Antimicrobial resistance due to carbapenemase production in Enterobacteriaceae clinical isolates is a global threat. Klebsiellapneumoniae harboring the blaKPC gene is one of the major concerns in hospital settings in Latin America.
The aim of this study was to characterize the antibiotic resistance mechanisms and to typify four carbapenem-resistant K. pneumoniae clinical isolates from the city of Manizales, Colombia.
We identified blaKPC-3 in all four isolates by polymerase chain reaction and subsequent sequencing. The plasmid-mediated quinolone resistance genes qnrB19-like and aac(6′)Ib-cr; fosfomycin resistance gene fosA and an insertion sequence IS5-like in mgrB (colistin resistance) were also detected. Sequence types ST11 with capsular type wzi75, and ST258 with wzi154, were characterized. The blaKPC-3 gene was mobilized in a 100-kb IncFIB conjugative plasmid with vagCD toxin–antitoxin system.
This work reports multiple resistance genes in blaKPC-producing K. pneumoniae and the first occurrence of ST11 clinical isolates harboring blaKPC-3 in Latin America.
La resistencia a antibióticos mediada por la producción de carbapenemasas en aislamientos clínicos de Enterobacteriaceae es una amenaza mundial. Klebsiellapneumoniae portador de blaKPC es uno de los mayores problemas a nivel hospitalario en Latinoamérica.
El objetivo de este estudio fue caracterizar los mecanismos de resistencia antibiótica y tipificar cuatro aislamientos clínicos de K. pneumoniae resistentes a carbapenems obtenidos en la ciudad de Manizales, Colombia.
Se identificó blaKPC-3 en todos los aislamientos mediante reacción en cadena de polimerasa y secuenciación. También se detectaron los genes de resistencia transferible a quinolonas qnrB19-like y aac(6’)Ib-cr y a fosfomicina fosA, y la secuencia de inserción IS5-like en mgrB (asociada a la resistencia a colistina). Se caracterizaron los secuenciotipos ST11 (cápsula wzi75) y ST258 (cápsula wzi154). Se comprobó que blaKPC-3 fue movilizado por un plásmido conjugativo IncFIB-vagCD de 100kb.
En este trabajo se reportan múltiples genes de resistencia en K. pneumoniae productor de blaKPC y se describen por primera vez aislamientos clínicos ST11 productores de blaKPC-3 en Latinoamérica.
Antimicrobial resistance due to carbapenemase production in Enterobacteriaceae clinical isolates is a global threat4. Recently, the WHO established a list of microorganisms to be prioritized in the research and development of new antimicrobials, placing carbapenem and/or third-generation cephalosporin-resistant enterobacteria within the microorganisms of maximum interest17.
Enterobacterial clinical isolates usually present not only β-lactam resistance due to either extended spectrum β-lactamase (ESBL) or carbapenemase production, but also resistance to both aminoglycosides and fluoroquinolones4. In this sense, plasmid-mediated quinolone resistance (PMQR) mechanisms could be present in these isolates; e.g. qnr alleles, the efflux pump oqxAB or the aac(6′)Ib-cr allele, which confer resistance to quinolones and the latter also to aminoglycosides (amikacin, kanamycin, tobramycin)10,19.
Occasionally, other antibiotics such as colistin or fosfomycin are necessary therapeutic options to treat clinical infections caused by these multiresistant microorganisms, particularly in carbapenemase-producers8. However, transferable fosfomycin and colistin resistance genes like fosA/fosB and different mcr alleles respectively have been detected in enterobacterial clinical isolates3,16.
All the aforementioned resistance determinants are mainly codified in conjugative plasmids considered epidemic. “Epidemic resistance plasmids” (ERP) play an important role in antibiotic resistance gene dissemination. Different features are described for those ERP, such as incompatibility groups, e.g. IncFII, IncA/C, IncL/M, IncN and IncI1, plasmid size larger than 50kb, conjugation properties, toxin–antitoxin systems related to plasmid maintenance in daughter cells, among others6.
