Optimised purification steps for concentrating trace target native antigens are needed. Combining the p-aminobenzamidine ligand with protease inactivation enables partial purification of mite non-protease allergens lacking proteases.
ObjectiveWe sought to analyse in detail proteins obtained using this method from eight species of synanthropic acaridid mites and tested IgE reactivity using pooled human sera.
Materials and methodsProteins affinity bound to p-aminobenzamidine as a ligand were identified by MALDI TOF/TOF. After electroblotting, the proteins were visualised using the fluorescent SYPRO-Ruby protein blot stain, and IgE reactivity was further analysed using pooled human sera collected from patients allergic to house dust mites.
ResultsMS/MS identification confirmed previous results that no proteases were purified. Protein patterns corresponding to the allergens Der f 7, Der f 30 and actins indicated that these proteins are purified using p-aminobenzamidine and are present across a wide spectrum of acaridid mites. When using Dermatophagoides farinae, apolipophorins (Der f 14), chitinase-like Der f 15 and 18, 70-kDa heat shock protein, and a Der f Alt a10 allergen homolog (gi|37958173) were also detected. The target antigens tropomyosins and paramyosins showed similar IgE binding among the mite species tested. IgE reactivity with miscellaneous D. farinae antigen was also observed.
ConclusionsPartial purification of mite non-protease antigens using a strategy combining p-aminobenzamidine with protease inactivation was verified by 1D-E and 2D-E analyses. IgE binding to p-aminobenzamidine-purified native non-protease mite antigens was tested using pooled sera. This preliminary study allows for further work on individual serum samples, allowing confirmation of immunoreactivity.
Compared to native forms of proteins, differences in biological activity can be observed with recombinant proteins.1–3 For example, variations in immunoreactivity have been found between native and recombinant antigens such as mite tropomyosin4 and paramyosin.5 Thus, there is a need to develop processes for preparation of native antigens, as only native forms of antigens can reveal the true immune response.
Synanthropic acaridid (astigmatid) mite species are typically classified into two artificial groups: house dust mites (HDMs) and stored-product mites (SPMs).6,7 Both groups are also called “domestic mites” because many SPMs of the families Acaridae, Glycyphagidae, and Chortoglyphydae are also found in house dust.8 Various mites have cross-reactive and species-specific antigens.9,10 Moreover, cross-reactivity with allergens from other invertebrates, including insects, molluscs, or crustaceans, has been observed for mites.10–12
Among the main groups (Grps) of mite allergens, tropomyosins (Grp 10) are considered pan-allergens because they are highly conserved and because these proteins of many invertebrates, including molluscs, parasitic worms and various orders of arthropods, exhibit cross-reactivity. In addition, tropomyosin from some invertebrates is considered a food allergen.11–17 To date, tropomyosin allergenicity has been identified for 28 invertebrate species (for detailed information, visit http://www.allergen.org), and the tropomyosins from six astigmatid (syn. acaridid) mites, Blomia tropicalis, Dermatophagoides farinae, D. pteronyssinus, Lepidoglyphus destructor, Tyrophagus putrescentiae and Chortoglyphus arcuatus, have been identified as allergens (http://www.allergen.org).4,15,18–20 In contrast to knowledge about tropomyosin, there is limited information regarding allergenicity related to paramyosin. Although only four allergenic paramyosins have been identified, three are of acaridid mite origin, i.e., B. tropicalis, D. farinae and D. pteronyssinus.21–23 In addition, some invertebrate muscle proteins are allergens, including troponin C from cockroaches (Blattella germanica),24Periplaneta americana, the stored-product mite T. putrescentiae and some other invertebrates (Homarus americanus, Crangon crangon and Penaeus monodon) for which these proteins are considered to be food allergens. Given that myosin light chains of B. germanica, several species of decapods, and D. farinae have been identified as allergens (http://www.allergen.org), it can be speculated that the entire muscle complex of certain invertebrates is allergenic. This assumption supports the recognition of α-actinin involved in muscle contraction and β-actin as new shrimp allergens.11
It has been reported that as a ligand, p-aminobenzamidine binds with high affinity to tropomyosins, paramyosins and actins from domestic mites.25 Binding by p-aminobenzamidine to these proteins has also been confirmed for B. germanica and Oryctolagus cuniculus. In this case, the methodology used employed a strategy for inactivating proteases that were not of interest using protease inhibitors during the purification process; thus, proteases were not purified, and only non-protease proteins were obtained.25 Additionally, the study showed that mite paramyosin is monomeric and dimeric and tropomyosin monomeric and tetrameric using non-reducing sodium dodecyl sulphate-polyacrylamide gel electrophoresis (SDS-PAGE).25 The applicability of this method using p-aminobenzamidine to concentrate trace target proteins of V. destructor has also been demonstrated.26
In this study, we show that the p-aminobenzamidine ligand combined with protease inactivation using inhibitors can concentrate mite non-protease proteins to enable analysis of native antigen immunoreactivity.
