Autoimmune mechanisms are considered to play a significant role in chronic urticaria pathophysiology. Additionally, clinical experience emphasises the coexistence of chronic urticaria manifestation with thyroid autoimmunity. As the role of CTLA-4 polymorphism in autoimmune thyroid diseases is well proven we speculated on the possible role of this polymorphism in the background of chronic urticaria.
Materials and methodsWe included 128 chronic spontaneous autoreactive urticaria patients (87 females and 41 males) and 101 healthy volunteers (71 females and 30 males). In all examined subjects CTLA-4 A49G polymorphism was analysed. Disease severity with Urticaria Activity Score as well as age of disease onset was also studied.
ResultsNo statistically significant differences in the allele or genotype distribution between urticaria patients and controls were observed. Furthermore, we found no association between CTLA4 polymorphism and urticaria severity as well as the age of disease onset.
ConclusionsOur data suggest that there is no contribution of CTLA-4 A49G polymorphism to chronic spontaneous autoreactive urticaria susceptibility. We recommend further research on other polymorphisms in chronic urticaria patients to explore in detail the potent role of the genetic background in the pathogenesis of this disorder.
Chronic urticaria (CU) is a debilitating skin disorder with poorly understood pathogenesis. Autoimmune origin should be considered in CU pathophysiology. Strong dependency between autoimmune CU and major histocompatibility complex allele suggests the role of genetic background in the pathogenesis of urticaria and indicates the association to other autoimmune diseases. Clinical experience emphasises the coexistence of CU with thyroid autoimmunity. Furthermore, autoimmune thyroid diseases (AITDs) can often manifest in association with other autoimmune disorders such as diabetes, vitiligo, multiple sclerosis, Addison's disease, rheumatoid arthritis and systemic lupus erythematosus. AITD is supposed to be a polygenic pathology with no major predisposition locus. The cytotoxic T-lymphocyte associated antigen 4 (CTLA-4) gene has been proven to be a susceptibility factor, separate from the HLA locus, in Graves’ disease and Hashimoto thyroiditis.1–3 The CTLA4 region and the HLA locus have been proposed as the loci that predispose to AITDs with an independent impact.4
CTLA4 is a regulatory molecule expressed on the surface of activated T lymphocytes providing a negative signal to the T cell, which limits immune responses.5–7CTLA4 induces antigen-specific apoptosis and limits T lymphocyte proliferation.6,8 In this mechanism it plays a role in regulating and maintaining self-tolerance. Human CTLA4 gene consists of four exons and three introns and its association with AITDs is suspected in relation to adenine to guanine transition at position 49 of exon 1 (A49G).9 This single nucleotide polymorphism results in the substitution of threonine with alanine at codon 17 of the leader sequence. Bearing in mind the relationship of the G49 allele to a weaker suppressing function of the Ala17 molecular variant of CTLA4, G/G homozygous patients potentially have a greater risk of AITD compared to the A/A genotype.
In our previous studies we proved the role of another autoimmunity related gene, namely PTPN22 in chronic spontaneous urticaria pathogenesis, meanwhile PDCD1 appeared not to be involved.10–12 The aim of the present study is to test the possible role of the CTLA4 gene in the susceptibility to CU.
Materials and methodsCharacteristics of study sampleThe study group consisted of 128 unrelated CU patients (87 females and 41 males, mean age 38.8 years, aged 20–60 years) with positive autologous serum skin test (ASST) result, without concomitant thyroid pathology. Precise medical history and physical examination resulted in establishing the diagnosis of spontaneous urticaria without known eliciting factors. The control group was composed of 101 unrelated healthy volunteers (71 females and 30 males, mean age 44.2 years, aged 19–59 years). All individuals were Caucasians and came from the Polish population.
Urticaria Activity Score (UAS) was used to detect disease severity. This symptom assessment tool analyses wheals and pruritus intensity.13,14 As recommended, we performed a seven-day assessment (UAS7) and yielded a total score of 0–42. All patients were treated according to current guidelines.15 The age of disease onset was also analysed.
Genomic DNA isolation and CTLA-4 genotypingGenomic DNA from blood leukocytes was obtained using MasterPure™ DNA Purification Kit (Epicentre Technologies, Madison, Wisconsin, USA), in accordance with the manufacturer's indications. In all examined subjects CTLA-4 A49G polymorphism was analysed. Genotyping by allelic discrimination using Custom TaqMan SNP Genotyping Assays and 7300 Real-time PCR System (Applied Biosystems, Foster City, CA, USA) was performed.
Statistical analysisThe differences in allele and genotype frequencies between groups were calculated with Chi-square test. Additionally, odds ratios (OR) with 95% confidence intervals (CI) were estimated. ANOVA test was used for UAS7 and age of disease onset comparisons between different genotype division subgroups (Statistica 8.0 PL, Statsoft Inc., USA).
The study was approved by the Bioethics Committee of the Medical University of Silesia in Katowice.