In relation to enterobacterial clinical isolates, Klebsiella pneumoniae is represented by the letter “K” in the acronym ESKAPE, among the six most significant and dangerous causes of hospital infections by antimicrobial-resistant microorganisms15.
Within the K. pneumoniae isolates, those who are KPC-producers are the main problem in hospital settings in Latin America9.
In particular, K. pneumoniae isolates belonging to clonal group CG258 (ST258, ST11, their single-locus variants and other closely related sequence types) are the most widely distributed in the world. ST258 was recognized as a successful clone, playing an important role in emergence and dissemination of blaKPC19, and ST11 is also significantly associated with the worldwide dissemination of this resistance mechanism, being similarly considered a successful clone1.
Identification of capsular types by amplification and sequencing of the wzi gene (Wzi protein anchors capsular polysaccharide to the cell surface) is often used to characterize these sequence types. In this sense, there are different wzi alleles that correlate with different capsule locus (KL)20, for example capsular type wzi154 (KL107), has been related to KPC-producing ST258 isolates16.
K. pneumoniae isolates belonging to ST258 were first reported in Latin America in Medellin, Colombia, carrying either blaKPC-2 carbapenemase in 200518 and blaKPC-3 in 200811. Subsequently, both enzymes have spread to other cities in Colombia and to other clonal complexes16.
Although there are reports of KPC-producing isolates recovered from the departments of Cundinamarca and Antioquia, where the cities of Bogotá and Medellin are respectively located16, there are so far no reports of this type of isolates from the Department of Caldas, geographically located between those previously mentioned departments.
In 72h, three carbapenem-resistant clinical isolates of K. pneumoniae were obtained from two hospitals located in the city of Manizales (capital of Caldas Department, Colombia) and a fourth isolate was obtained 30 days later, also displaying colistin resistance.
The aim of this study was to characterize the antibiotic resistance mechanisms and to typify four carbapenem-resistant K. pneumoniae clinical isolates from Manizales city.
Four K. pneumoniae isolates (Kpn1–4) were collected from the microbiology laboratories of two hospitals (A and B) in the city of Manizales between May and June 2016. Identification was determined by MALDI-TOF MS (Matrix-assisted laser desorption/ionization time-of-flight mass spectrometry). Susceptibility testing was evaluated by the automatized system MicroScan and the agar dilution test was performed to determine fosfomycin susceptibility. All the results were interpreted in accordance with the EUCAST guidelines (www.eucast.org/clinical_breakpoints). Carbapenemase production was suspected by the phenotypic test KPC+MBL Confirm ID Kit (Rosco, Taastrup, Denmark). Resistance mechanisms were investigated by polymerase chain reaction (PCR) using primers to amplify the following determinants (Table S1): ESBL (blaCTX-M-group-1, blaCTX-M-group-2, blaCTX-M-group-3, blaCTX-M-group-4, blaCTX-M-group-25, blaTEM, blaSHV, blaOXA-1, blaOXA-2, blaPER-2); carbapenemases (blaKPC, blaOXA-48, blaGES); transferable-resistance determinants for: aminoglycosides (aac(6′)Ib, methylases: armA, rmtA-D, npmA), quinolones (aac(6′)Ib-cr, qnrA, B, C, D, S, VC, qepA), fosfomycin (fosA, fosA3) and colistin (mcr-1 to 3). Positive results were confirmed by Sanger sequencing, and sequences were analyzed using BLASTn (https://blast.ncbi.nlm.nih.gov).
Capsular type was determined by PCR and sequencing for wzi gene, alleles were identified in the Klebsiella sequence typing database (https://bigsdb.pasteur.fr/klebsiella/klebsiella.html). mgrB alterations were searched by both PCR and sequencing in order to determine other mechanisms involved in colistin resistance (Table S1).
