Abnormal expression of long non-coding RNAs (lncRNAs) plays a prominent role in glioma progression. However, the biological function and mechanism of lncRNA DLGAP1 antisense RNA 1 (DLGAP1-AS1) in gliomas are still unknown.
MethodsThe authors assessed DLGAP1-AS1 and miR-628-5p expression in glioma tissues and cell lines using quantitative real-time polymerase chain reaction (qRT-PCR) and evaluated their effects on glioma cell proliferation, migration, invasion, and epithelial-mesenchymal transition (EMT) using the cell counting kit-8 (CCK-8) assay, 5-Ethynyl-2′-deoxyuridine (EdU) assay, Transwell assay, and western blot, respectively. The expression of DEAD-box helicase 59 (DDX59) was quantified using western blotting, and a dual-luciferase reporter gene assay was performed to detect the interaction between DLGAP1-AS1 and miR-628-5p.
ResultsThe authors observed increased DLGAP1-AS1 expression in glioma tissues and cell lines with higher WHO grades and shorter survival time. DLGAP1-AS1 promoted the proliferation, migration, invasion, and EMT of glioma cells, while miR-628-5p counteracted these effects. The authors identified DLGAP1-AS1 as a molecular sponge of miR-628-5p in glioma cells as the biological functions of DLGAP1-AS1 are partially mediated via miR-628-5p. In addition, DLGAP1-AS1 upregulated DDX59 expression by inhibiting miR-628-5p expression.
ConclusionThe DLGAP1-AS1/miR-628-5p/DDX59 axis regulates glioma progression.
Glioma – a cancer of the brain and spinal cord – has a high recurrence rate, morbidity and mortality, and poor prognosis.1-3 Elucidating the mechanism of glioma progression has great significance with respect to improving the prognosis of patients with this deadly disease.
Long non-coding RNAs (lncRNAs) are ≥ 200 nt in length, lack protein-coding capabilities but are involved in regulating biological processes, such as gene imprinting, RNA splicing, and chromatin modification.4-6 LncRNAs can control gene expression at the transcriptional and post-transcriptional levels and thus play key roles in cancer biology.7 In addition, as lncRNA expression exhibits tissue specificity,8 they may serve as biomarkers and potential therapeutic targets. For example, lncRNA ATB promotes the growth of gastric cancer by regulating the miR-141-3p/TGF-β2 axis.9 LncRNA SIK1-LNC inhibits the proliferation and metastasis of lung cancer cells, and its expression is downregulated in lung cancer.10 Recently, lncRNA DLGAP1 antisense RNA 1 (DLGAP1-AS1) has been found to act as an oncogenic lncRNA with its expression upregulated in hepatocellular carcinoma.11,12 However, little is known about the role of DLGAP1-AS1 in gliomas.
MicroRNAs (miRNAs) – small non-coding RNAs approximately 21–25 nt in length – are crucial players in cancer biology.13-16 They can repress gene expression by binding to the 3ʹ-untranslated region (3’-UTR) of an mRNA, regulating various physiological and pathological processes.17 Reportedly, miR-628-5p represses the malignant phenotypes of glioma cells by targeting high mobility group protein B3 (HMGB3) and DEAD-box helicase 59 (DDX59). It is downregulated in glioma tissues and cells, indicating that miR-628-5p is a tumor suppressor in the brain.18,19
In the present study, the authors investigated the expression patterns, biological functions, and mechanisms of action of DLGAP1-AS1 in gliomas. The authors found that DLGAP1-AS1 expression was significantly upregulated in glioma tissues and cell lines, and it promoted the proliferation, migration, invasion, and Epithelial-Mesenchymal Transition (EMT) of glioma cells. Mechanistically, DLGAP1-AS1 functions as a competitive endogenous RNA (ceRNA) to sponge miR-628-5p and upregulate DDX59 expression. The present study proposes a novel ceRNA network for glioma progression.
Material and methodsTissues collectionTissue samples were obtained from patients who underwent surgery in the Department of Neurosurgery, Xiangyang Central Hospital, from May 2014 to July 2018, including 59 glioma tissue samples and 59 corresponding adjacent non-tumor tissue samples. After removal, the samples were stored in liquid nitrogen at -196°C. This study followed the 2007 World Health Organization (WHO) classification of tumors of the central nervous system (WHO grade I, n = 12: pilocytic astrocytomas (n = 8) and myxopapillary ependymomas (n = 4); grade II, n = 23: diffuse astrocytomas (n = 17), oligoastrocytomas (n = 3), and oligodendrogliomas (n = 3); grade III, n=14, anaplastic astrocytomas (n=6), anaplastic oligodendrogliomas (n = 5), and anaplastic oligoastrocytomas (n = 3); grade IV, n = 10: glioblastomas). The tumor samples were divided into low-grade tumors (grade I and II, n = 35) and high-grade tumors (grade III and IV, n = 24).20 This study was approved by the Research Ethics Committee of the Xiangyang Central Hospital. Written informed consent was obtained from each patient.
Cell lines and cell cultureThe Cell Bank of Shanghai Institutes for Biological Sciences, Chinese Academy of Sciences (Shanghai, China) provided the glioma cell lines (U-118MG, U251, U87MG, and LN229 cells), astroglia cell line (HA cells), and human embryonic kidney cell line (HEK-293 cells). The cells were maintained in Dulbecco's modified Eagle's medium (DMEM, Thermo Fisher Scientific, Waltham, MA, USA) supplemented with 10% fetal bovine serum (FBS, Gibco, Carlsbad, CA, USA), 100 U/mL penicillin, and 100 μg/mL streptomycin (Gibco, Carlsbad, CA, USA) in 5% CO2 at 37°C.
