Special issue on hepatocellular carcinoma (HCC) and hepatitis B and C as its main causes worldwide
More infoHepatocellular carcinoma (HCC) is one of the most common and fatal cancers in the world. This study aims to investigate the mechanism by which miR-221-3p regulates HCC cell proliferation, migration and invasion, so as to provide a new idea for targeted therapy towards HCC.
Materials and MethodsExpression quantification data including mature miRNA and mRNA were accessed from TCGA-LIHC dataset, and matched clinical information was obtained as well, which helped identify the miRNA of interest. Thereafter, effect of the miRNA on HCC cell biological functions was assessed with a series of in vitro experiments, such as qRT-PCR, MTT, wound healing assay and Transwell. To gain more insight into the mechanism of the miRNA in HCC, bioinformatics method was conducted to predict downstream target gene. The potential targeting relationship between the miRNA and the predicted mRNA was validated by dual-luciferase reporter assay. Western blot was performed to test protein expression.
ResultsMiR-221-3p identified by differential expression analysis was found to be significantly elevated in HCC tissue. Overexpressing miR-221-3p noticeably enhanced HCC cell proliferative, migratory and invasive abilities. Leukemia inhibitory factor receptor (LIFR), confirmed as a downstream target of miR-221-3p in HCC by dual-luciferase reporter assay, was poorly expressed in HCC tissue and cells. Additionally, the expression of LIFR was decreased following the targeted binding between miR-221-3p and LIFR 3’-UTR, while increasing the expression of LIFR attenuated the promoting effect of miR-221-3p on HCC cells.
ConclusionMiR-221-3p is an oncogene in HCC cells, and it exerts its role in HCC cell viability and motility via targeting LIFR.
According to statistics [1], hepatocellular carcinoma (HCC) accounts for 75-85% of primary liver cancers, and globally, it has become the sixth most common cancer, accounting for approximately 5% of new cancer cases. Additionally, it is also the fourth leading cause of cancer death, taking up approximately 8% of all cancer deaths in 2018. Surgical resection is the main treatment for early HCC patients, but most of the cases are diagnosed at advanced stages, with frequent metastasis, recurrence, chemotherapy resistance, and poor prognosis [2,3]. In addition, the mechanism targeting HCC tumorigenesis is still unclear, and there is a lack of effective targeted drugs. In view of these, this study aims to provide a new insight into HCC pathogenesis, clinical diagnosis and treatment.
MicroRNAs (miRNAs) are single-stranded, non-coding, endogenous and evolutionarily conserved RNA molecules with about 18–25 nucleotides in length [4]. Previous research proved that miRNAs play a key role in various cell biological processes by serving as tumor suppressor genes or oncogenes [5]. MiRNAs have molecular functions in regulating cell differentiation, proliferation and apoptosis [6]. As regulatory factors that negatively regulate gene expression, miRNAs are able to completely or incompletely base-pair with the 3’-untranslated region (3’-UTR) of target mRNAs, thereby leading to the direct mRNA degradation or translational interruption [7,8]. The aberrant expression of miRNAs in human tumors has been widely reported. For instance, expression levels of several miRNAs, such as miR-106b [9], miR-657 [10] and miR-372 [11], are differentially up-regulated or down-regulated in tumor cells, which is likely to regulate HCC occurrence via affecting cell proliferation, migration, invasion and apoptosis.
MiR-221 is aberrantly expressed in various cancers, such as HCC [12], gastric cancer [13] and breast cancer [14]. Hence, miR-221 is likely to be closely associated with tumorigenesis, and may become a potential target for the diagnosis and treatment of tumors. Additionally, miR-221-3p as an antiangiogenic gene-regulating miRNA is encoded by a gene on X chromosome. It is proven that miR-221-3p plays a crucial role in cell proliferation, migration, invasion and apoptosis of various human malignant tumors [15–17]. However, there are few studies about the functional mechanism of miR-221-3p in HCC.