Conjugation assays were carried out using tetracycline-resistant E. coli CAG12177 [F– λ–zej-298::Tn10(Tetr) gyrA261(Nalr) rph-1] (E. coli Genetic Stock Center) as recipient. Transconjugants were selected on Luria–Bertani agar plates supplemented with tetracycline (32mg/l) and ceftazidime (2mg/l)10.
Incompatibility groups and toxin–antitoxin systems were detected by PCR using transconjugant genomic DNA as template5,13.
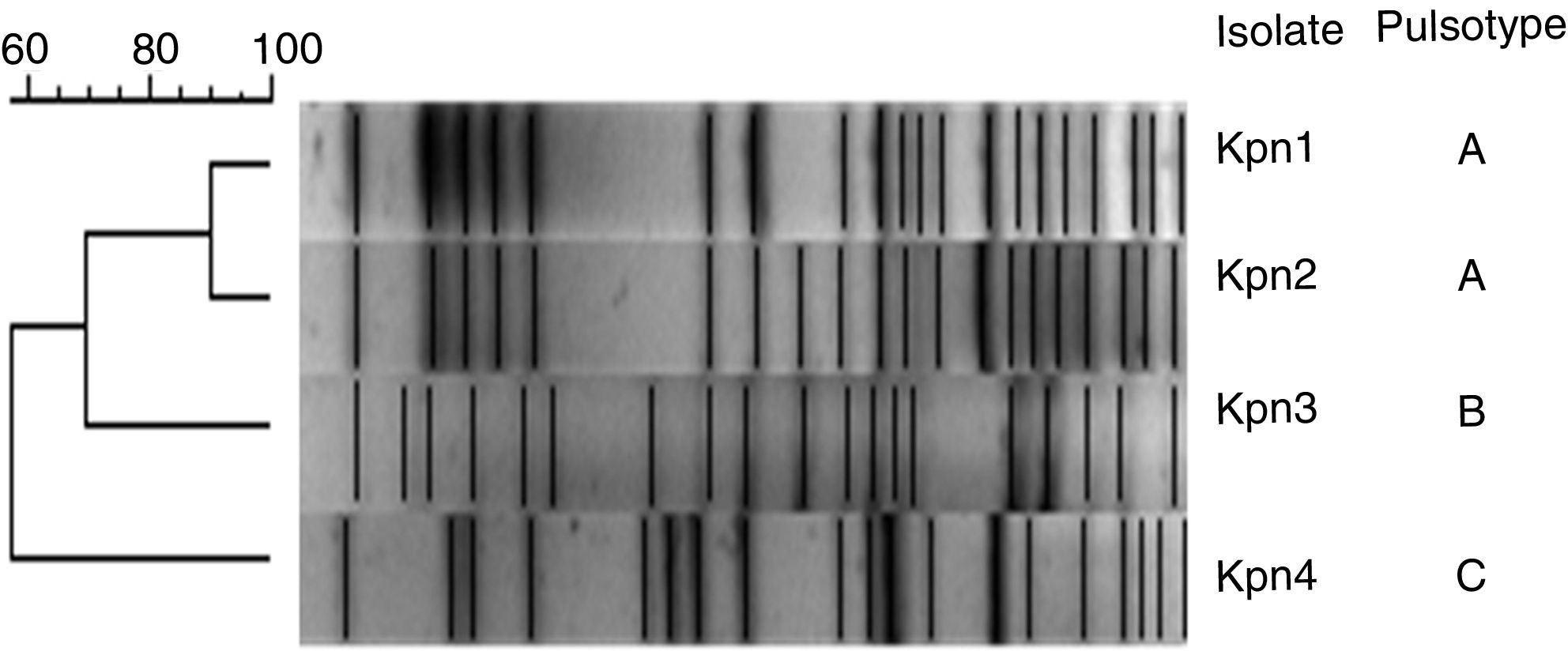
XbaI-pulsed-field gel electrophoresis (PFGE) analysis was performed and results were analyzed using the unweighted pair-group method with an arithmetic mean (UPGMA) as previously reported10.