Quantitative real-time polymerase chain reaction (qRT-PCR)To determine DLGAP1-AS1 and DDX59 expression, the authors extracted the total RNA from tissues or cells using TRIzol reagent (Invitrogen, Carlsbad, CA, USA), reverse transcribed cDNA using a reverse transcriptase kit (Takara, Dalian, China), and performed qRT-PCR with SYBR Green Master Mix (Takara, Dalian, China). The NormFinder program was used to determine the appropriate housekeeping genes for the normalization of qRT-PCR data. After evaluating GAPDH (M-value = 0.518) and ACTB (M-value=0.646), the authors used them as the endogenous controls.21 To determine miR-628-5p expression, qRT-PCR was performed using a TaqMan miRNA reverse transcription kit (Applied Biosystems, Grand Island, NY) with U6 and U48 as endogenous controls. Relative expression levels of DLGAP1-AS1, DDX59, and miR-628-5p were estimated using the 2−△△CT method, △Ct=Ct (target gene)−Ct (endogenous control), △△Ct=△Ct (test group)−△Ct(normal group).22 The primer sequences were as follows: DLGAP1-AS1 forward, 5′-TATGATGATATCAAGAGGGTAGT-3′ and reverse, 5′-TGTATCCAAACTCATTGTCATAC-3′. DDX59 forward, 5′-GATGTTCCCGTTGATGCTGT-3′ and reverse, 5′-GAGCTTTATTCGAGAGCAAAACT-3′. GAPDH forward, 5′-TGGGTGTGAACCATGAGAAG-3′ and reverse, 5′-GTGTCGCTGTTGAAGTCAGA-3′. ACTB forward, 5′-GTCAGGTCATCACTATCGGCAAT-3′ and reverse, 5′-AGAGGTCTTTACGGATGTCAACGT-3′. miR-628-5p primers, U6 and U48 were provided in the TaqMan miRNA reverse transcription kit.
Cell transfectionSpecific short hairpin RNAs (shRNAs) against DLGAP1-AS1 (sh-DLGAP1-AS1#1, sh-DLGAP1-AS1#2, and sh-DLGAP1-AS1#3), negative control shRNA (sh-NC), pcDNA3.1 vector overexpressing DLGAP1-AS1, and the empty vector were all purchased from GeneChem (Shanghai, China). miR-628-5p mimics, miR-628-5p inhibitor, negative control mimic (NC mimic), and negative control inhibitor (NC inhibitor) were obtained from GenePharma (Shanghai, China). U251, U87MG, or LN229 cells were transfected with Lipofectamine 3000 (Invitrogen, Carlsbad, CA, USA) according to the manufacturer's instructions.
Cell counting kit-8 (CCK-8) assayThe transfected glioma cells were transferred into a 96-well plate (1 × 103 cells/well), and CCK-8 solution (Dojindo Laboratories, Kumamoto, Japan) was loaded into each well at 0, 24, 48, 72, and 96h, respectively, followed by incubation for 3h. The absorbance of each well was recorded at 450 nm using a microplate reader (BioTek, Winooski, VT, USA).
5-Ethynyl-2′-deoxyuridine (EdU) assayThe EdU kit was obtained from RiboBio (Guangzhou, China). The transfected cells were cultured in a 96-well plate (5 × 103 cells/well) for 24h and then incubated with 50 mM EdU reagent for 4h. After discarding the medium, the cells were fixed with 4% paraformaldehyde and incubated for 30 min in the dark with the Apollo fluorescent staining solution. The authors then washed them twice with PBS and incubated with Hoechst staining solution for 20 min. Finally, the authors rinsed the cells three times with PBS and observed and counted them under a fluorescence microscope.
Transwell assayTranswell assays were performed using a Transwell system with 8 μm pore size (Corning Incorporated, Corning, NY, USA). In the migration assay, 100 μL of cell suspension (approximately 1 × 104 cells) prepared in serum-free medium was added to the upper chamber, and 500 μL of medium containing 10% FBS was added to the lower chamber. After the cells were cultured for 12h, the cells on the upper side of the membrane were scraped off, and the remaining cells were fixed with 4% paraformaldehyde and then stained with 0.1% crystal violet for 30 min. Subsequently, the number of stained cells was counted under a microscope. For the cell invasion assay, 50 μL of diluted Matrigel (1:8, Sigma-Aldrich, St Louis, MO, USA) was dripped into the upper chamber of the Transwell system to cover the membrane before the inoculation of the cells, and the remaining steps were the same as in the migration assay.
Dual-luciferase reporter assayWild-type and mutant type DLGAP1-AS1 sequences containing miR-628-5p binding sites were synthesized and inserted into the pGL3 vector (Promega, Madison, WI, USA) to construct a wide-type reporter plasmid (DLGAP1-AS1-WT) and mutant reporter plasmid (DLGAP1-AS1-MUT). HEK-293 cells were then co-transfected with the reporter plasmids, miR-628-5p mimic, or control miRNA using Lipofectamine 3000 (Invitrogen, Carlsbad, CA, USA). After 48h, the cells were harvested, and the luciferase activity of each group was measured using a Dual-Luciferase Reporter Assay kit (Promega, Madison, WI, USA).
Western blotTotal protein was extracted using RIPA buffer (Beyotime, Shanghai, China). Protein samples were quantified using a BCA Protein Assay Kit (Pierce, Rockford, IL, USA), suspended in loading buffer, and denatured. Subsequently, the protein samples were separated via SDS-PAGE and transferred to polyvinylidene fluoride membranes (Life Technologies, Gaithersburg, MD, USA). After blocking with 5% skimmed milk for 1h at room temperature, the PVDF membranes were incubated with primary antibody [Anti-DDX59 (Abcam, ab109592, 1:200) or anti-GAPDH (Abcam, ab8245, 1:2000)] at 4°C overnight and then with secondary antibody (HRP-labeled, Beyotime, 1:2000) for 1.5h at room temperature. Finally, the protein bands were visualized using an ECL Plus kit (Life Technologies, Gaithersburg, MD, USA). GAPDH was used as an endogenous control.