Materials and methodsMitesEight species of synanthropic acaridid mites, Acari: Acaridida (alternatively Astigmata), were selected for this study because of their medical and economic importance. The mites were cultivated on an IWAKI 25-cm2 surface area in 70-mL-capacity tissue culture flasks (IWAKI flasks; Cat No. 3100-025; Sterilin, Newport, UK).27 Two different rearing diets were used to cultivate the wide collection of species. A wheat-derived rearing diet (Acarus siro, Tyrophagus putrescentiae, Aleuroglyphus ovatus) consisted of a mixture of oat flakes, wheat germ and dried yeast extract (Rapeto, Bezdruzice, Czechia) (10:10:1 w/w). A fish food-derived rearing diet (Tyroborus lini, Blomia tropicalis, Dermatophagoides farinae, Glycyphagus domesticus, Lepidoglyphus destructor) was composed of dog food (Ontario-pet, Placek, Podebrady, Czechia), wheat germ, dried fish food (LonBio, AquaTropic Lonsky, Praha, Czechia), Pangamin and gelatin (Serva Electrophoresis, Heidelberg, Germany) (10:10:3:2:1 w/w). All the biological samples originated from the rearing facility at Crop Research Institute (http://www.vurv.cz), Prague, Czechia. Mites were reared in small amounts of diet analogously to described.28 The mites (minimal purity: 95% pure mites) were separated from the spent growth medium, allowed to defecate for 24h, and then collected.
Protein extraction using protease-blocking extraction bufferA 0.1g sample of pure mite bodies was homogenised in a sterilised glass Potter-Elvehjem homogeniser (Art. No. 6305; Kartell Labware division, Noviglio, Italy) in 1mL cold 0.01M phosphate-buffered saline with 1% CHAPS (w/w) and 20μL protease inhibitor mixture (Cat No. 80-6501-23, GE Healthcare Life Sciences, Uppsala, Sweden). Samples were homogenised three times using a drilling machine for 2min each, followed by 20min of cooling on ice. Next, 0.5mL extraction buffer was added, and homogenisation was repeated three times for 1min each. The homogenate was allowed to stand for 10min on ice, and the supernatant was transferred to a centrifuge tube (Orange Scientific, Braine-l’Alleud, Belgium) and centrifuged for 15min at 10,000×g and 4°C using an MR 23i centrifuge (Jouan Industries, France). The supernatant was filtered using a glass Luer-lock syringe and 0.45-mm regenerated cellulose filter (TR-200435, Teknokroma, Barcelona, Spain).
Affinity chromatographyA 1-mL HiTrap Benzamidine FF column (Cat No. 17-5143-01, GE Healthcare Life Sciences) was used for purification of mite proteins. The column was equilibrated with 12 column volumes of 0.01M phosphate saline buffer. The filtered supernatant was diluted to a final volume of 3mL and passed through the column dropwise. The unbound fraction was removed with 20 volumes of 0.01M phosphate saline buffer. The affinity-bound proteins were eluted using 12 column volumes of 0.05M Tris-glycine buffer pH 3.0. The purified proteins were desalted and cleaned using a PD MidiTrap G-25 column (Cat No. 17-5143-01; GE Healthcare Lifesciences) according to the manufacturer's instructions. The protein content was measured using the Bradford reagent (Cat No. B6916; Sigma-Aldrich, Saint Louis, MO, USA). The purified protein was divided into 1–2mL aliquots and added to 15-mL centrifuge tubes; the tubes were covered with a 0.22-mm polytetrafluoroethylene (PTFE) filter, which was fixed using a cap with a vent hole. The samples were frozen and lyophilised in a PowerDry LL3000 lyophiliser (Thermo, Shanghai, China) and stored frozen for later use.
Serum collectionSera were obtained from patients at Medical Centre Prague (http://www.mc-praha.cz/en), Prague, Czechia, with written consent using the protocol approved by the institutional human ethics committee of the institute. Serum from 20 patients with a history of allergy and IgE reactivity to HDMs (routine medical analysis of sIgE against D. pteronyssinus and D. farinae) was pooled and used for screening the immunoreactivity of the purified proteins. All sera used were found to contain IgEs specific to HDMs by routine diagnostics. The average age of the patients was 33±9 (years±SD). Seven females and 13 males were included in the study.
SDS-PAGE and western blottingThe protein samples were separated using a Tris-glycine electrophoresis system according to the manufacturer's instructions (Sigma-Aldrich). For SDS-PAGE, proteins were diluted in SDS sample buffer containing dithiothreitol. Electrophoresis was performed using a constant voltage with a MiniPROTEAN® Tetra Cell (Bio-Rad, Shanghai, China). The proteins were then transferred to a Hybond-ECL nitrocellulose membrane (Cat. No. RPN203D, GE Healthcare Bioscience, Germany) using a TE77 PWR semi-dry blotter (Cat. No. 11-0013-42, Mfg for GE by Hoefer, USA). After electroblotting, the membranes were completely immersed in 7% acetic acid and 10% methanol and incubated at room temperature for 15min. The membranes were then washed three times with ddH2O water for 10min each and stained using the SYPRO Ruby protein blot stain (Cat. No. S11791; Invitrogen, USA) according to the manufacturer's instructions. The stained proteins were analysed using the InGenius documentation system with GeneSnap software (Syngene, Cambridge, UK); imaging was accomplished using a UV transilluminator (∼302nm) and a 16-bit charge-coupled device (CCD) camera (1024×1024 pixels) with an SG emission filter.