Accordingly, we investigated the functional mechanism by which miR-221-3p regulates HCC cell proliferation, migration and invasion via a series of experiments to further explore HCC pathogenesis. Our study may provide a new idea for the clinical diagnosis and treatment of HCC.
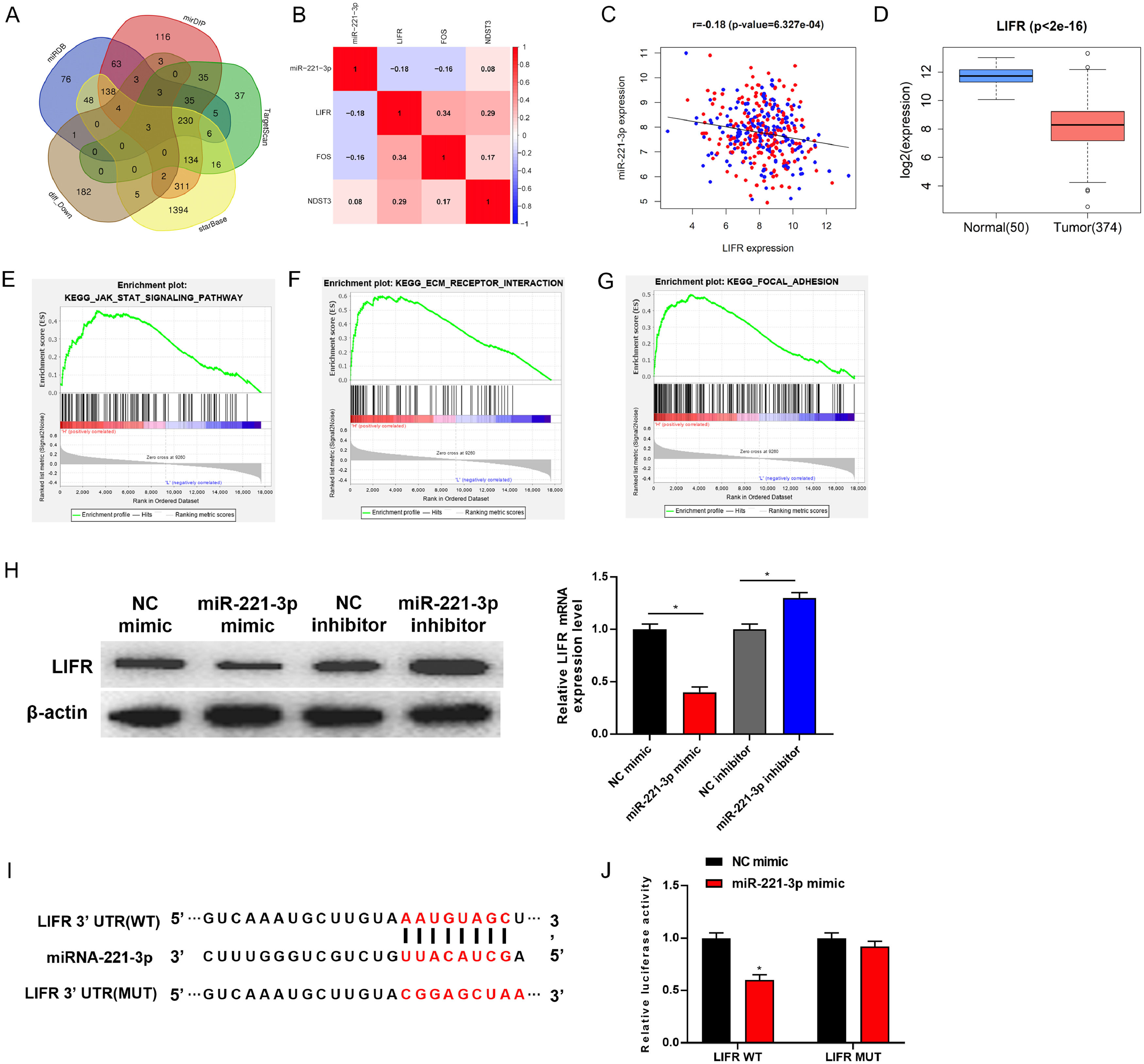
2Materials and methods2.1Bioinformatics analysisExpression data of relevant mature miRNAs (including 50 normal samples, 375 cancer samples) and mRNAs (including 50 normal samples, 374 cancer samples) were acquired from The Cancer Genome Atlas-Liver Hepatocellular Carcinoma (TCGA-LIHC) dataset. Package edgeR was used to perform differential expression analysis on miRNAs (|logFC|>1.5, padj<0.01) and mRNAs (|logFC|>2, padj<0.01), respectively. Combined with the differential analysis results, miR-221-3p was ultimately determined as the research target. The miRDB, mirDIP, TargetScan and starBase databases were used to perform target prediction for miR-221-3p. Then, the predicted mRNAs were mapped into a Venn diagram with the mRNAs differentially downregulated in tumor tissue in TCGA-LIHC, and the mRNA had the highest correlation with miR-221-3p was selected as the research object.