Statistical analysisThe data are shown as the mean ± standard deviation. The normality of the data was evaluated using the Kolmogorov-Smirnov test. For normally distributed data, the Student's t-test or one-way analysis of variance (ANOVA) was employed to analyze the differences between two or multiple groups. For skewed data, comparisons between two groups were performed using the Wilcoxon signed-rank test. In survival analysis, glioblastoma (GBM) patients were divided into two groups: DLGAP1-AS1 high expression (group cutoff: 50%) and DLGAP1-AS1 low expression (group cutoff: 50%) (n=81 in each group), and the overall survival rate of GBM patients was analyzed using the Kaplan-Meier method and log-rank test. GraphPad Prism 6.0 was used for drafting, and SPSS software (version 18.0; SPSS, Inc., Chicago, IL, USA) was used for statistical analysis. Statistical significance was set at p < 0.05.
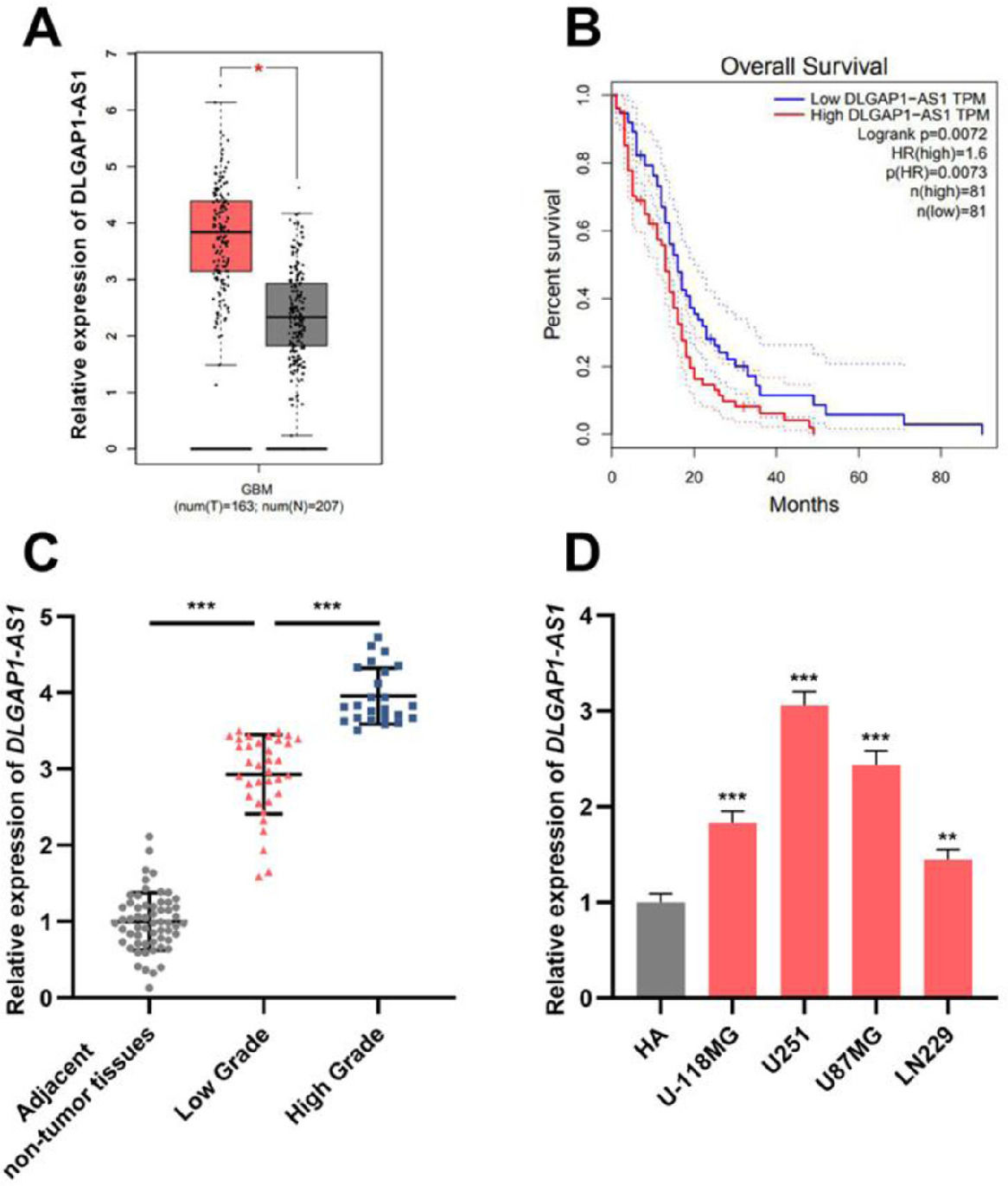
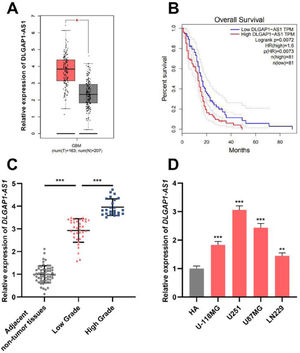
ResultsDLGAP1-AS1 expression is elevated in glioma tissues and cell linesThe GEPIA database showed that DLGAP1-AS1 expression in GBM tissues was higher than that in normal tissues (Fig. 1A). Additionally, Kaplan-Meier survival analysis revealed that high DLGAP1-AS1 expression was associated with poor survival in GBM patients (Fig. 1B). Next, the authors performed qRT-PCR to determine DLGAP1-AS1 expression in gliomas and adjacent non-tumor tissues of 59 glioma patients. The results indicated that DLGAP1-AS1 expression in glioma tissues was remarkably higher than that in adjacent non-tumor tissues, and DLGAP1-AS1 expression was higher in high-grade tumor samples than in low-grade tumor samples (Fig. 1C and Supplementary Fig. 1A). Consistently, DLGAP1-AS1 expression was significantly elevated in glioma cell lines (compared to that in HA cells) (Fig. 1D and Supplementary Fig. 1B). In U251 and U87MG cells, DLGAP1-AS1 was highly expressed, and therefore, U251 and U87MG cells were chosen for the follow-up experiments.