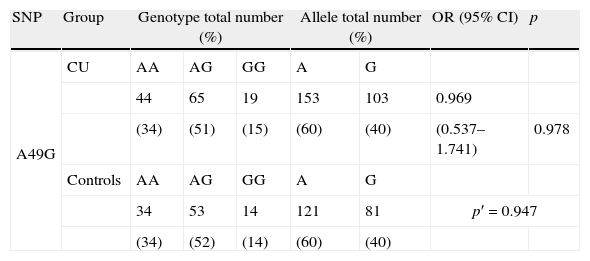
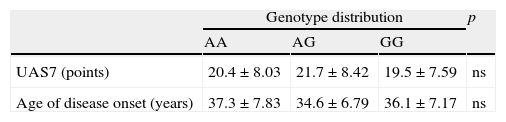
ResultsThe assay for Hardy–Weinberg equilibrium revealed no deviation in both analysed groups. In the allele or genotype distribution no statistically significant differences between CU patients and controls were found (Table 1). Furthermore, we found no association between CTLA4 and urticaria severity as well as the age of disease onset when analysed in different genotype division subgroups (Table 2).
CTLA4 A49G genotype and allele distribution in chronic urticaria (CU) patients and healthy controls.
| SNP | Group | Genotype total number (%) | Allele total number (%) | OR (95% CI) | p | |||
| A49G | CU | AA | AG | GG | A | G | ||
| 44 | 65 | 19 | 153 | 103 | 0.969 | |||
| (34) | (51) | (15) | (60) | (40) | (0.537–1.741) | 0.978 | ||
| Controls | AA | AG | GG | A | G | |||
| 34 | 53 | 14 | 121 | 81 | p′=0.947 | |||
| (34) | (52) | (14) | (60) | (40) | ||||
SNP: single nucleotide polymorphism; OR: odds ratio; CI: confidence interval; p: statistical significance (p′: allele frequency statistical significance); the odds ratio was calculated for patients homozygous or heterozygous carrying risk allele vs. homozygous.
Urticaria Activity Score one-week assessment (UAS7) and age of disease onset in chronic urticaria patients with A49G different genotypes.
| Genotype distribution | p | |||
| AA | AG | GG | ||
| UAS7 (points) | 20.4±8.03 | 21.7±8.42 | 19.5±7.59 | ns |
| Age of disease onset (years) | 37.3±7.83 | 34.6±6.79 | 36.1±7.17 | ns |
p: statistical significance; ns: statistically non-significant; data are presented as mean±standard deviation.
Autoimmune diseases share a common set of susceptibility genes. AITDs are among the most common autoimmune conditions. CTLA4 gene is related to autoimmunity in general without organ-specificity, and its role in AITDs is rather non-specific.16–18 The relationship between the CTLA4 variation and the downregulation of T cell activation is well proven.19,20 Nucleotide variation, which is an adenine to guanine transition, occurs at location 49 (A49 G) of exon 1 and results in amino acid substitution of alanine to threonine of codon 17 of the leader peptide.21 The exon 1 polymorphism markedly influences an inhibitory function of CTLA4. G49 allele of codon 17 dimorphism is suspected to be related to the reduced control of T cell proliferation and was proven to be associated with the overproduction of autoantibodies, including production of thyroglobulin and thyroid peroxidase autoantibodies.19,22,23
Besides HLA, CTLA4 is the only locus for which dependency with Graves’ disease has been presented in different populations. Interestingly, the area of relation to Graves’ disease on chromosome 2q31–q33 is close to the region of susceptibility to insulin-dependent diabetes mellitus.21,24 Additionally, the association between this locus and other autoimmune diseases such as multiple sclerosis, Addison's disease, rheumatoid arthritis and systemic lupus erythematosus was presented.25 Taken together, CTLA4 is proposed to be the HLA-independent locus of human autoimmunity.
Because of the frequent coexistence of CU and AITDs as well as proven genetic susceptibility to AITDs, we hypothesised CU genetic pathogenesis. Bearing in mind that CTLA4 polymorphism is generally connected with the production of autoantibodies, we selected patients with positive ASST as manifesting autoreactive variant of CU. In our study no statistically significant differences in the CTLA4 allele and genotype distribution between CU patients and controls were found. This finding is interesting in the light of the frequent coexistence of CU and Hashimoto thyroiditis. Undoubtedly, polymorphisms of the CTLA4 gene show strong association with Graves’ disease whereas the findings in Hashimoto thyroiditis are contrary. Additionally, A49G dimorphism allele frequencies can vary in relation to ethnic differences. Although A49 allele dominates in the Caucasian population, whereas G49 allele is more frequent in Asian population,26 our study group was ethnically homogenous. Altogether the study data suggest no contribution of this polymorphism to CU susceptibility.
Autoimmune disorder is an effect of genetic susceptibility loci cooperation, although those loci independently present a rather small risk of defect. In the chromosome 2q33–34 there is a cluster of homologous genes (CTLA4, CD28 and ICOS) related to immune activation and considered promising candidate genes for susceptibility to autoimmune diseases.27 We conclude that further genetic studies on other polymorphisms and other loci in this area in CU patients are necessary to explore deeply the potent role of the genetic background in its pathogenesis.
Ethical disclosuresPatients’ data protectionThe authors declare that no patient data appear in this article.
Right to privacy and informed consentThe authors declare that no patient data appears in this article.
Protection of human subjects and animals in researchThe authors declare that the procedures followed were in accordance with the regulations of the responsible Clinical Research Ethics Committee and in accordance with those of the World Medical Association and the Helsinki Declaration.
Conflict of interestAuthors declare no conflict of interest.
Study was supported by a research Grant from the Medical University of Silesia.