The membranes were washed with ddH2O water and blocked with BSA-T20 (3% bovine serum albumin and 0.1% Tween-20) in PBS (0.01M phosphate-buffered saline, pH 7.4). For immunological detection, the membranes were incubated overnight at 4°C with the pooled human sera (see above) diluted 1/10 with BSA-T20. The membranes were washed three times for 10min in PBS-T20. A goat anti-human IgE (¿-chain specific)-peroxidase antibody (Cat No. A9667; Sigma-Aldrich) was used as the secondary antibody. After 1h of incubation with the secondary antibody, the membranes were washed three times for 10min in PBS-T20. After washing the membranes in ddH2O water, the immunoreactive bands were visualised using a TMB (3,3′,5,5′-tetramethylbenzidine)-membrane peroxidase substrate (Cat No. 50-77-00; KPL, USA). The membranes were scanned using a Scanjet G4050 (Hewlett Packard, USA).
Two-dimensional electrophoresis (2D-E)Isoelectric focusing (IEF) was performed using an Ettan IPG Phor 3 instrument (GE Healthcare Life Sciences). Separation was performed using 13-cm ceramic strip holders and Immobiline dry strips with pH ranges of 3–10 (Cat No. 17-6001-14, GE Healthcare Life Sciences) and 4–7 (Cat No. 17-6001-13, GE Healthcare Life Sciences). A DeStreak Rehydration solution containing 0.5% immobilised pH gradient (IPG) buffer (pH 3–10 or 4–7, Cat No. 17-6000-87 or Cat No. 17-6000-86 from GE Healthcare Life Sciences) was used for active rehydration. The separation programme was as follows: (1) Step, 30V, 10h (active rehydration); (2) Step, 500V, 500Vh; (3) Grad, 1,000V, 800Vh; (4) Grad, 6000V, 15,000 Vh; and (5) Step, 6000V, 16,000Vh. The isoelectric focusing programme with active rehydration period was 19h, for a total of 32,600Vh. Immediately following IEF, the strips were equilibrated for 15min in an equilibration buffer containing dithiothreitol (Cat No. 43817, Sigma-Aldrich), followed by 15min in a buffer containing iodoacetamide (Cat No. 57670, Sigma-Aldrich). The strips were placed over an SDS-PAGE gel and fixed with 1% agarose. Electrophoresis was performed at a constant voltage of 30V for 50min, after which the proteins were separated at a constant voltage of 300V in a cooled apparatus. After electrophoresis, the gel was processed using the SYPRO Ruby (Cat. No. S1200, Invitrogen) staining method. The gel was fixed overnight in fixing solution (50% liquid chromatography-mass spectrometry (LC-MS)-grade methanol, 7% ice acetic acid, and 43% nanopure water). After removing the fixing solution, the gels were washed (10% LC-MS grade methanol, 7% ice acetic acid, and 83% nanopure water) for 30min. Finally, the results were visualised using the G:BOX documentation system (Syngene, Cambridge, UK) in automatic capture mode.
Protein identificationFor mass spectrometry identification, 1D SDS-PAGE of the proteins purified from L. destructor, D. farinae, B. tropicalis, G. domesticus and A. ovatus was performed. In addition, 2D-E separation and spot analysis of the D. farinae sample was performed. After Coomassie staining, bands or spots were excised, destained and digested with trypsin. The MS analysis followed methodology previously described.29 The spectra were searched using local Mascot v. 2.1 (Matrix Science, Boston, MA, USA) against the metazoan and entire non-redundant protein sequence databases of NCBI (23,214,025 sequences; 7,977,717,942 residues). The output of the metazoan search was selected for data presentation. The database search criteria were as follows: enzyme – trypsin; taxonomy – Metazoa; fixed modification – carbamidomethylation; variable modification – methionine oxidation; peptide tolerance – 80ppm, allowing one missed cleavage; and MS/MS tolerance – 0.3Da. Hits scoring p<0.05 were considered significant. Matrix-assisted laser desorption/ionisation time-of-flight tandem (MALDI TOF/TOF) MS analysis was performed at Service Laboratories of the Biology Section, Laboratory of Mass Spectrometry, Faculty of Science, Charles University in Prague (https://www.natur.cuni.cz/eng), Prague, Czechia.