DLGAP1-AS1 expression is upregulated in glioma tissues and cell lines. A, Bioinformatic analysis was used to analyze DLGAP1-AS1 expression in GBM and normal brain tissues. B, The GEPIA database was used to perform survival analysis of GBM patients with high and low DLGAP1-AS1 expression. C, The expression of DLGAP1-AS1 in glioma tissues and adjacent non-tumor tissues was detected using qRT-PCR (n = 59). D, DLGAP1-AS1 expression in glioma cell lines (U-118MG, U251, U87MG, and LN229 cells) and normal cell lines (HA cells) was detected using qRT-PCR. The experiments were repeated three times, and the average was recorded. *p < 0.05, **p < 0.01, and ***p < 0.001.
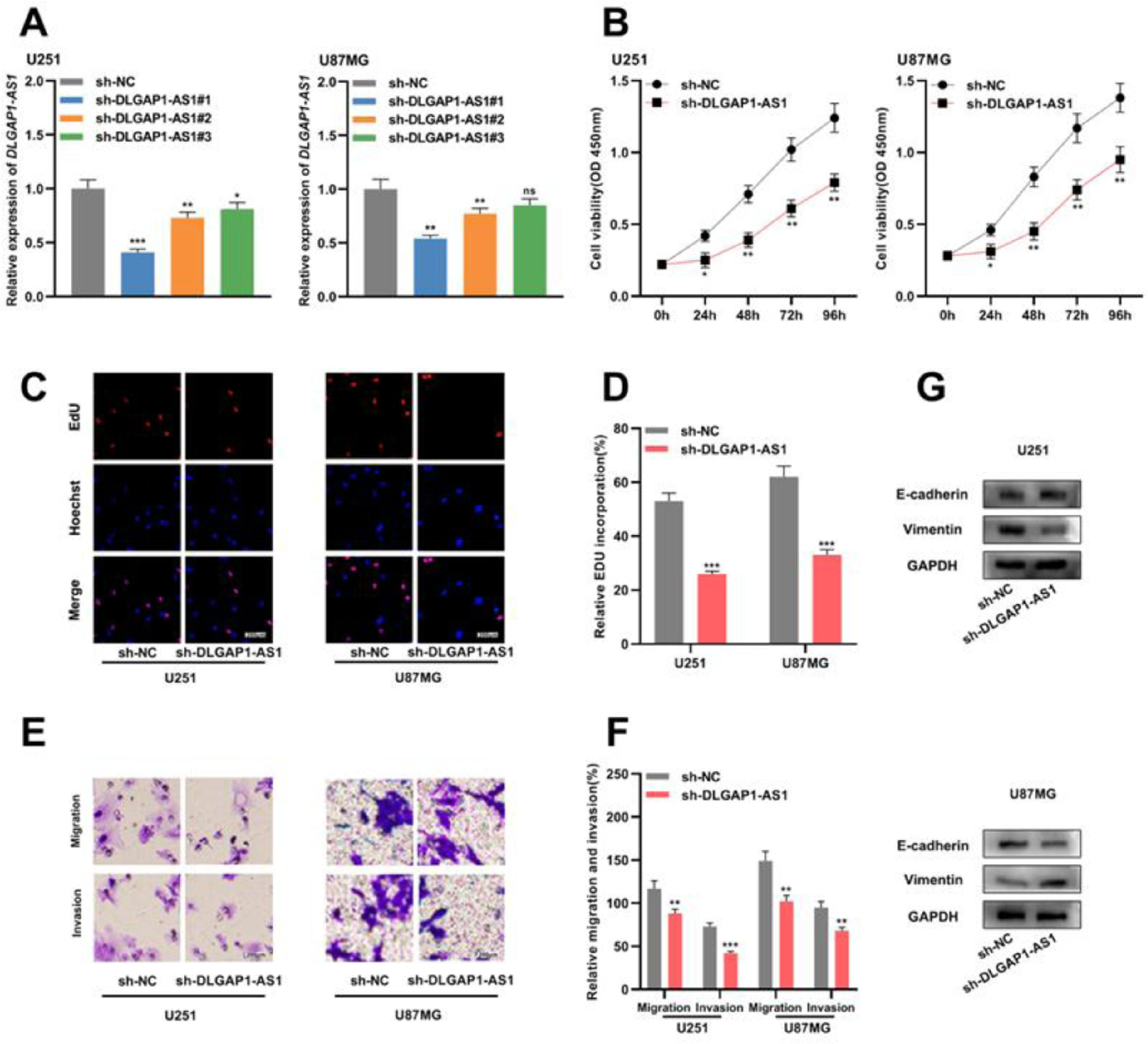
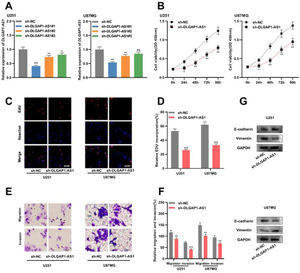
To determine the biological function of DLGAP1-AS1 in glioma cells, the authors used shRNAs to knockdown DLGAP1-AS1 expression in U251 and U87MG cells and determined the transfection efficiency using qRT-PCR. It was found that sh-DLGAP1-AS1#1 had the highest efficiency and used it for subsequent experiments (Fig. 2A and Supplementary Fig. 1C). CCK-8 and EdU assays suggested that DLGAP1-AS1 knockdown significantly reduced the proliferation of U251 and U87MG cells compared to the control group (Fig. 2B-D). The authors performed Transwell assays to evaluate the migration and invasion of glioma cells and found that DLGAP1-AS1 knockdown markedly reduced the migration and invasion of U251 and U87MG cells (compared with the control group) (Fig. 2E-F). Additionally, western blotting suggested that DLGAP1-AS1 knockdown increased E-cadherin expression and decreased vimentin expression in glioma cells (Fig. 2G). These findings highlight that DLGAP1-AS1 knockdown could repress the malignancy of glioma cells.