Multi locus sequence typing characterization of K. pneumoniae isolates was conducted according to the K. pneumoniae MLST database. (https://bigsdb.pasteur.fr/klebsiella/klebsiella.html).
Plasmid size was estimated for transconjugants by treatment with S1 nuclease followed by PFGE10.
This study was approved by the Direction Board of the involved hospitals, and the patients’ data were obtained from clinical records.
Kpn1 and Kpn2 isolates were recovered from urine cultures of catheterized patients in intermediate care units from Hospital A (39-year-old woman, May 13th; and 72-year-old man in May 15th, respectively); meanwhile Kpn3 was obtained from a midstream urine sample from a 70-year-old patient in Hospital B, May 14th. Finally, Kpn4 was recovered from a catheter tip from an adult patient in Hospital A, June 17th. Antibiotic susceptibility results are shown in Table 1.
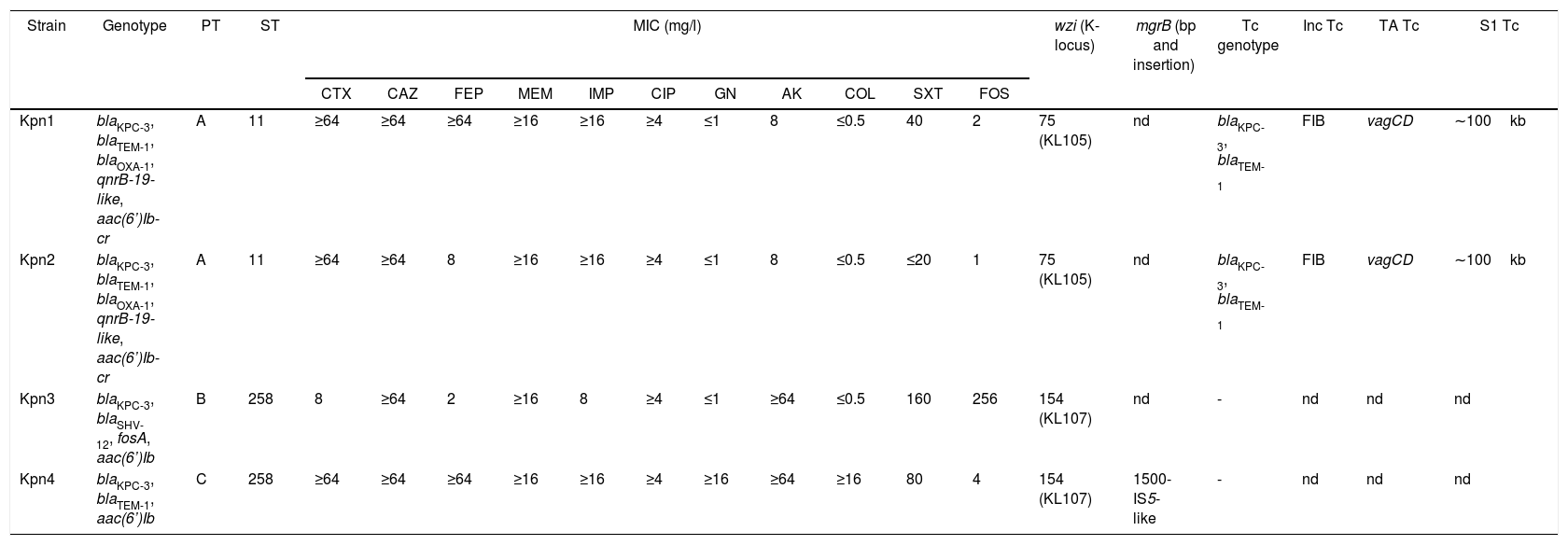
Characteristics of K. pneumoniae isolates.