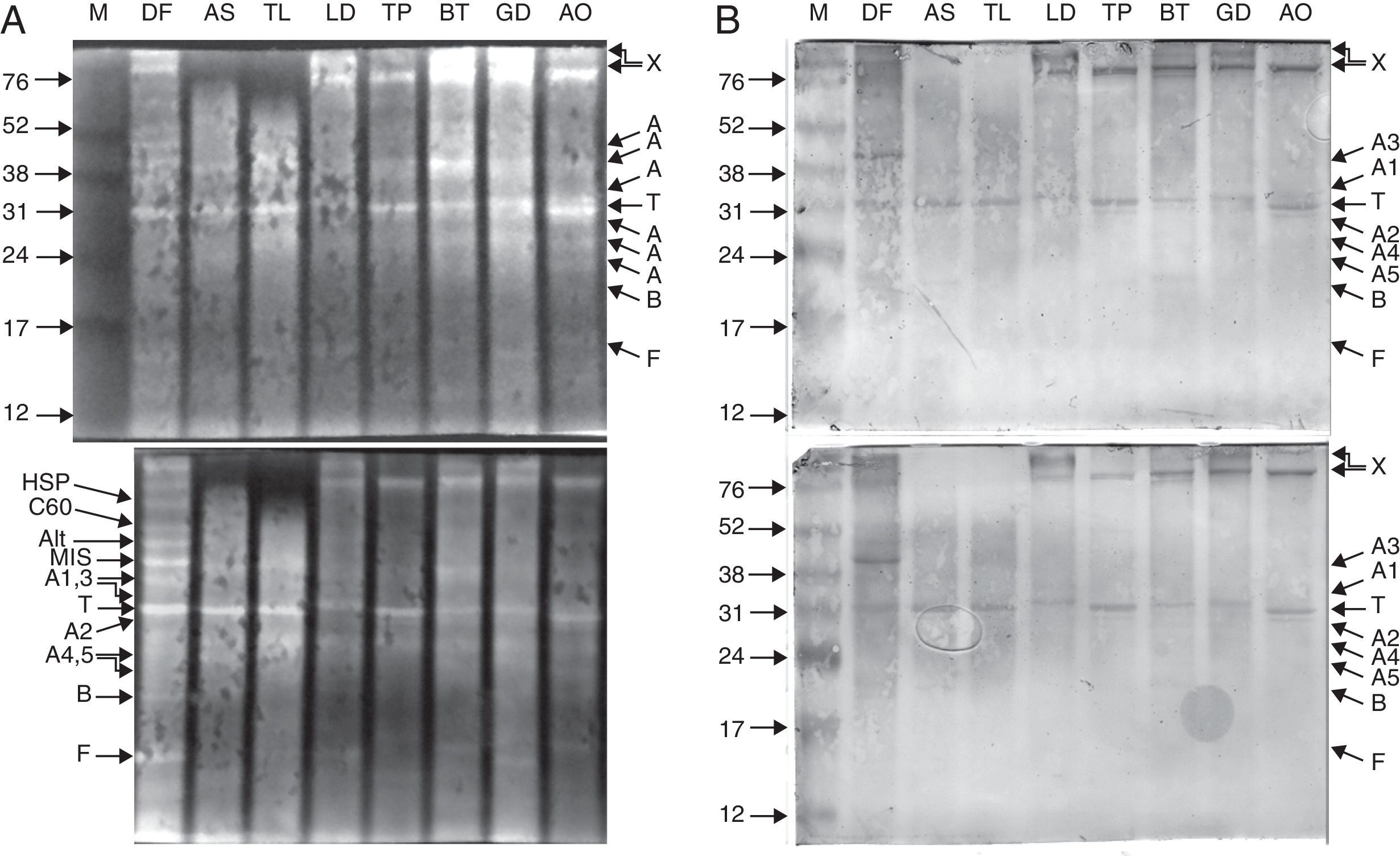
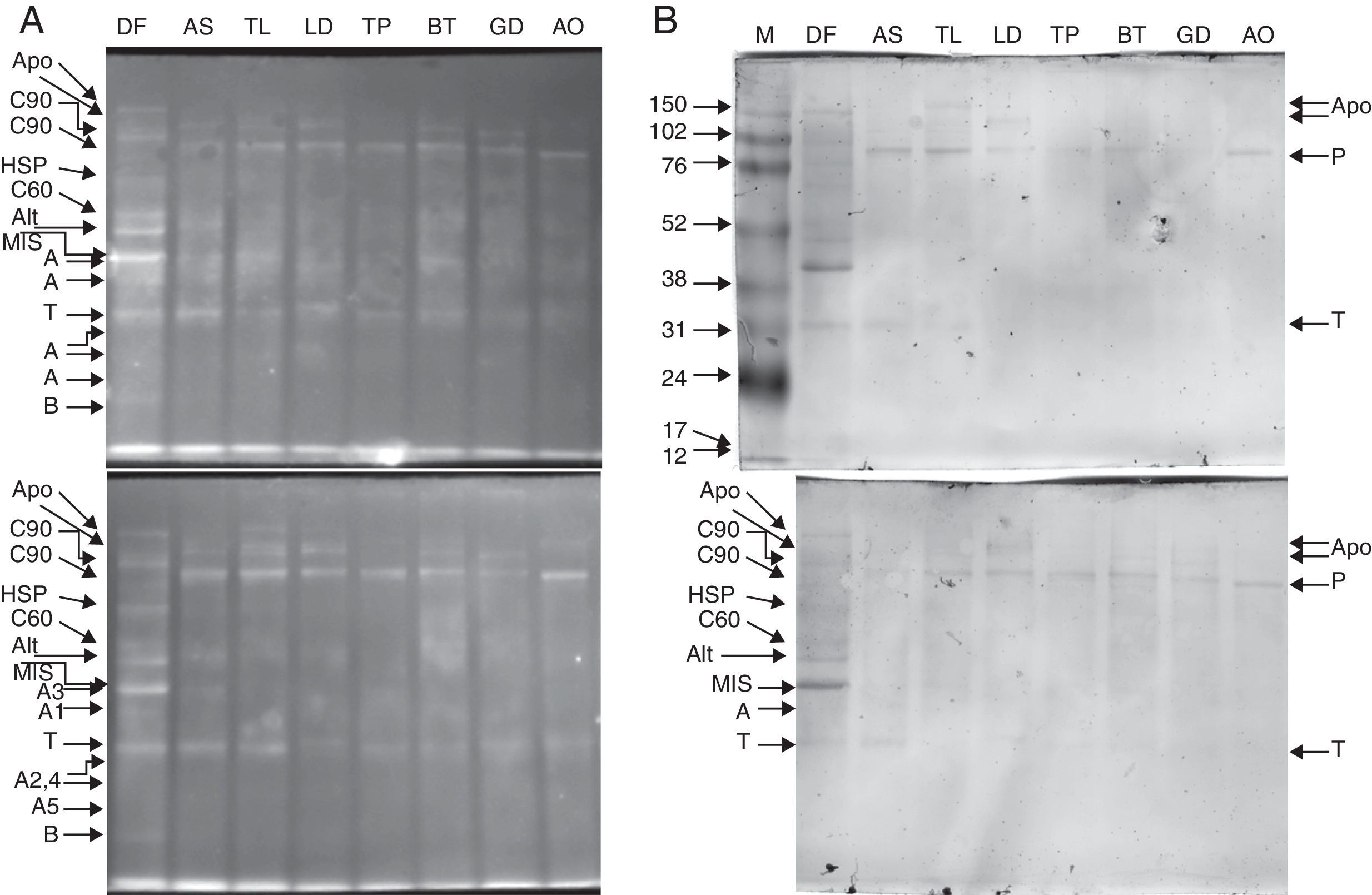
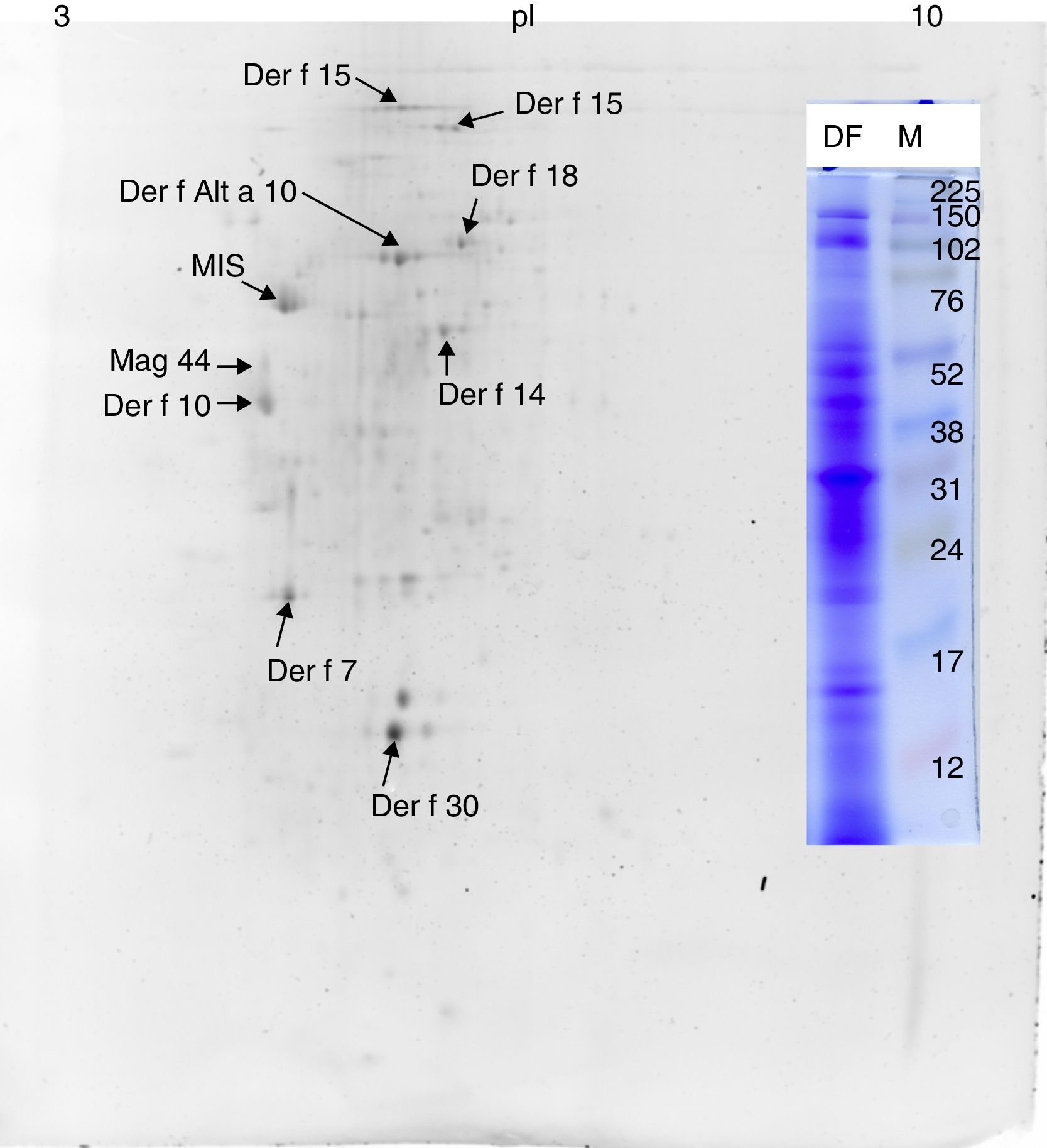
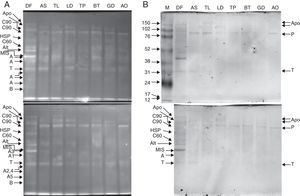
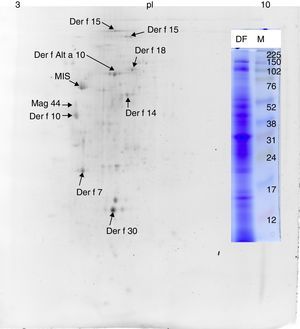
ResultsMS/MS protein identificationsIn total, 170 protein samples of bands from D. farinae, L. destructor, A. ovatus, G. domesticus and B. tropicalis were excised after 1D SDS-PAGE electrophoresis and further analysed by MALDI TOF/TOF MS. The protein patterns of 1D-E-MS/MS are shown in Figs. 1A and 2A, and the results of 2D-E-MS/MS are presented in Fig. 3. For details of the protein identifications, see Supplementary Material Table S1.
Purified proteins separated by 15% Tris-glycine electrophoresis, blotted onto nitrocellulose membranes and stained with SYPRO ruby protein blot stain (A). The antigens were further probed using sera from 20 patients allergic to house dust mites, followed by anti-human IgE antibody reactivity assessment. IgE-reactive proteins were visualised using the TMB substrate (B).