DLGAP1-AS1 knockdown suppresses the proliferation, migration, invasion, and EMT of glioma cells. A, sh-NC, sh-DLGAP1-AS1#1, sh-DLGAP1-AS1#2, and sh-DLGAP1-AS1#3 were transfected into U251 and U87MG cells to construct low expression models of DLGAP1-AS1, and the transfection efficiency was determined using qRT-PCR. (B–D) The proliferation of U251 and U87MG cells transfected with sh-NC or sh-DLGAP1-AS1#1 was detected using CCK-8 (B) and EdU assays (C–D). (E–F) Transwell assay was used to detect the migration and invasion of U251 cells (E) and U87MG cells (F) transfected with sh-NC or sh-DLGAP1-AS1#1. G, The expression levels of EMT-related proteins E-cadherin and vimentin in U251 and U87MG cells transfected with sh-NC or sh-DLGAP1-AS1#1 were detected using western blotting. The experiments were repeated three times, and the average was recorded. *p < 0.05, **p < 0.01, and ***p < 0.001, ns was not statistically significant.
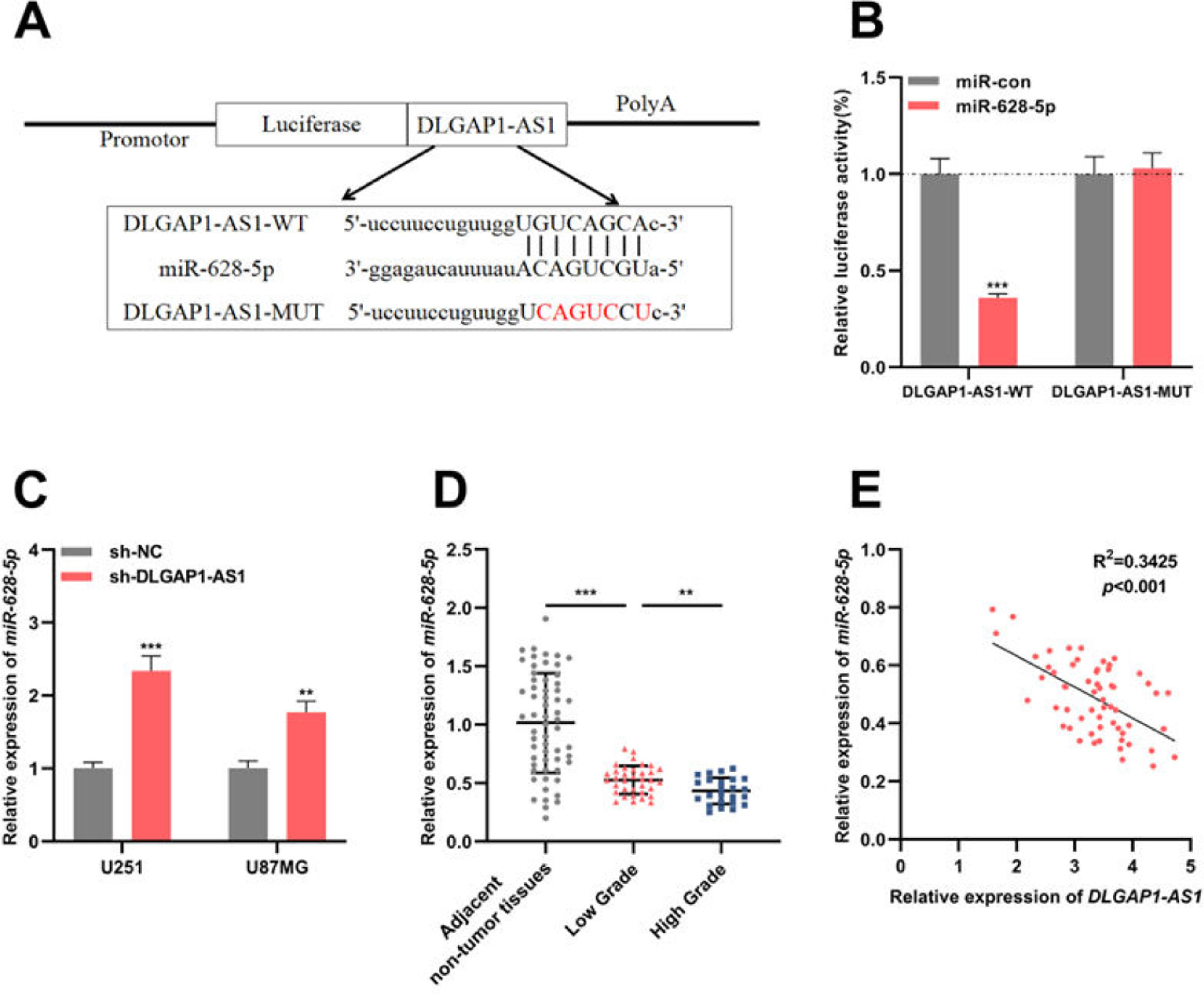
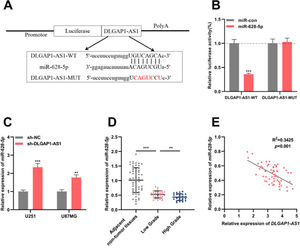
To identify the candidate miRNAs that could interact with DLGAP1-AS1, the authors searched the StarBase database (version 2.0) and found that the DLGAP1-AS1 sequence had a binding site for miR-628-5p (Fig. 3A). The results of the dual-luciferase reporter assay revealed that miR-628-5p restrains the luciferase activity of the DLGAP1-AS1-WT reporter but had no significant effect on the luciferase activity of the DLGAP1-AS1-MUT reporter (Fig. 3B). qRT-PCR showed elevated expression of miR-628-5p in U251 and U87MG cells transfected with sh-DLGAP1-AS1, indicating that DLGAP1-AS1 negatively regulates miR-628-5p expression (Fig. 3C and Supplementary Fig. 1D). Additionally, consistent with a previous report,18miR-628-5p expression was significantly decreased in glioma tissues (compared to that in adjacent non-tumor tissues) (Fig. 3D and Supplementary Fig. 1E). Next, the authors analyzed the correlation between DLGAP1-AS1 and miR-628-5p expression in glioma tissues using Pearson's correlation analysis, and the authors demonstrated that DLGAP1-AS1 expression was negatively correlated with miR-628-5p expression in glioma tissues (Fig. 3E, R2=0.3425), further implying that DLGAP1-AS1 targets miR-628-5p and represses its expression in gliomas.