2.2Cell cultureNormal human liver cell line HL-7702 [L-02] (No. 3131C0001000200006) was purchased from Shanghai Institutes for Biological Sciences, CAS. Human HCC cell lines Hep3b (No. 3111C0001CCC000376), HepG2 (No. 3111C0001CCC000035) and Huh7 (No. 3111C0001CCC000679) were ordered from Cell Source Center, Institute of Basic Medical Sciences, CAMS. All the cell lines were cultured in Dulbecco's Modified Eagle's Medium (DMEM) supplemented with 20% fetal bovine serum (FBS), 100 U/mL penicillin and 100 μg/ml streptomycin. All the cell lines were incubated in a humidified atmosphere of 5% CO2 at 37°C.
2.3Cell transfectionMiR-221-3p mimic, miR-221-3p inhibitor, leukemia inhibitory factor receptor (LIFR) over-expression vector (oe-LIFR), LIFR siRNA (si-LIFR) and negative control purchased from GeneChem (Shanghai, China) were transfected into cells by using the Lipofectamine 2000 reagent (Invitrogen, Carlsbad, CA, USA).
2.4Real-time quantitative PCR (qRT-PCR)Total RNA was extracted using TRIzol reagent (Invitrogen, Carlsbad, CA) according to the manufacturer's instructions. Afterward, 1 μg RNA was taken and transcribed into cDNA using universal primers or miR-221-3p-specific stem-loop primers, respectively. qRT-PCR was performed on ABI 7500 (Applied Biosystems, Mannheim, Germany) using specific primers under the following thermocycling conditions: pre-denaturation at 95°C for 10 min, 94°C for 2 min, followed by 30 cycles of 94°C for 30 s, 58°C for 30 s and 72°C for 30 s. GAPDH and U6 were used as internal references for detection of LIFR mRNA and miR-221-3p expression level, respectively. The 2−ΔΔCt method was used to compare the difference in relative expression of target genes between the control group and test group. The experiment was repeated three times. The primer sequences were listed in Supplementary Table 1.
2.5MTT assayMTT assay was performed to assess cell proliferation. HepG2 or Huh7 cells were seeded into a 96-well plate at a density of 5 × 103 or 1 × 104 cells/well. After cells were grown to 70–80% in confluence, the constructed plasmids or gene fragments were transfected into the above cells using Lipofectamine 2000. At 0 h, 24 h, 48 h and 72 h, the cells were treated using 20 µl of MTT solution (5 mg/ml PBS) (Sigma, St Louis, MO, USA) at 37°C for 4 h. Subsequently, the supernatant was removed and 150 µl dimethyl sulfoxide (DMSO) was added to each well. Finally, absorbance of each well at 490 nm was identified by a microplate reader (TECAN, Männedorf, Switzerland). The experiment was repeated three times.