Legend: A – actin; T – tropomyosin monomer; B – Der f 7 allergen (identified only in the DF sample); F – ferritin Der f 30 (identified only in the DF sample); Alt – Der f Alt a 10 allergen homolog (gi|37958173) (identified only in the DF sample); Apo – apolipophorin, group 14 mite allergen (identified and corresponding bands present only in the DF sample); C60 – 60kDa chitinase-like allergen (identified only in the DF sample); C90 – 90kDa chitinase-like allergen (identified only in the DF sample); MIS – miscellaneous IgE-reactive protein of Dermatophagoides farinae; X – accumulated proteins of higher molecular weight; AO – Aleuroglyphus ovatus; GD – Glycyphagus domesticus; BT – Blomia tropicalis; TP – Tyrophagus putrescentiae; LD – Lepidoglyphus destructor; TL – Tyroborus lini; AS – Acarus siro; DF – Dermatophagoides farinae; M – marker.
Purified proteins separated by 10% Tris-glycine electrophoresis, blotted onto nitrocellulose membranes and stained with SYPRO ruby protein blot stain (A). The antigens were further probed using sera from 20 patients allergic to house dust mites, followed by anti-human IgE antibody reactivity assessment. IgE-reactive proteins were visualised using the TMB substrate (B).
Legend: A – actin; T – tropomyosin monomer; B – Der f 7 allergen (identified only in the DF sample); F – ferritin Der f 30 (identified only in the DF sample); Alt – Der f Alt a 10 allergen homolog (gi|37958173) (identified in the DF sample); Apo – apolipophorin, group 14 mite allergen (identified in the DF sample); C60 – 60kDa chitinase-like allergen (identified in the DF sample); C90 – 90kDa chitinase-like allergen (identified in the DF sample); HSP – heat shock protein (identified in the DF sample); MIS – miscellaneous IgE-reactive protein of Dermatophagoides farinae; AO – Aleuroglyphus ovatus; GD – Glycyphagus domesticus; BT – Blomia tropicalis; TP – Tyrophagus putrescentiae; LD – Lepidoglyphus destructor; TL – Tyroborus lini; AS – Acarus siro; DF – Dermatophagoides farinae; M – marker.
