Common variable immunodeficiency (CVID) is a heterogeneous disorder characterized by low serum levels of immunoglobulins (Igs) and recurrent infection. In most CVID patients, a defect in the differentiation of B cells into plasma cells has been observed. Several factors play an important role in the proliferation and differentiation of B cells, including IRF4 and XBP1 transcription factors.
MethodsIn the present study we investigated the expression of IRF4 and XBP1 in the B-cells of CVID and healthy controls (HCs). For this purpose, we assessed the expression of IRF4 and XBP1 at both mRNA and protein levels by real time-PCR and flow cytometry, respectively.
ResultsWe found that IRF4 expression was significantly increased in CVID patients compared with controls. Although the XBP1 protein level was lower in patients in comparison to controls, this difference was not significant.
ConclusionTaken together, increased IRF4 expression could be involved in defective functions of B cells in CVID patients.
Common variable immunodeficiency (CVID) is the most common primary immunodeficiency disease (PID) with a prevalence of around 1:25,000 to 1:50,000 individuals.1 CVID patients present heterogeneous clinical manifestations such as recurrent bacterial infections, gastrointestinal disease, lymphoproliferative disorders, autoimmune phenomena and allergic diseases.2–5 Various mutations in genes involved in B-cell activation and differentiation have been identified in some proportion of CVID patients and, as indicated in the OMIM database, at least 14 genetic variants of the disease have been identified so far.6,7 Intravenous immunoglobulin (IVIG) is routinely utilized to decrease chronic infections in patients with CVID.8
In recent years, several immunological defects in the innate and adaptive immune systems have been identified in CVID.7,9–11 In most patients, despite the normal number of peripheral blood B cells, memory B cells are decreased and they have the defect in differentiation of B cells to plasma cells, antibody production and immunoglobulin isotype switching.12,13 Defects in B-cell differentiation could be due to an increase in apoptosis of B-cells or defect in B cell activation in CVID patients,7,14 although the exact causes of that remain unclear. B cells that properly respond to antigen, differentiate into plasma blasts (short-lived) and finally plasma cells producing high affinity immunoglobulins.15 B-cell differentiation to plasma cells is regulated by repression or up-regulation of different transcription factors including repression of paired box 5 (PAX5), B-cell lymphoma 6 protein (BCL6), melanogenesis associated transcription factor (MITF), metastasis-associated protein (MTA3), BACH2 or up regulation of X-box binding protein 1 (XBP-1), interferon regulatory factor 4 (IRF-4), and B lymphocyte-induced maturation protein-1 (BLIMP-1).16
XBP1 is a part of the bZip transcriptional activator that is encoded by the XBP-1 gene located on chromosome 22. XBP-1 acts downstream of PRDM1 (BLIMP1) and IRF4 transcription factors17 and is the main factor in the unfolded protein response (UPR). UPR responds to endoplasmic reticulum (ER) stress inspired by the accumulation of unfolded proteins and is pivotal for antibody-producing cells such as plasma cells. Although plasmablasts are generated in the absence of XBP-1,18 expression of XBP-1 excites antibody secreting cells (ASCs) for the production of the high level of immunoglobulins.19 Therefore, XBP-1 plays an essential function in the terminal differentiation of plasma cells.16 On the other hand, while plasmablasts are generated in the absence of XBP-1, their differentiation depends on the IRF4 and BLIMP-1 transcription factors.18
Interferon-regulatory factor-4 (IRF-4) is another essential regulator of plasma-cell development which is a member of the family of transcription factors. It is possible that IRF-4 induces massive transcription of immunoglobulin genes, through binding enhancers to the Ig κ- and λ light chains genes. IRF-4 expression at low concentration stimulates reactions of germinal center (GC) and at high concentration promotes differentiation of B cells into plasma cells.15 In mice, the defect of IRF-4 has been significantly associated with decreased immunoglobulin levels. Hence, according to the roles of IRF-4 and XBP-1 in the differentiation of antibody secreting cells and because the most frequent defect in CVID patients is the lack of B cell maturation into the functional plasma cell, in the present study, we assessed the expression of these transcription factors in CVID patients compared to healthy controls (HCs).