2.6Cell migration and invasion assaysWound healing assay was performed for evaluation of cell migratory ability. HepG2 cells were inoculated into a 3.5-cm culture plate. After cells were grown to 70–80% in confluence, a scratch was made on cell monolayer using a 200 μl pipette tip. The cell migration distance at 0 h and 48 h were measured under a microscope, respectively.
Transwell assay was conducted to assess cell invasive capability using a 24-well plate. HepG2 cells (5 × 104) were seeded into the upper chamber pre-coated with Matrigel, while the lower chamber contained 10% FBS-supplemented medium to stimulate cell invasion. After cells were cultured in a humidified atmosphere of 5% CO2 at 37°C for 24 h, a cotton swab was used to remove non-invasive cells in the upper chamber. Invasive cells in the lower chamber were stained with 0.5% crystal violet, photographed and observed under a microscope.
2.7Dual-luciferase reporter assayIn order to assess the probability of the binding of miR-221-3p to LIFR 3’-UTR, wild-type (WT) and mutant (MUT) LIFR luciferase vectors were generated by cloning WT and MUT LIFR 3’-UTR into pGL3 luciferase vectors (Promega Corporation). By using Lipofectamine 2000 reagent (Invitrogen), HepG2 cells were co-transfected with LIFR WT/LIFR MUT vectors and NC mimic/miR-221-3p mimic. At last, luciferase activity was determined using the Dual-Luciferase Reporter Assay kit (Promega, Fitchburg, WI, USA).
2.8Western blotFirstly, HepG2 cells were lysed with radio-immunoprecipitation (RIPA) lysis buffer (SOLAIBO, CHINA) 48 h after transfection. Then, total proteins were extracted. Concentration of the proteins was measured by a bicinchoninic acid (BCA) protein assay kit. For sodium dodecyl sulphate-polyacrylamide gel electrophoresis (SDS-PAGE), each lane was added with 50 μg total proteins. Following electrophoresis, proteins were transferred onto poly-vinylidene difluoride (PVDF) membranes (Bio-Rad, Her-cules, California). After blocked with 5% non-fat milk at room temperature for 1 h, the membranes were incubated overnight with primary antibodies and then washed with TBST 3 times. Goat anti-rabbit secondary antibody IgG labeled with horseradish peroxidase was then incubated with the membranes at room temperature for 2 h. The membranes were sequentially washed 3 times with TBST. Enhanced chemiluminescence reagent was used for 60 s of signal development. Analysis of relative protein expression was performed with Image J (National Institutes of Health, Bethesda, Maryland) software. The experiment was repeated three times. The antibodies were listed in Supplementary Table 2.
2.9Statistical analysisAll statistical analyses were performed using SPSS 22.0 software. All measurement data were expressed as mean ± standard deviation (SD). In addition, Student's t-test was used to analyze the data differences between two groups, while one-way analysis of variance was used to analyze data differences among multiple groups. * represents p < 0.05.
3Results3.1MiR-221-3p is up-regulated in HCC tissue and cellsDifferential analysis was performed on TCGA-LIHC data to obtain differentially expressed miRNAs in HCC (Fig. 1A). From the data analyzed, miR-221-3p was markedly elevated in HCC tumor tissue (Fig. 1B). Research suggested that miR-221 is aberrantly expressed in various cancers and is closely related to tumor malignant progression [12,15–17]. Hence, miR-221-3p was then focused in this study. qRT-PCR was carried out to determine miR-221-3p expression in normal human liver cell line and human HCC cell lines (Hep3b, HepG2 and Huh7). The result demonstrated that miR-221-3p was highly expressed in HCC cell lines with the highest up-regulation observed in HepG2 cells (Fig. 1C). Collectively, these results unveiled that miR-221-3p was up-regulated in HCC tissue and cell lines.
MiR-221-3p is up-regulated in HCC tissue and cells
(A) Volcano map of differential miRNAs in TCGA-LIHC dataset. Red represents differential up-regulation, and blue represents differential down-regulation; (B) Relative expression of miR-221-3p in TCGA-LIHC dataset; (C) qRT-PCR was performed to detect miR-221-3p expression in normal human liver cell line HL-7702 and human HCC cell lines (Hep3b, HepG2 and Huh7); p < 0.05.