The 15% SDS-PAGE separation selected optimal for Grp 10 mite allergenic tropomyosins of approximately 31kDa was successful, and the western blot demonstrated the IgE reactivity those antigens across the mites tested (Fig. 1A, B). The pattern of bands of ∼100-kDa Grp 11 mite allergenic paramyosins was also well observed after optimal 10% SDS-PAGE separation. In the case of paramyosin, Mascot revealed similarity of the paramyosins of SPMs L. destructor, G. domesticus and A. ovatus (paramyosins have not been sequenced in these mites) with B. tropicalis (see the list of paramyosin identifications in Supplementary Material Table 1). However, paramyosin was not identified in the investigated sample of D. farinae. Furthermore, a pattern of five different bands corresponding to different forms of actins was identified, three with molecular masses lower than that of tropomyosin and two with molecular masses higher than that of tropomyosin.
Proteins identified only in the Dermatophagoides farinae sampleThe Der f Alt a 10 allergen homolog (gi|37958173; GenBank Accession No. AAP35081.1) belonging to the aldehyde dehydrogenase family was identified exclusively in the D. farinae sample. Blast30 search showed that this protein has the highest identity (69%) with an aldehyde dehydrogenase-like protein of T. putrescentiae (Accession AOD75396.1), and high identity with aldehyde dehydrogenases from other mite species was also observed. In addition, the group 14 allergen or Mag 3 (syn. group 14 allergen) allergen was detected in the D. farinae sample. Heat shock protein 70, Der f 7 and Der f 30 (Figs. 1 and 2) were also detected in the D. farinae sample, and corresponding bands in other species suggest the possible presence of homologous proteins. Analysis of 12 spots in the D. farinae 2D-E (Fig. 3) pattern enabled identification of nine proteins corresponding to described mite allergens. One protein with high IgE reactivity by 1D western blot is an un-described (miscellaneous) protein. Moreover, 2D-E-MS/MS analysis identified mite allergenic chitinase-like Der f 15 and Der f 18, tropomyosin Der f 10 and tropomyosin, denoted in databases as Mag 44, apolipophorin Der f 14, Der f 7, Der f 30 and Der f Alt a 10 allergen.
Western blotting results for tropomyosin, paramyosin and actin human IgE bindingThe results of western blotting using a pool of serum samples from 20 human patients diagnosed with HDM allergies by enzyme-linked immunosorbent assay (ELISA; routine medical analysis) are shown in Figs. 1B and 2B, which represent the results for 15% and 10% SDS-PAGE protein separations, respectively. Figs. 1A and 2A demonstrate the abundance of proteins bound to the nitrocellulose membrane before immunoreactivity. The antigens were blotted onto the membrane with similar patterns, and the immunostaining pattern indicated that the immunoreactive proteins of the mite species used in the study were, in many cases, similar between species with respect to the strength of reactivity. The best immunoreactivity result after 15% SDS-PAGE separation was observed for tropomyosins (Fig. 1B); the best immunoreactivity for the paramyosin pattern was observed after 10% SDS-PAGE (Fig. 2B). The actin bands denoted A1 and A2 showed good IgE binding by western blotting (Fig. 1B).
Western blotting results for non-muscle proteinsHuman IgE reactivity was also detected for bands corresponding to the Der f Alt a10 allergen homolog and an miscellaneous protein (MIS in Figs. 1–3). Furthermore, a pattern corresponding to Der f 7 and Der f 14 apolipophorin-like (Mag 3) exhibited immunoreactivity. However, we did not observe a signal corresponding to Der f 30, which exhibited a relatively abundant protein content by both 1D-E and 2D-E separations.
DiscussionAllergy to HDMs and SPMs, formerly referred to collectively as domestic mites, is the most prevalent cause of allergic sensitisation; indeed, this source of antigens affects up to 85% of asthmatics worldwide.31 It is believed that native mite proteins may be used for therapeutic purposes, and many recombinant proteins have been prepared and tested.17 In the present study, the identity of p-aminobenzamidine-partially purified antigens25 was examined in detail, and the biological activity of these antigens was shown. IgE binding to a wide spectrum of native mite pan-allergenic tropomyosins and paramyosins was assessed. Preliminary results based on IgE reactivity testing with pooled human serum indicated the immunoreactivity of pan-allergens purified from eight species of domestic mites (L. destructor, T. lini, D. farinae, A. siro, T. putrescentiae, B. tropicalis, G. domesticus and A. ovatus). In addition, isoforms of actin were found to bind human IgE, suggesting the allergenic potential of mite actins. IgE reactivity of the native Der f Alt a10 homolog (gi|37958173; GenBank Accession No. AAP35081.1), Der f 14, Der f 7, Der f 15 and Der f18 allergens was also observed. Protein patterns corresponding to Der f 7 and Der f 30 indicated that these proteins are present across a wide spectrum of acaridid mites. Moreover, we observed immunoreactivity with an MIS antigen from D. farinae with an unknown function.