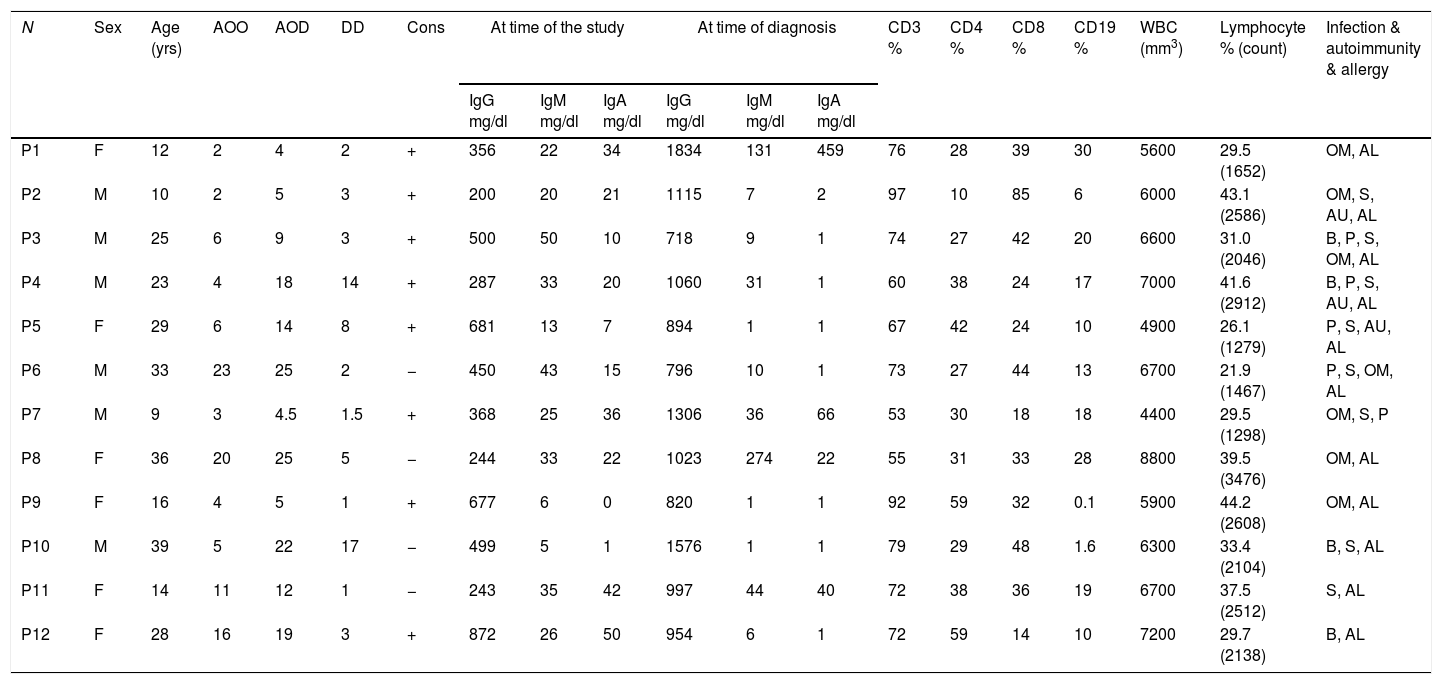
Materials and methodsSubjectsTwelve CVID patients who attended Al-Zahra hospital, Isfahan, Iran, were diagnosed according to the new diagnostic criteria of the European Society for Immune Deficiencies (ESID) including a significant reduction of IgG and strongly reduced level of IgA with or without low IgM levels (at least two standard deviations [SDs] below the mean for age), the exclusion of defined causes of hypogammaglobinemia in patients with age >4 years, poor antibody response to vaccines and no evidence of profound T-cell deficiency. Peripheral blood samples were collected just before gamma globulin replacement. The demographic information, age of onset (AOO), age of diagnosis (AOD) and laboratory findings of the patients are summarized in Table 1. Age- and sex-matched healthy blood donors were included as controls. The protocol for the present study was approved by the Ethics Committee of the Isfahan University of Medical Sciences, Iran. After explaining the purposes of the study, oral and written informed consent was taken.
Sex, age and immunological laboratory characteristic of the CVID patients.