DLGAP1-AS1 targets miR-628-5p in glioma. A, DLGAP1-AS1-WT luciferase reporter vector and DLGAP1-AS1-MUT luciferase reporter vector were constructed. B, DLGAP1-AS1-WT or DLGAP1-AS1-MUT luciferase reporter vector and miR-628-5p mimics or control miRNA were co-transfected into HEK-293T cells, and the luciferase activity of the cells in each group was determined. C, qRT-PCR was used to detect the expression of miR-628-5p in U251 and U87MG cells transfected with sh-NC or sh-DLGAP1-AS1#1. D, The expression of miR-628-5p in glioma cell lines (U-118MG, U251, U87MG, and LN229 cells) and normal cell lines (HA cells) was detected using qRT-PCR. E, Pearson's correlation analysis showed that the expression of DLGAP1-AS1 and miR-628-5p was negatively correlated in glioma tissues. The experiments were repeated three times, and the average was recorded. **p < 0.01 and ***p < 0.001.
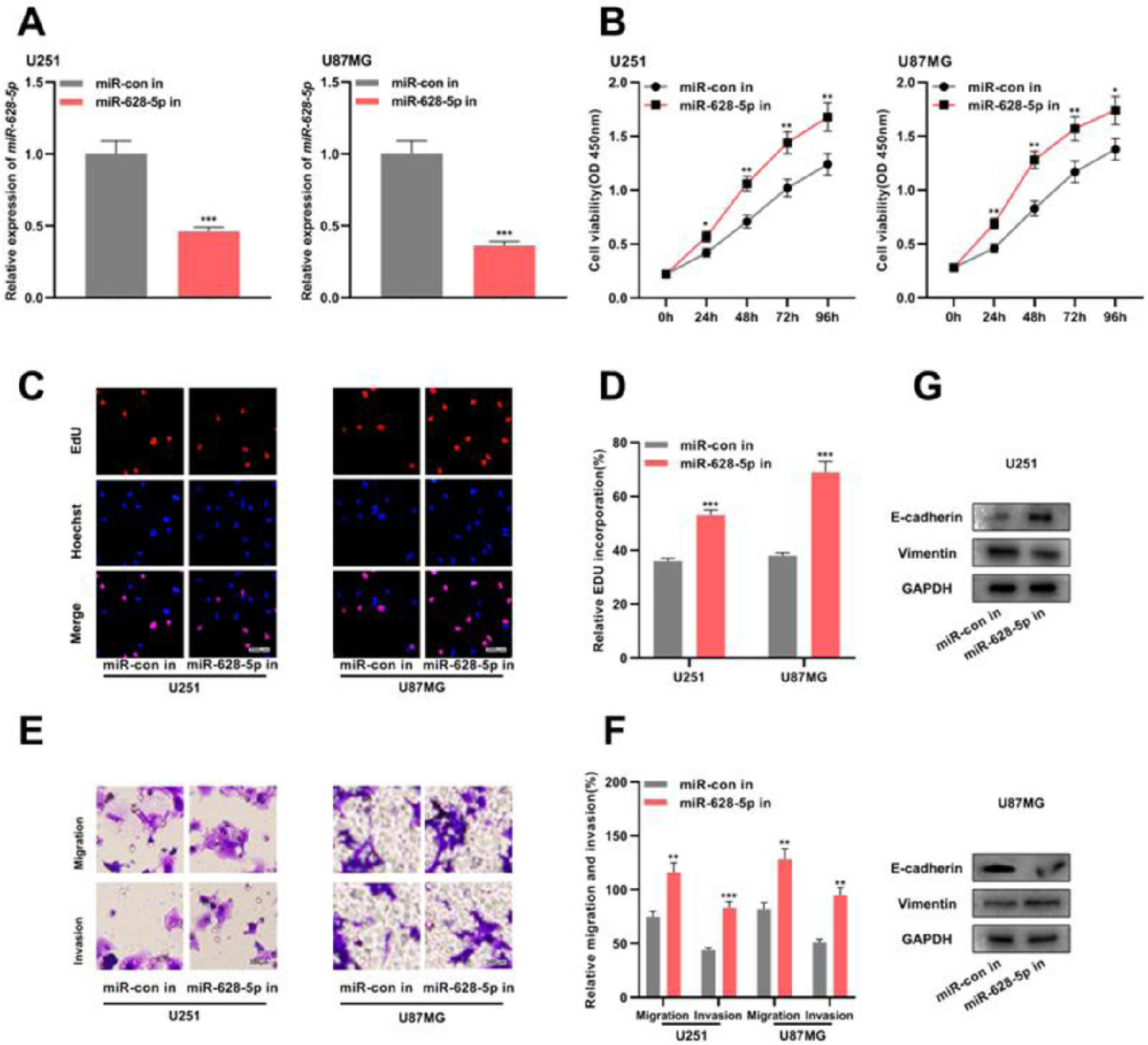
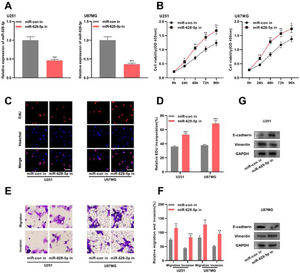
Previous studies have shown that miR-628-5p overexpression inhibits the proliferation of glioma cells.18,19 In the present study, the authors transfected miR-628-5p inhibitor into U251 and U87MG cells and verified the transfection efficiency using qRT-PCR (Fig. 4A and Supplementary Fig. 1F). Subsequently, CCK-8, EdU, and Transwell assays and western blotting revealed that the inhibiting miR-628-5p promoted the proliferation, migration, invasion, and EMT of U251 and U87MG cells, indicating that miR-628-5p exerted tumor-suppressive functions in gliomas (Fig. 4B-G).