To identify the functional role of miR-221-3p in HCC cell progression, HepG2 and Huh7 cell lines which had relatively high miR-221-3p expression were used to overexpress and knock down miR-221-3p expression. qRT-PCR uncovered that the expression of miR-221-3p in the two cell lines transfected with miR-221-3p mimic was noticeably increased, while decreased after cells transfected with miR-221-3p inhibitor (Fig. 2A). In order to explore the effect of miR-221-3p on HCC cell biological functions, MTT assay was firstly conducted to analyze cell proliferative ability, finding that cell proliferation of these two cell lines was remarkably promoted by over-expressed miR-221-3p. Additionally, the cell proliferative ability was reduced when miR-221-3p was suppressed (Fig. 2B).
MiR-221-3p fosters HCC cell proliferation, migration and invasion
(A) qRT-PCR was conducted to detect the expression of miR-221-3p in HepG2 and Huh7 cell lines upon over-expression or inhibition of miR-221-3p; (B) Proliferative ability of HepG2 and Huh7 cell lines was examined by MTT; (C-D) Migratory and invasive capacities of HepG2 cells were evaluated via (C) wound healing assay and (D) Transwell assay; p < 0.05.
To better understand the effect of miR-221-3p on human HCC cell migratory and invasive capacities, HepG2 cell line was chosen for wound healing assay and Transwell assay. Wound healing assay indicated that over-expressing miR-221-3p markedly promoted HepG2 cell migration, while silencing it produced an opposite effect (Fig. 2C). Similarly, Transwell assay demonstrated that HepG2 cell invasive ability was significantly raised upon miR-221-3p over-expression but markedly weakened by down-regulated miR-221-3p (Fig. 2D).
Taken together, the above results showed that miR-221-3p is likely to be an oncogene in HCC to foster HCC cell proliferation, migration and invasion.
3.3LIFR is directly targeted and down-regulated by miR-221-3p in HCCIn order to gain deeper insight into the molecular mechanism by which miR-221-3p regulates human HCC cell proliferation, migration and invasion, bioinformatics analysis was performed to find potential target genes of miR-221-3p. The miRDB, mirDIP, TargetScan and starBase databases were consulted to find potental target genes of miR-221-3p. The predicted target genes were then intersected with differentially down-regulated mRNAs in TCGA-LIHC dataset (Fig. 3A). Three target genes (LIFR, FOS and NDST3) were obtained ultimately. By using correlation analysis, LIFR having the highest correlation coefficient with miR-221-3p was confirmed as the ultimate target gene (Fig. 3B-C). As TCGA data revealed, LIFR was lowly expressed in HCC tumor tissue (Fig. 3D). Additionally, GSEA pathway enrichment analysis results of LIFR showed that LIFR was enriched in JAK-STAK, KEGG_ECM_RECEPTOR_INTERACTION, KEGG_FOCAL_ADHESION and other pathways related to cell development, proliferation, migration and invasion (Fig. 3E–G). This indicated that LIFR may be involved in regulating the proliferation, migration and invasion of HCC. In addition, studies verified that LIFR is able to serve as a tumor metastasis suppressor gene and prognostic biomarker for HCC sufferers [18–20]. Taken together, we speculated that miR-221-3p may foster HCC cell proliferation, migration and invasion via targeting LIFR. The specific bioinformatics analysis process was shown in Supplementary Fig. 1.
LIFR is directly targeted and down-regulated by miR-221-3p in HCC
(A) Target genes of miR-221-3p were predicted by miRDB, mirDIP, TargetScan and starBase databases, and were then intersected with differentially down-regulated mRNAs in TCGA-LIHC dataset; (B) Pearson correlation analysis of miR-221-3p and 3 candidate target genes; (C) Pearson correlation analysis of miR-221-3p and LIFR gene; (D) Relative expression of LIFR in TCGA-LIHC dataset; (E-G) GSEA pathway enrichment analysis of LIFR; (H) Western blot and qRT-PCR were performed to determine the protein and mRNA expression of LIFR in HepG2 cells upon miR-221-3p over-expression or silence, respectively; (I) starBase database was employed to predict the binding site between miR-221-3p and LIFR 3’-UTR; (J) Dual-luciferase reporter assay was conducted to detect whether miR-221-3p targets LIFR; p < 0.05.