| N | Sex | Age (yrs) | AOO | AOD | DD | Cons | At time of the study | At time of diagnosis | CD3 % | CD4 % | CD8 % | CD19 % | WBC (mm3) | Lymphocyte % (count) | Infection & autoimmunity & allergy | ||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| IgG mg/dl | IgM mg/dl | IgA mg/dl | IgG mg/dl | IgM mg/dl | IgA mg/dl | ||||||||||||||
| P1 | F | 12 | 2 | 4 | 2 | + | 356 | 22 | 34 | 1834 | 131 | 459 | 76 | 28 | 39 | 30 | 5600 | 29.5 (1652) | OM, AL |
| P2 | M | 10 | 2 | 5 | 3 | + | 200 | 20 | 21 | 1115 | 7 | 2 | 97 | 10 | 85 | 6 | 6000 | 43.1 (2586) | OM, S, AU, AL |
| P3 | M | 25 | 6 | 9 | 3 | + | 500 | 50 | 10 | 718 | 9 | 1 | 74 | 27 | 42 | 20 | 6600 | 31.0 (2046) | B, P, S, OM, AL |
| P4 | M | 23 | 4 | 18 | 14 | + | 287 | 33 | 20 | 1060 | 31 | 1 | 60 | 38 | 24 | 17 | 7000 | 41.6 (2912) | B, P, S, AU, AL |
| P5 | F | 29 | 6 | 14 | 8 | + | 681 | 13 | 7 | 894 | 1 | 1 | 67 | 42 | 24 | 10 | 4900 | 26.1 (1279) | P, S, AU, AL |
| P6 | M | 33 | 23 | 25 | 2 | − | 450 | 43 | 15 | 796 | 10 | 1 | 73 | 27 | 44 | 13 | 6700 | 21.9 (1467) | P, S, OM, AL |
| P7 | M | 9 | 3 | 4.5 | 1.5 | + | 368 | 25 | 36 | 1306 | 36 | 66 | 53 | 30 | 18 | 18 | 4400 | 29.5 (1298) | OM, S, P |
| P8 | F | 36 | 20 | 25 | 5 | − | 244 | 33 | 22 | 1023 | 274 | 22 | 55 | 31 | 33 | 28 | 8800 | 39.5 (3476) | OM, AL |
| P9 | F | 16 | 4 | 5 | 1 | + | 677 | 6 | 0 | 820 | 1 | 1 | 92 | 59 | 32 | 0.1 | 5900 | 44.2 (2608) | OM, AL |
| P10 | M | 39 | 5 | 22 | 17 | − | 499 | 5 | 1 | 1576 | 1 | 1 | 79 | 29 | 48 | 1.6 | 6300 | 33.4 (2104) | B, S, AL |
| P11 | F | 14 | 11 | 12 | 1 | − | 243 | 35 | 42 | 997 | 44 | 40 | 72 | 38 | 36 | 19 | 6700 | 37.5 (2512) | S, AL |
| P12 | F | 28 | 16 | 19 | 3 | + | 872 | 26 | 50 | 954 | 6 | 1 | 72 | 59 | 14 | 10 | 7200 | 29.7 (2138) | B, AL |
N: number; yrs: years; M: male; F: female; AOO: age of onset; AOD: age of diagnosis; DD: diagnosis delay; Cons: consanguinity; OM: otitis media; B: bronchiectasis; P: pneumonia; S: sinusitis; AU: autoimmunity; AL: allergy.
Blood samples were collected from all subjects and processed within 3h (freshly isolated PBMC) at room temperature. PBMC were isolated from EDTA blood samples by Lymphodex (BIOZ, Los Altos, CA, USA) density gradient centrifugation. CD19+ B lymphocytes (positive selection) were purified using magnetic-activated cell sorting (MACS) following the manufacturer's protocol (Miltenyi Biotec, Gladbach, Germany). MACS-isolated B cells were typically ≥90% pure, calculated by flow cytometry. Finally, CD19+ B cells were counted by a hemocytometer.
Cell cultureFor the differentiation of B cell to plasma blast, isolated CD19+ B cells were cultured in RPMI 1640 medium (BIO-IDEA, USA) supplemented with 100μg/ml penicillin (BIO-IDEA, USA), 100μg/ml streptomycin (BIO-IDEA, Tehran, Iran) and 100μg/ml FBS (Gibco, Waltham, MA, USA) for 24h, in a humidified atmosphere containing 5% CO2 at 37°C. B cells were cultivated in the presence of anti-IgM (100μg/ml) and anti-CD40 (10μg/ml) antibodies (Biolegend, San Diego, CA, USA) for BCR stimulation. Twenty-four hours after stimulation, the B cells were collected for flow cytometry analysis and reverse transcriptase polymerase chain reaction (RT-PCR).
Flow cytometryThe expression of XBP-1and IRF-4 transcription factors were analyzed in isolated B cells after stimulation with anti-IgM and anti-CD40 antibodies (Biolegend, San Diego, CA, USA) for 24h. The B-cells were fixed and permeabilized using an Intra Stain kit (Eskan Teb Asia, Tehran, Iran) according to the manufacturer's protocol. To assay XBP-1, the cells were incubated with anti-XBP-1 (Abcam, Cambridge, MA, USA) as the primary antibody, and were then incubated with a PE-conjugated monoclonal goat anti-rabbit IgG as the secondary antibody (Abcam, Cambridge, MA, USA). For the IRF-4 assay, the cells were incubated with Anti-human IRF4 FITC (eBioscience, Waltham, MA, USA). Normal goat serum was used as a blocking agent. Flow cytometry acquisitions and analysis were carried out on a FACSCalibur™ analyzer (BD Biosciences, San Jose, CA, USA) equipped to detect three fluorescent parameters with the assistance of BD CellQuest Software.