The inhibition of miR-628-5p expression promotes the proliferation, migration, invasion, and EMT of glioma cells. A, miRNA inhibitor control (miR-con in) and miR-628-5p inhibitor (miR-628-5p in) were transfected into U251 and U87MG cells to construct models of the inhibition of miR-628-5p expression, and the transfection efficiency was detected using qRT-PCR. (B–D) The proliferation of U251 and U87MG cells was detected using CCK-8 (B) and EdU assays (C-D). (E-F) Transwell assay was used to detect the migration and invasion of U251 (E) and U87MG cells (F). G, Western blot assay was used to detect the expression of EMT-related proteins E-cadherin and vimentin in U251 and U87MG cells. The experiments were repeated three times, and the average was recorded. *p < 0.05, **p < 0.01, and ***p < 0.001.
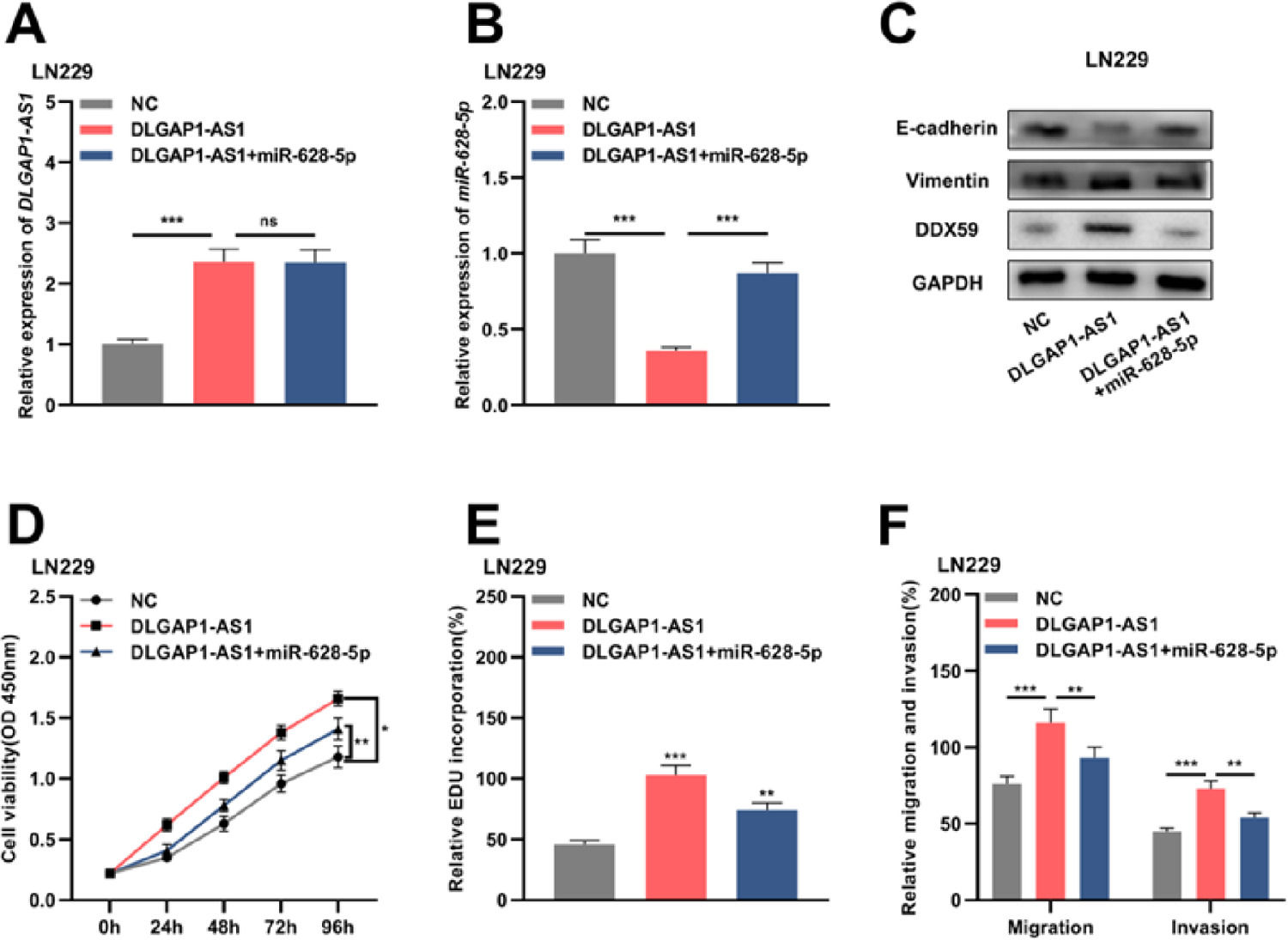
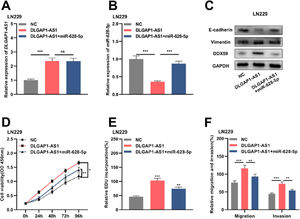
Reportedly, miR-628-5p impedes the proliferation of glioma cells by negatively regulating DDX59 expression.19 To elaborate on the mechanism of DLGAP1-AS1 in the biology glioma cells, the authors performed compensation experiments. The authors divided LN229 cells into three groups and transfected them with either the empty vector+NC mimics, pcDNA3.1 overexpressing DLGAP1-AS1+NC mimics, or pcDNA3.1 overexpressing DLGAP1-AS1+miR-628-5p mimics (Fig. 5A-B and Supplementary Fig. 1G). Western blotting was performed to detect DDX59, E-cadherin, and vimentin expression after transfection. The authors found that DLGAP1-AS1 overexpression promoted the expression of DDX59 and vimentin and repressed the expression of E-cadherin, while miR-628-5p overexpression partially reversed these effects (Fig. 5C). Furthermore, DLGAP1-AS1 overexpression promoted the proliferation, migration, invasion, and EMT of LN229 cells, and co-transfection with miR-628-5p mimics partially counteracted the functions of DLGAP1-AS1 (Fig. 5D-F). These experiments indicated that DLGAP1-AS1 could promote the proliferation, migration, invasion, and EMT of glioma cells by sponging miR-628-5p and upregulating DDX59 expression.