Afterwards, the relationship between miR-221-3p and LIFR was validated. Western blot and qRT-PCR revealed that over-expression of miR-221-3p silenced LIFR, while silenced miR-221-3p reversely activated LIFR (Fig. 3H). To confirm the probability of the binding of miR-221-3p to LIFR 3’-UTR (Fig. 3I), LIFR-WT and LIFR-MUT vectors were constructed and co-transfected with NC mimic or miR-221-3p mimic into HepG2 cells. Dual-Luciferase reporter assay indicated that over-expressing miR-221-3p inhibited the luciferase activity of cells with LIFR WT, but showed no effect on that of cells with LIFR MUT (Fig. 3J). Collectively, these experiments demonstrated that LIFR was directly targeted and down-regulated by miR-221-3p.
3.4LIFR suppresses HCC cell proliferation, migration and invasionTo gain deeper insight into the underlying functions of LIFR in HCC, HepG2 cells were transfected with oe-LIFR, si-LIFR and their negative controls (oe-NC and si-NC), respectively. qRT-PCR and western blot were carried out to evaluate the mRNA and protein expression levels of LIFR in each group, respectively (Fig. 4A-B). MTT assay unveiled that cell proliferation was promoted by silencing LIFR, but inhibited by over-expressing LIFR (Fig. 4C). Wound healing assay uncovered that cell migration was fostered by silencing LIFR, but repressed upon LIFR over-expression (Fig. 4D). Transwell assay indicated that cell invasive capability was enhanced by silencing LIFR, but suppressed upon LIFR over-expression (Fig. 4E). Collectively, these results verified that LIFR could inhibit HCC cell proliferation, migration and invasion.
LIFR suppresses HCC cell proliferation, migration and invasion
(A-B) qRT-PCR and western blot were carried out to evaluate the mRNA and protein expression levels of LIFR in HepG2 cells overexpressing or silencing LIFR; (C-E) HCC cell proliferative, migratory and invasive abilities were assessed via (C) MTT assay, (D) wound healing assay and (E) Transwell assay, respectively; p < 0.05.
In order to explore the effect of miR-221-3p targeting LIFR on HCC cells, LIFR over-expression vector (or empty vector) and miR-221-3p mimic (or NC mimic) were co-transfected into HepG2 cells.
Western blot suggested that after co-transfection of miR-221-3p mimic and oe-LIFR, the inhibition of LIFR induced by miR-221-3p was reversed by oe-LIFR (Fig. 5A). MTT assay indicated that LIFR overexpression reversed the promoting effect of miR-221-3p on the proliferative ability of HCC cells (Fig. 5B). Wound healing and Transwell assays revealed that the restoration of LIFR markedly repressed miR-221-3p-induced HCC cell migration and invasion. Besides, overexpression of miR-221-3p also reduced the inhibitory effect of overexpressed LIFR on HCC cell migration and invasion (Fig. 5C-D). Taken together, it could be concluded that miR-221-3p facilitated HCC cell growth via targeting and down-regulating LIFR, while this promoting effect could be attenuated by over-expressing LIFR.
MiR-221-3p promotes HCC cell growth via targeting LIFR
(A) Western blot was performed to determine LIFR protein expression in HepG2 cells after co-transfection of miR-221-3p mimic and oe-LIFR; (B-D) HCC cell proliferative, migratory and invasive capacities were assessed via (B) MTT assay, (C) wound healing assay and (D) Transwell assay, respectively; p < 0.05.