RNA isolation and quantitative real-time PCRTotal RNA was extracted from isolated B cells using the Total RNA Mini Kit (Yekta Tajhiz Azma, Tehran, Iran) and reverse transcription was performed using the cDNA Reverse Transcription Kit (Thermo Scientific, Waltham, MA, USA) according to the manufacturer's protocol. Quantitative real-time polymerase chain reaction (qPCR) was performed in duplicate with 500ng of complementary DNA (cDNA), using the 7900HT Fast Real-Time PCR System (Applied Biosystems, Foster City, CA, USA). Quantitative RT-PCR for the detection of IRF-4 and XBP-1 mRNA expression was performed with SYBR Green (Thermo Scientific, Waltham, MA, USA).
The IRF-4, XBP-1 and ACTB specific primers sequences are listed in Table 2. The geometric mean of the housekeeping gene ACTB was used as an internal control to correct the raw values for the genes of interest. Mean cycle threshold (Ct) values were standardized by calculating ΔCt using the housekeeping gene β-actin and were calculated using the 2 −ΔΔCT method.
Target transcripts and specific primer sequences.
| Gene symbol | Target transcript(s) | Forward primer | Reverse primer |
|---|---|---|---|
| IRF4 | ENST00000493114.1 ENST00000380956.8 | GCCCAACAAACTGGAGAGAG | AGGTTCTACGTGAGCTGTGATG |
| XBP1 | ENST00000216037.10 ENST00000611155.4 | GGAAGCCAAGGGGAATGAAGTG | GGTCCAAGTTGTCCAGAATGCC |
| ACTB | ENST00000646664.11 | AGCACAGAGCCTCGCCTTT | GTTGTCGACGACGACCG |
Statistical analyses of the data were performed using GraphPad Prism 7.03 software. Using Kolmogorov–Smirnov and Shapiro–Wilk tests, we estimated whether data were normally distributed. Parametric and non-parametric analyses were performed based on the finding of these tests. Independent sample t-test was performed for data with a normal distribution, while the Mann–Whitney U test was used for data with abnormal distribution.