| Strain | Genotype | PT | ST | MIC (mg/l) | wzi (K-locus) | mgrB (bp and insertion) | Tc genotype | Inc Tc | TA Tc | S1 Tc | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| CTX | CAZ | FEP | MEM | IMP | CIP | GN | AK | COL | SXT | FOS | ||||||||||
| Kpn1 | blaKPC-3, blaTEM-1, blaOXA-1, qnrB-19-like, aac(6’)Ib-cr | A | 11 | ≥64 | ≥64 | ≥64 | ≥16 | ≥16 | ≥4 | ≤1 | 8 | ≤0.5 | 40 | 2 | 75 (KL105) | nd | blaKPC-3, blaTEM-1 | FIB | vagCD | ∼100kb |
| Kpn2 | blaKPC-3, blaTEM-1, blaOXA-1, qnrB-19-like, aac(6’)Ib-cr | A | 11 | ≥64 | ≥64 | 8 | ≥16 | ≥16 | ≥4 | ≤1 | 8 | ≤0.5 | ≤20 | 1 | 75 (KL105) | nd | blaKPC-3, blaTEM-1 | FIB | vagCD | ∼100kb |
| Kpn3 | blaKPC-3, blaSHV-12, fosA, aac(6’)Ib | B | 258 | 8 | ≥64 | 2 | ≥16 | 8 | ≥4 | ≤1 | ≥64 | ≤0.5 | 160 | 256 | 154 (KL107) | nd | - | nd | nd | nd |
| Kpn4 | blaKPC-3, blaTEM-1, aac(6’)Ib | C | 258 | ≥64 | ≥64 | ≥64 | ≥16 | ≥16 | ≥4 | ≥16 | ≥64 | ≥16 | 80 | 4 | 154 (KL107) | 1500-IS5-like | - | nd | nd | nd |
PT: pulsotype, ST: sequence type, MIC: minimum inhibitory concentration CTX: cefotaxime, CAZ: ceftazidime, FEP: cefepime, MEM: meropenem, IMP: imipenem, CIP: ciprofloxacin, GN: gentamicin, AK: amikacin, COL: colistin, SXT: trimethoprim-sulfamethoxazole, FOS: fosfomycin, wzi: capsular gene and K-locus, mgrB: colistin resistance gene, Tc: transconjugant (−: negative results), Inc: incompatibility group, TA: toxin–antitoxin systems, S1: plasmid size in blaKPC positive transconjugants (kb), nd: not determined
All isolates showed positive synergy between boronic acid and meropenem in the phenotypic test. We confirmed the presence of blaKPC-3 in all four isolates. Additionally, we found: blaTEM-1 and blaOXA-1 in both Kpn1 and Kpn2, blaTEM-1 in Kpn4, and the ESBL coding gene blaSHV-12 in Kpn3 (Table 1).
The aac(6′)Ib gene responsible for amikacin resistance was detected in Kpn3 and Kpn4. With regard to plasmid-mediated quinolone resistance genes, the combination qnrB19-like and aac(6′)Ib-cr was detected in both Kpn1 and Kpn2. The fosfomycin-resistance gene fosA was characterized in Kpn3, which displayed in vitro resistance to this antibiotic. Colistin resistance was observed in Kpn4 due to the presence of the insertion sequence IS5-like in the mgrB gene at position 74–75 (Table 1).
Three pulsotypes (PT) were identified by PFGE: Kpn1 and Kpn2 belonged to PT-A, Kpn3 to PT-B and Kpn4 to PT-C. Additionally, PT-A belonged to sequence type ST-11 and both PT-B and PT-C belonged to ST-258 (Fig. 1).
Capsular typing by wzi sequences showed different alleles: wzi75 (KL105) was identified in isolates belonging to ST11 and wzi154 (KL107) in the ST258 isolates (Table 1).
Conjugation assays were positive to Kpn1-2, TcKpn1 and TcKpn2 presented blaKPC-3/blaTEM-1. Incompatibility group IncFIB and the toxin–antitoxin system vagCD were characterized in transconjugants. The transferred plasmid size was approximately 100- kb in both transconjugants (Table 1).