DLGAP1-AS1 regulates glioma cell proliferation, migration, invasion, and EMT via the miR-628-5p/DDX59 axis. A, The DLGAP1-AS1 overexpression vector and DLGAP1-AS1 overexpression vector+miR-628-5p mimic were transfected into LN299 cells, and the expression of DLGAP1-AS1 was detected using qRT-PCR. B, qRT-PCR was used to detect the expression of miR-628-5p in LN299 cells. C, Western blotting was used to detect the expression of DDX59 and EMT-related proteins E-cadherin and vimentin in LN299 cells. (D–E) The proliferation of LN229 cells was detected using CCK-8 (D) and EdU assays (E). F, Transwell assay was used to detect the migration and invasion of LN229 cells. The experiments were repeated three times, and the average was recorded. *p < 0.05, **p < 0.01, and ***p < 0.001, ns was not statistically significant.
LncRNAs play vital roles in human diseases, including tumors.23 They are abnormally expressed in diverse tumors, including glioma, and help regulate the malignant biological behaviors of tumor cells, such as proliferation, migration, invasion, apoptosis, and drug resistance.24 Many lncRNAs are associated with the pathogenesis and progression of gliomas. For example, increased LINC00689 expression in glioma tissues and cell lines is associated with rapid deterioration and poor prognosis of patients.25 LncRNA PLAC2 inhibits the nuclear translocation of STAT1, thus reducing RPL36 expression, inhibiting glioma cell proliferation, and inducing cell cycle arrest.26 LncRNA PVT1 promotes the expression of BMP2 and BMP4 by regulating GREM1 expression and promoting glioma progression.27 Herein, the authors confirmed the elevated DLGAP1-AS1 expression in glioma, which was linked to unfavorable pathological characteristics and poor prognosis of the patients. Functionally, DLGAP1-AS1 overexpression promoted the proliferation, migration, invasion, and EMT of glioma cells, while its knockdown exerted opposite effects, indicating that DLGAP1-AS1 is a novel oncogenic lncRNA in glioma and might be a promising therapeutic target.
miR-628-5p is a well-known regulator in cancer biology. In most cancers, miR-628-5p functions as a tumor suppressor. However, the elevated levels of miR-628-5p in osteosarcoma are related to the adverse prognosis of the patients.28 The serum of patients with prostate cancer has low levels of circulating miR-628-5p, suggesting that miR-628-5p is a promising non-invasive biomarker for the diagnosis and prognostic evaluation of prostate cancer. Functionally, miR-628 reduces the proliferation and invasion of prostate cancer cells by repressing FGFR2 expression.29,30 In pancreatic ductal adenocarcinoma, miR-628-5p suppresses the migration and invasion of cancer cells by repressing Akt/NF-κB signaling.31 In gastric cancer, miR-628-5p targets PIN1 to inhibit cancer progression.32 In glioma, miR-628-5p represses the malignant behavior of glioma cells.18,19 In the present study, the authors found that reduced miR-628-5p expression in glioma tissues promoted the proliferation, migration, invasion, and EMT of glioma cells, further confirming the anti-tumor effects of miR-628-5p on glioma cells.
DDX59 is a member of the DEAD/Deah box RNA helicase family.33,34 Reportedly, DDX59 is highly expressed in lung adenocarcinoma tissues and contributes to the growth of EGFR⁻ lung cancer cells.35,36DDX59 knockdown restrained the proliferation of glioma cells, and DDX59 overexpression partially weakened the inhibitory effects of miR-628-5p, suggesting that it is also an oncogene in gliomas.19 In the present study, the authors proposed a ceRNA network of DLGAP1-AS1, miR-628-5p, and DDX59. LncRNAs, like ceRNAs, can modulate gene expression by competitively binding to miRNAs, and an imbalance in the ceRNA network can cause diseases.37,38 For example, in gastric cancer, LINC01133 sponges miR-106-3p to upregulate APC expression and inhibit cancer progression.37 In hepatocellular carcinoma, as a ceRNA, DLGAP1-AS1 elevates the level of the carcinogenic cytokine IL-6 by sponging miR-26a/b-5p and activating the Wnt/β-catenin pathway.12 In the present study, through bioinformatics analysis, a dual-luciferase reporter assay, and qRT-PCR, the interaction between DLGAP1-AS1 and miR-628-5p was predicted and validated in glioma cells. The authors also demonstrated that DLGAP1-AS1 promotes the malignant phenotypes of glioma cells by sponging miR-628-5p and elevating DDX59 expression. These results not only partly explain the mechanism underlying the dysregulation of miR-628-5p and DDX59 in gliomas but also elucidate the mechanism by which DLGAP1-AS1 participates in glioma progression.
To briefly recapitulate, the present study confirms that DLGAP1-AS1 is overexpressed in glioma and promotes aggressive cancer progression by regulating the miR-628-5p/DDX59 axis. These findings help clarify the mechanism of glioma progression and provide potential targets for molecular therapy of gliomas. In future studies, animal experiments are needed to verify these results, and it is necessary to enroll more patients from different medical centers to evaluate the potential value of DLGAP1-AS1 as a biomarker to predict the prognosis of the patients.
Author contributionsKe-qi Hu and Xiang-sheng Ao contributed equally to the experimental design and execution, statistical analysis, and manuscript writing.
The authors thank Hubei Yican Health Industry Co., Ltd (Wuhan, China) for its linguistic assistance during the preparation of this manuscript.