ResultsDemographic and clinical manifestation of patientsTwelve CVID patients (six male and six female) were investigated in this study. The mean age of patients at the time of the study was 22.8 years. The mean age at onset of symptoms was 8.49 years and the mean diagnostic age was 13.53 years. Demographic and immunological data of CVID patients are summarized in Table 1. The most frequent clinical manifestations among patients with CVID were sinusitis, pneumonia, bronchiectasis and otitis media (Table 1).
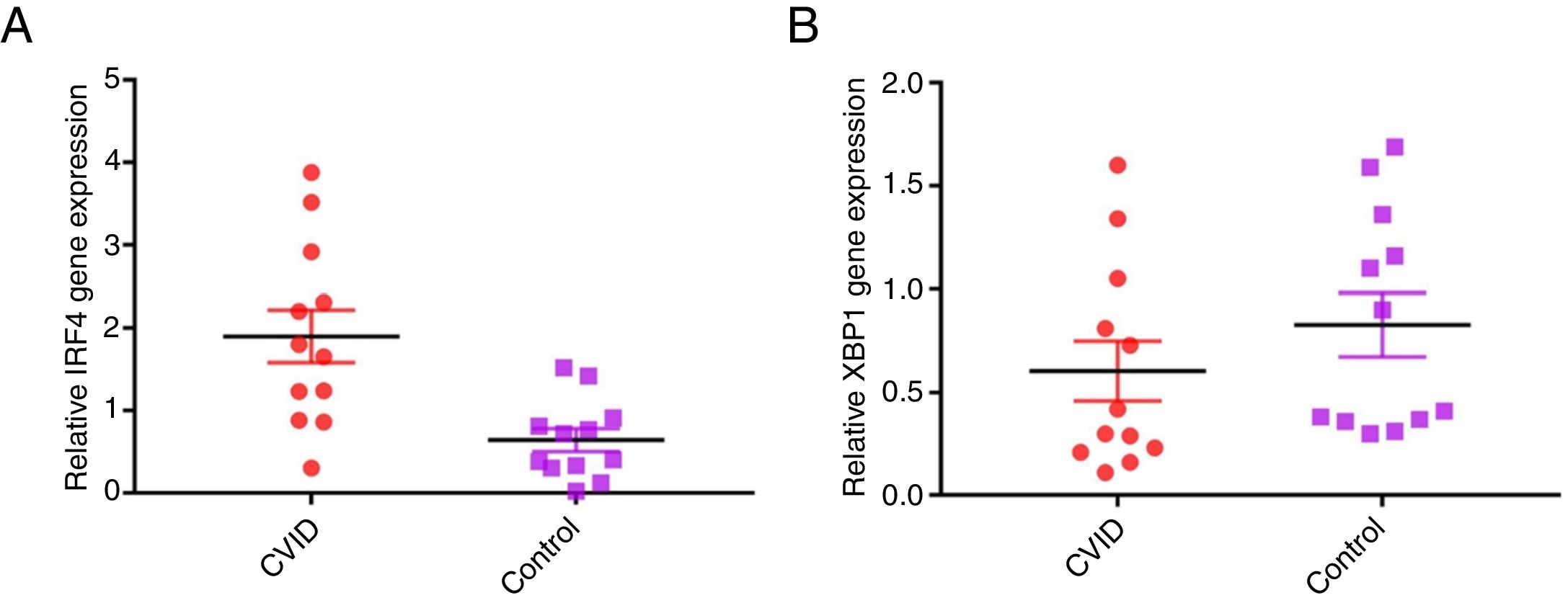
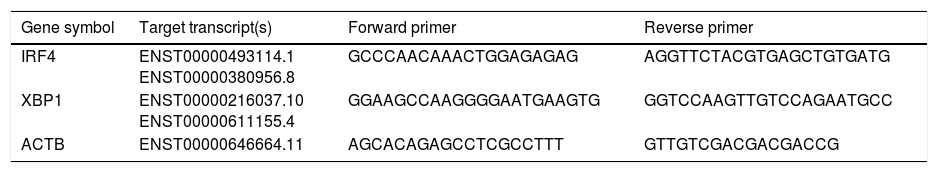
Increased IRF4 gene expression in isolated B cells of CVID patients compared with controlsIn order to evaluate possible changes in mRNA expression of IRF4 and XBP1 in patients compared with controls, Real time PCR was applied. We found that IRF-4 gene expression was significantly increased in CVID patients in comparison with controls group (p=0.0015) (Fig. 1A). Additionally, we observed that the expression of XBP1 gene was slightly lower in the patients than in the controls; however, this reduction was not significant (p=0.302) (Fig. 1B).
Comparison of XBP1 and IRF4 gene expressions in isolated B cells of the CVID patients and controls by the quantitative reverse transcriptase-polymerase chain reaction. The bars represent gene expressions in CVID patients (black bars) and controls (white bars). (A) IRF4 gene expression was significantly increased in CVID patients and (B) XBP1 gene expression was slightly lower in patients in comparison with controls. *p<0.05; error bars correspond to mean±SEM, statistical significance between patients (n=12) and controls (n=12). Analyses were done with independent sample t-test.
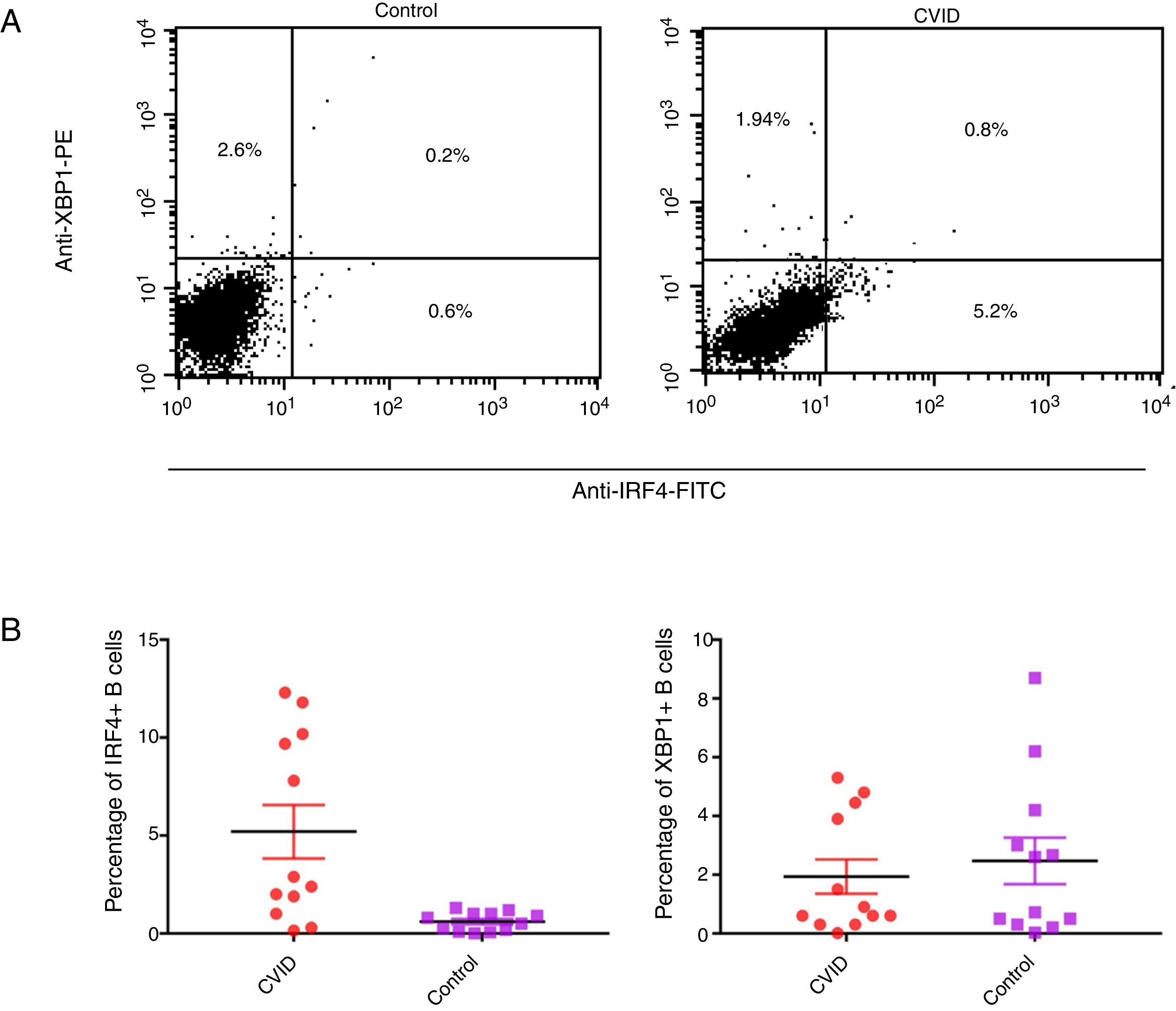
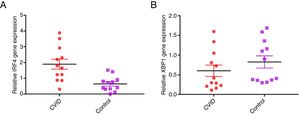