In this work, we describe the first K. pneumoniae ST11 isolates harboring the blaKPC-3 gene in Latin America. Sequence types characterized in this study are closely related, since it is believed that K. pneumoniae ST258 was originated by a recombination process between ST11 and ST442 clones. In this sense, ST11 is a single locus variant from ST258 and belongs to the same clonal complex (CC258)7.
There are different reports highlighting the relevance of K. pneumoniae from ST258 or ST11 with blaKPC-2 or extended spectrum β-lactamases in Latin America1,9,10,12.
Andrade et al. recently described in Brazil the circulation of K. pneumoniae ST11 harboring blaKPC-2 or blaCTX-M-2, with different capsular types (K27, K64 and KL202) from those reported in this work1. Taking these results into consideration, the strains reported in our work would not be related to those circulating in Brazil.
With regard to publications on this topic from Colombia, Rojas et al. described ST11 in K. pneumoniae by whole genome sequencing. However, those strains were non-carbapenemase producers and their capsular types were not available. Interestingly, those isolates did present the combination of transferable quinolone resistance genes aac(6′)Ib-cr and qnrB16.
Although information concerning the capsular type of these KPC-producing isolates is not available, it is possible to hypothesize that ST11 isolates reported by Rojas et al. could have acquired the conjugative IncFIB-blaKPC-3-vagCD plasmid, becoming resistant to carbapenems.
This incompatibility group was previously associated with blaKPC-3 plasmids, but principally IncFIB(K) type16. In regard to toxin–antitoxin systems, information in databases is scarce and we were not able to find any results about vagCD presence in this kind of plasmids. We believe that the report of IncFIB-blaKPC-3-vagCD plasmids is a novel contribution.
Kpn1 and Kpn2 belonged to the same pulsotype and were recovered (only two days apart) from patients with urinary catheters, hospitalized in the same nosocomial unit. For these reasons, we believe that horizontal transmission of the strain could have occurred between these patients, probably through inadequate urine catheter manipulation.
In relation to ST258, the strains belonging to this sequence type corresponded to different pulsotypes and were isolated from different hospitals. Although the identified capsular type was wzi154, such as the capsular type reported in Rojas et al. in ST258 strains16, the resistance profiles of the two isolates were dissimilar.
With regard to KL107 (wzi154) isolates, Arena F et al. reported blaKPC-3-producing ST258 K. pneumoniae as not hypermucoviscous or hypervirulent clones2. It is remarkable that there is scarce information about the capsule locus KL105 (wzi75) identified in ST11 isolates in the databases visited to date.
On the other hand, fosfomycin resistance was detected in Kpn3 due to the presence of fosA, while, colistin resistance was identified in Kpn4 due to the insertion of IS5-like in mgrB. The presence of fosA and mutations in mgrB was recently reported in ST258 K. pneumoniae strains from Colombia. However, in the work by Poirel L. et al., blaKPC-2 was the carbapenemase detected in ST258 isolates, and the strain that harbored blaKPC-3 was from France and not from Colombia14,16.
It has been extensively reported that blaKPC dissemination is a growing problem worldwide. This phenomenon occurs in different K. pneumoniae lineages, due to a wide variety of plasmids, and is intimately linked to CC258 as we previously mentioned19.
As far as we know, ST11 belonging to CC258 and harboring blaKPC-3 has not already been reported in Latin America.
In this sense, we believe that new studies are required to verify if the ST11 clone harboring blaKPC-3 had already been circulating or if this is actually the first occurrence of a clone which could become successful in Colombia.
Conflict of interestThe authors declare that they have no conflicts of interest.
The authors would like to thank the team of curators at Institute Pasteur MLST and genome databases for curating the sequences generated during this study and making them publicly available at http://bigsdb.web.pasteur.fr.
We would like to express our gratitude to Araci Martinez for the implementation of MALDI-TOF MS for strains identification.