Many studies concerning determination of the allergenic potential of tropomyosins and paramyosins from invertebrate sources have been conducted, but none have compared a wide spectrum of species. Most of the tropomyosins analysed to date for IgE reactivity were recombinant Der p 10,18 Blo t 10,15 Lep d 1019 and Tyr p 10.20 One study on mite tropomyosin, which is included on the IUIS list of allergens (http://www.allergen.org), examined both native and recombinant tropomyosins of D. farinae.4 These authors found that the native tropomyosin showed 80.6% reactivity, which is comparable to the reactivity of major native allergens such as Der f 1 and Der f 2, whereas the recombinant protein was less reactive.4 IgE binding to native mite tropomyosins partially purified using p-aminobenzamidine as a ligand was verified in this study, allowing future testing of the allergenicity of collections of these native antigens.
In this study, we observed human serum IgE reactivity with isoforms of actin, one of the primary proteins in muscle and the cytoskeleton. Because eukaryotes display a variety of highly conserved isoforms of actin,32,33 the total number of mite actin isoforms in mites is most likely higher than five. The number of purified isoforms and the IgE reactivities of concrete isoforms should be assessed by 2D electrophoresis analysis and 2D immunoblot analysis, respectively, in the future. The fact that human serum exhibits reactivity with mite actin isoforms indicates that these isoforms could be novel invertebrate allergens. Furthermore, this result suggests that the components of the actin-linked regulatory system formed by actins, tropomyosin, paramyosin, myosin and troponin C have similar pan-allergenic epitopes because these proteins are highly conserved and are present in one invertebrate complex.
We found a pattern of purified proteins that is different from that of the proteins previously purified from D. farinae.25 Tandem MS analysis of the bands excised from 1D-E and 2D-E gel revealed the presence of the Der f Alt a10 allergen homolog (gi|37958173; GenBank Access No. AAP35081.1), Der f 14 (apolipophorin-like ∼40kDa or ∼177/190kDa), chitinase-like Der f 15 and Der f 18, Der f 30, Der f 7 and heat shock protein (∼70kDa) allergens in the D. farinae sample. In addition, an unknown allergen with strong binding with human serum IgE was found. However, a band corresponding to paramyosin was not identified in this D. farinae sample, nor was it identified by MS/MS. The reason for the lack of a paramyosin band in this sample is not clear; however, it is possible that the D. farinae mites culture used for the preparation of antigens in this study were at a stage during which different protein abundances were present. To date, the identity of the Der f Alt a10 allergen homolog, which is according to Blast30 an aldehyde dehydrogenase, has been determined only at the transcript level.34 We identified the native protein and its reactivity with human serum IgE binding. Similar findings that bands corresponding to D. farinae Mag 1 and Mag 3 high-molecular-weight (177kDa) allergen (Der f 14) are apolipophorin-like mite allergens have been reported by Epton et al.,35 whose study included a D. pteronyssinus antigen.35 Analogous to previously published results, we also suggest that smaller derivatives of apolipophorin may be present after electrophoresis, because a smaller unit of Der f 14 was detected by 2D-E. These different results for the D. farinae sample suggest the necessity of verification of the antigen, especially in cases when the antigens would be used for therapeutic purposes against specific allergenic epitopes. The finding that the p-aminobenzamidine ligand binds chitinase-like proteins and apolipophorins agrees with the fact that these proteins are able to bind calcium.36,37 We propose that p-aminobenzamidine is useful for analysis of non-protease calcium-binding proteins such as Der f 15 and Der p 15 in high abundance in D. farinae28 and D. pteronyssinus38 mite faecal extract, respectively. Specifically, elimination of highly abundant proteases in mite faeces will result in concentration of other proteins of interest in a partially purified fraction.
Among all of the analysed partially purified native antigens, the targets tropomyosin and paramyosin were found to be the most abundant and most reproducible after affinity chromatography processing. Furthermore, a variety of proteins obtained in the p-aminobenzamidine-bound fraction exhibited human serum IgE binding. These results should be broadened in future studies, such as those using 2D western blotting to analyse IgE reactivity of particular antigens and their isoforms. Additional separation steps, i.e., gel chromatography, can also aid in the fractionation of antigens.
Ethical disclosuresConfidentiality of dataThe authors declare that they have followed the protocols of their work centre on the publication of patient data and that all the patients included in the study have received sufficient information and have given their informed consent in writing to participate in that study.
Right to privacy and informed consentThe authors declare that no patient data appears in this article.
Protection of human subjects and animals in researchThe authors declare that the procedures followed were in accordance with the regulations of the responsible Clinical Research Ethics Committee and in accordance with those of the World Medical Association and the Helsinki Declaration.
Conflict of interestThe authors have no conflict of interest to declare.
This work was supported by the project no. OC10019 (COST CM0804 action Chemical Biology with Natural Products) of the Ministry of Education, Youth and Sports of the Czech Republic (http://www.msmt.cz) and by the project no. RO0416 of the Grant Agency of the Ministry of Agriculture of the Czech Republic (http://eagri.cz). We would like to thank Martin Markovic for valuable help.