Changes in mRNA expressions disclosed by RT-qPCR do not always translate into changes in protein expression. Therefore, we performed flow cytometry analysis on isolated B cells after 24 activations and stimulation by anti-IgM and anti-CD40 antibodies. Consistent with the changes in IRF4 and XBP1 gene expression, protein expression of IRF4 was significantly increased in the CVID patients than in the controls (p=0.0014), although the XBP1 protein level was lower in the patients in comparison to the controls; however, these differences were not significant (p=0.596) (Fig. 2A and B).
Expression of XBP1 and IRF4 transcription factors in B-cell of controls and patients. (A) Dot plots show XBP1 and IRF4 expression in B-cells of a representative control and a CVID patient. After 24h stimulation the cells were stained with anti-XBP-1 as primary antibody, and then incubated with a PE-conjugated monoclonal goat anti-rabbit IgG as the secondary antibody and anti-IRF4 FITC. The plots are based on gating of viable cells in the forward and side scatter plots. (B) Bar chart shows the mean percentage of XBP1 and IRF4 expression in B-cells of controls (white bars) and patients (black bars). *p<0.05; error bars correspond to mean±SEM, statistical significance between patients (n=12) and controls (n=12). Analyses were done with Mann–Whitney U test for IRF4 and independent sample t-test for XBP1 values.
CVID patients are associated with some defects in activation, survival, and differentiation of B-cells. Since abnormalities in essential regulators of B cell activation and plasma cell development could be involved in the pathogenesis of CVID, we investigated the expression of two important transcriptional factors in the development and differentiation of B cells (IRF4 and XBP1) in CVID patients. On the one hand, we identified a significant increase in the mRNA and protein expression of IRF4 in B cells of patients compared with controls. On the other hand, although we found that the mRNA and protein expression of XBP1 were lower in the B-cells of the patients, this difference was not statistically significant.
