fig. 5C, * P<0.05).
In addition, double-transfected Huh7 and Hep3B were examined by the 5-FU chemoresistance assay. The results showed that upregulating hsa-miR-155-5p significantly increased 5-FU chemoresistance in LINC01189-overexpessed HCC cells (Fig. 5D, * P< 0.05).
4DiscussionsEmerging evidence has demonstrated that, lncRNAs may play critical roles in various aspects of human liver diseases, including chronic HCV infection, fibrosis and HCC [12–14,23]. In the current study, the main focus was to investigate the expression and functions of a novel lncRNA, LINC01189 in HCC. In a very recent clinical study, it was demonstrated that LINC01189 was aberrantly regulated in peripheral blood mononuclear cells of patients with rheumatoid arthritis, suggesting that LINC01189 may be a potential biomarker for rheumatoid arthritis [24]. Other than that, the expressing pattern or regulatory effects of LINC01189 in other human diseases have never been elucidated, thus very much highlighting the clinical significance and importance of our study.
Firstly in the current study, we applied quantitative qRT-PCR to measure LINC01189 expression pattern in HCC tumors, as well as HCV-infected HCC tumors and cell lines. We discovered that LINC01189 was predominantly downregulated in HCC, especially in HCV-infected HCC tumors and cell lines. Particularly, we discovered that ectopically overexpressing LINC01189 could functionally regulation proliferation in HCV-infected HCC cancer cells. In an early study, Diaz and colleagues indicated that epigenetic transcripts, miRNAs were closely correlated with HCV-related HCC [13]. In another study, it was demonstrated that lncRNA of LINC01419 was drastically upregulated in Hepatitis B Virus- and HCV-associated HCCs [13]. Along with the discovery in our study, it’s becoming more obvious that epigenetic factors, including miRNAs and lncRNAs, may have profound roles in the development of HCV-related HCC.
An interesting observation in the current study is that, we were not able to knock down LINC01189 expression in HCC cell lines, either with or without HCV infection. One may suspect that the reason we did not observe LINC01189 downregulation after siRNA transfection was due to technique difficulty. Several lines of evidence argue against it. First, while we manufactured the LINC01189-specific siRNA, we did not use single siRNA. Instead, we mixed four different siRNAs aiming at different targets on LINC01189, which was confirmed by the manufacturer to be able to efficiently knock down >∼85% gene expression of LINC01189 in various types of hosting cells (data not shown). Second, besides siRNA, we also tried other downregulating strategies, such as lentiviral transfection of short hair-pin RNAs (shRNAs). However, we discovered a large portion of transfected HCC cells was apoptotic or dead that we could not efficiently maintain them in the culture. Thus, while we could not rule out the possibility that other technique manipulations may achieve the goal of LINC01189 downregulation in HCC cell lines, the possible explanation for the obtained results might be that LINC01189 was already maximally suppressed (or at the minimal level of expression), that any downregulation approach may not be sustainable to maintain cell health. On the other hand, it is worth-noting that, our strategy of overexpressing LINC01189 was a success, which allowed us to reveal functional roles of LINC01189 in regulating cancer cell development in both HCV-related and HCV-unrelated manners, a novel discovery of biological implication of LINC01189 in human diseases.
Another important finding of our study is that we demonstrated that LINC01189 acted as a ceRNA to regulate downstream target of hsa-miR-155-5p in HCC. Dual-luciferase assay and qRT-PCR assay demonstrated that LINC01189 did attach to hsa-miR-155-5p, and inhibited hsa-miR-155-5p in HCC cells. Moreover, we showed that LINC01189 -mediated inhibition on cancer cell proliferation and 5-FU chemoresistance were reversed by hsa-miR-155-5p upregulation, thus indicating the functional role of epigenetic axis of LINC01189 / hsa-miR-155-5p in regulating HCC development. In previous studies, the prognostic implication and functional roles of hsa-miR-155 in HCC were well-documented [25–27]. However, the current work is the first study to reveal a possible upstream regulatory mechanism of hsa-miR-155-5p. Future works, possibly focusing on the downstream upregulated or downregulated signaling pathways of LINC01189 / hsa-miR-155-5p axis may further broaden our understanding on underlying mechanisms of modulations in HCC.
Ethics statementIn this study, the ethics approval was obtained from the Medical Research and Ethics Committees at the Xi’an Jiaotong University Medical College Red Cross hospital in Xi’an, Shaanxi, China. Consent forms were signed by all participating patients. Clinical and experimental procedures were performed according to the guideline of the WMA Declaration of Helsinki — ethical principles for medical research involving human subjects (https://www.wma.net/policies-post/wma-declaration-of-helsinki-ethical-principles-for-medical-research-involving-human-subjects/).
Conflict of InterestNone.
The project was supported by the key research and development program of Shaanxi (Grant No: 2019SF-156).