HCC is a type of primary liver cancer. MiRNAs are noncoding RNA molecules which are able to regulate gene expression [21,22]. Apart from important roles in healthy individuals, miRNAs also get involved in carcinogenesis and occurrence of other human diseases [23]. For example, miRNAs are over-expressed and play a carcinogenic role in most epithelial malignancies, such as papillary thyroid carcinoma [24], pancreatic cancer [25] and lung cancer [26]. As underlying tumor markers, miRNAs have been discussed in many fields of oncology, and a study indicated that miRNAs play important roles in HCC development [27].
Previous research revealed that miR-221 is aberrantly expressed in HCC tissue. Although its functional mechanism in tumors remains unclear [4], the significance of miR-221 is obvious as it is likely to be a breakthrough target for tumor prevention, diagnosis and treatment. Besides, miR-221-3p plays different roles in different cancers. For instance, miR-221-3p acts as a tumor suppressor gene in epithelial ovarian cancer and directly targets ARF4, which unveils that it may be helpful for the clinical prognosis and treatment of epithelial ovarian cancer [17]. Additionally, miR-221-3p plays an oncogenic role in gastric cancer by silencing PTEN to promote the metastasis of gastric cancer via progesterone [28]. However, the function and underlying molecular mechanism of miR-221-3p in human HCC are poorly understood. In the present study, miR-221-3p was highly expressed in HCC tissue and cell lines. Accordingly, we speculated that miR-221-3p may play a regulatory role in HCC as an oncogene. To verify this speculation, we performed MTT, wound healing and Transwell assays to prove that over-expressing miR-221-3p facilitated the proliferation, migration and invasion of HCC cells.
Bioinformatics analysis was employed to predict that miR-221-3p may foster the occurrence and development of HCC through targeting and regulating LIFR. Besides, we conducted dual-luciferase reporter assay and western blot to validate that LIFR was a direct target of miR-221-3p. LIFR as a member of the family of type I cytokine receptors plays different roles in different cancers. Studies uncovered that LIFR as a marker of poor prognosis is up-regulated in various tumor tissues, such as melanoma [29], nasopharyngeal carcinoma (NPC) [30] and prostate cancer [31]. It was found that LIFR is able to foster nasopharyngeal carcinoma tumor growth and repress DNA damage response by responding to extracellular ligand leukemia inhibitory factor (LIF). In addition, it was also proved that feedback activation of LIFR limits response to histone deacetylase inhibitors [32]. In addition to this, LIFR also plays a tumor-suppressive role in many cancers. For instance, LIFR acts as a tumor suppressor gene and tumor metastasis suppressor gene in breast cancer, and the aberrant methylation of LIFR promoter in India breast cancer sufferers is closely related to patient's clinical features [33]. Additionally, research reported that LIFR is able to inhibit the migration and invasion of pancreatic cancer [34] and breast cancer [35] cells. Due to the functional specificity of LIFR in different cancers and few studies existed about LIFR in HCC, exploration of the functions and the potential molecular mechanisms of LIFR in HCC is highly significant. In this study, LIFR was found decreased in HCC, and played a part in suppressing HCC cell progression.
In conclusion, we found that LIFR was poorly expressed, while miR-221-3p was highly expressed in HCC tissue and cells. After miR-221-3p was elevated in HCC cell lines, miR-221-3p could target and down-regulate LIFR expression, thereby promoting cell growth. Besides, this effect could be inhibited upon LIFR activation. Therefore, it could be concluded here that miR-221-3p promoted HCC cell proliferation, migration and invasion by targeting LIFR, which may provide a novel underlying target for the treatment of HCC.
Ethical approvalEthical approval is not applicable for this article. This article does not contain any studies with human or animal subjects.
Authors' contributionsConceptualization: Wei Tan;
Methodology: Zhuokai Li;
Formal analysis and investigation: Zhuokai Li and Weifen Xia;
Writing - original draft preparation: Weifen Xia and Wei Tan;
Writing - review and editing: Jinde Zhu and Rengen Fan;
Not applicable.