In the process of B cell differentiation, germinal centers (GCs) are formed in the absence of XBP1, but several transcription factors including Bcl-3, Bcl-6, NF-kB/p52 and IRF-4 are essential for GCs formation.20 Warnatz et al. have demonstrated that GCs are normal in CVID patients, although there is a different degree of imprecision and hyperplastic GCs in most of them.21 High expression of IRF4 in CVID patients in our study suggests that hyper plastic GCs in these patients could be induced by stimulation of IRF4 signaling pathways. On the other hand, Julkunen et al. have suggested that IRF4 protein bounds to a regulatory element in its own promoter and controls IRF4 mRNA expression by an auto-regulatory loop in dendritic cells. This might be a justification for increased IRF4 expression in CVID-derived B cells.22 IRF proteins have the ability to form homo and heterodimers with other IRFs and interact with other transcription factors in a cell type-specific manner which regulates cell-specific gene expression.23–26 Therefore, high expression of IRF4 in our CVID patients may be due to the impact of other transcription factors.27–30
Additionally, IRF4 positively regulates the expression of BLIMP1 and activation of induced deaminase (AID) molecules. AID is essential for class switch recombination (CSR) and somatic hyper mutation (SHM).31 Indrevaer et al. have indicated that improper and high expression of IRF4 in some patients leads to reduced expression of AICDA/AID and subsequently results in the impairment of CSR and low levels of IgG production, similar to CVID phenotype.32 In addition, there is a double-positive feedback between IRF4 and BLIMP1 in B cells. Histon modification in DNA is very important for the proliferation and differentiation of B cells, as histon deastylases by inhibiting the BLIMP1 and AID decrease the expression of IRF4.26,33,34 Since IRF4 plays an important role in the proliferation and differentiation of B cells, increased IRF4 expression under the influence of stimulation of the signaling pathways or positive feedback mechanisms could be involved in the transition of B cells from proliferation step to differentiation. Another important point is that IRF4 has been detected in several types of cancer and malignancies such as skin cancer, bladder cancer (high expression) and chronic lymphocytic leukemia (CLL) (low expression).35–37 In addition, it is involved in the pathogenesis of many autoimmune disorders including systemic lupus erythematosus (SLE), multiple sclerosis (MS), inflammatory bowel disease (IBD) and type I diabetes.38 Thus, the incidence of malignancies and autoimmune diseases in CVID patients might be related to the different expression of IRF4.4,39
In the current study, we also observed a slight, but not significant, decrease in mRNA and protein expression level of XBP1 in the B-cells of patients. This result was unexpected because IRF4 is located upstream of XBP1 and increased expression of IRF4 plus other factors that are located upstream of XBP1 should have promoted XBP1 expression in CVID patients. Stimulation of B cells with anti-CD40 and IgM leads to high level activation of signaling pathways and subsequently increases the expression of factors that are involved in the differentiation of B cells, including XBP1, IRF4, etc. One possibility which may justify decreased XBP-1 expression in CVID patients is that, accumulation of misfolded immunoglobulins and dysfunction of proteins may have an impact on the XBP1 expression by increased XBP1 activity. XBP1 is an important part of the unfolded protein response (UPR) and endoplasmic reticulum (ER) to remove Igs and finally apoptosis of impaired plasma cells which may simultaneously cause an over-activation of XBP. Nevertheless, Taubenheim et al. have indicated that XBP1 is not necessary for migration of plasma cells to the bone marrow.18 They reported that in the absence of XBP1, the expression of IRF4, BLIMP1 and differentiation of plasma cells were normal. Indeed, XBP1 is not the main player in the gene regulatory network, differentiation of B cells into antibody secreting cells (ASCs), expression of IRF4, BLIMP1 and generation of plasmablasts. It has been reported that the generation of ASCs may happen in the absence of XBP1, however, there is a low level of Igs and changes in the morphology of plasma cells in absence of XBP1, but not in the sense that the ASCs have a malfunction.18
It is noteworthy that the low number of registered patients who were entered in the study was an unintended limitation of this work which may affect the interpretation of the outcomes. For instance, we were unable to analyze possible correlations between the studied genes and concomitant diseases like autoimmunity.
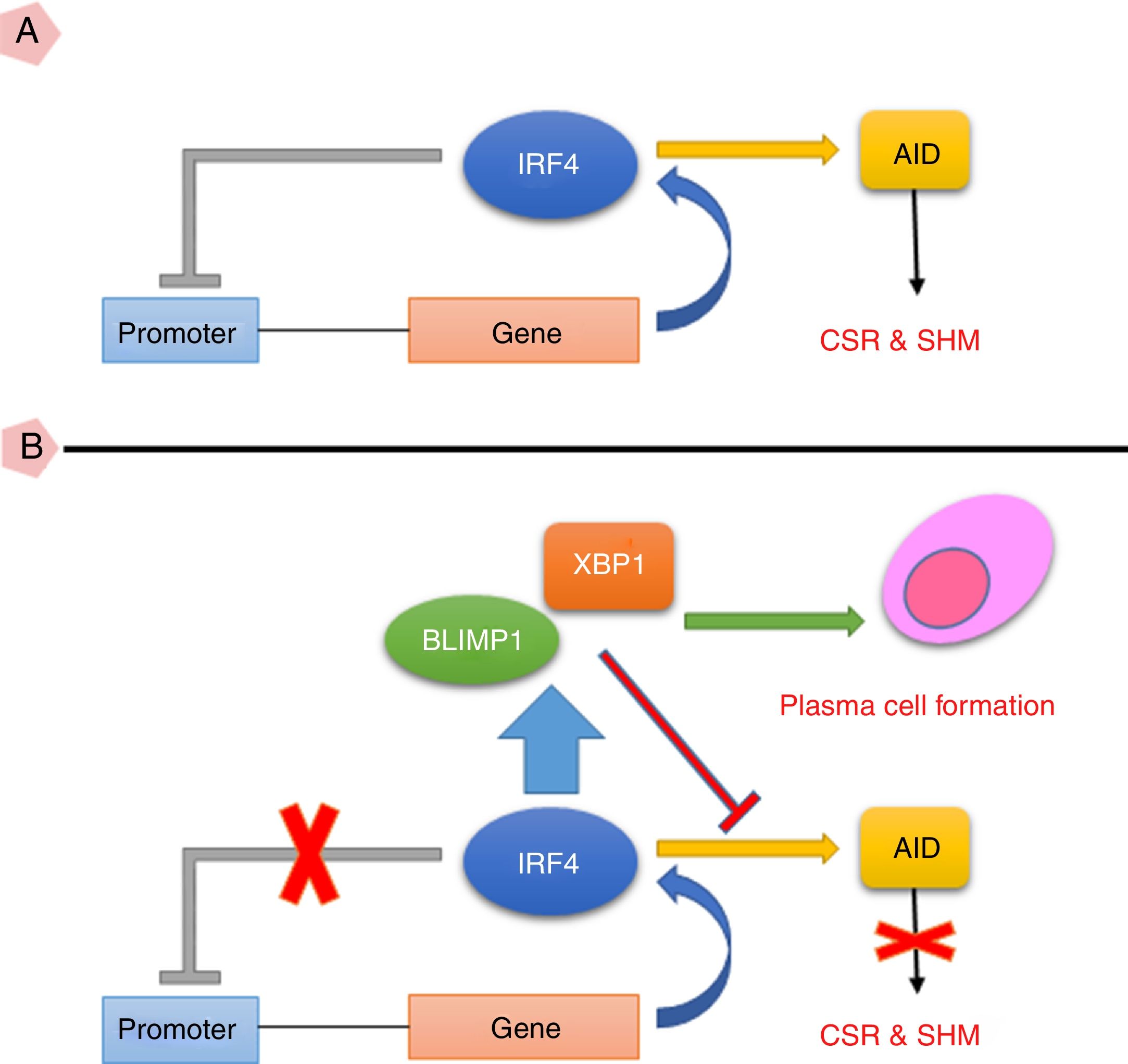
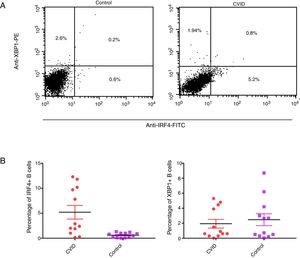
Taken together, the current study suggests that high expression of IRF4 in CVID patients may play important roles in defective B cell functions. As IRF4 expression in our patients was not able to activate negative feedback on itself and also was not able to elevate XBP1 level, the existence of an intrinsic or extrinsic defect in IRF4 in these patients could be speculated (Fig. 3). However, further investigations are required to identify any possible defect in IRF4, XBP1 and other upstream related transcription factors in the CVID patients.
(A) IRF4 can bind to its promoter and regulate its expression level via a negative feedback manner. In the early steps of final differentiation of activated B cells, expression of IRF4 is adjusted at an intermediate level which lead to run the expression of AID and to promote CSR and SHM in B cells. (B) At the end steps of final differentiation toward plasma cells, IRF4 expression level elevates and induces expression of BLIMP1 and XBP1. This in turn, inhibits AID expression and promotes plasma cell formation. A probable impairment in IRF4 function might be reciprocated by corruption of the negative feedback and increase of its expression. However, this might not be sufficient for plasma cell formation.
AID: activation induced deaminase; CSR: class switch recombination; SHM: somatic hypermutation.
There is no conflict of interest to declare.
This work was financially supported as a grant in aid (grant number: 394678) by Isfahan University of Medical Sciences, Isfahan, Iran.