Autosomal dominant hyper-IgE syndrome (AD-HIES) is a primary immunodeficiency mainly caused by mutations in STAT3, a signalling molecule implicated in the development of appropriate immune responses. We aimed to characterise the innate immune response in AD-HIES.
MethodsThe frequency of innate immune cells in peripheral blood (PB) from seven AD-HIES patients and healthy controls were determined. CD80/CD86 surface expression and cytokine levels in supernatants from PBMC after stimulation with TLR-2, -4 and -9 agonists were also measured by flow cytometry. In addition, several SNPs within these TLR genes in genomic DNA samples from patients and controls were examined.
ResultsA significantly reduced number of PB iNKT cells was observed in the AD-HIES group. CpG-stimulated pDC and mDC from patients exhibited a lower increase in the expression of the costimulatory molecule CD80. We also observed an increase in the secretion of IL-12p70, TNF-alpha and IL-10 in PBMC from HIES patients after LTA or LPS stimuli. No association was found between the different SNPs detected and the HIES phenotype.
ConclusionsThese findings demonstrate that important mediators of the innate immunity responses are affected in AD-HIES. More studies are necessary to investigate how the STAT3 function interferes with development of iNKT cells and TLR-mediated responses.
The hyper-IgE syndrome (HIES) is a multi-systemic disorder usually first diagnosed in infancy, affecting the immune but also the musculoskeletal, dental, craniofacial and vascular systems. Nevertheless, HIES is considered a primary immunodeficiency due to the increased susceptibility of patients to infections. Chronic mucocutaneous candidiasis and respiratory tract infections mainly by fungi and encapsulated bacteria resulting in pneumonia with pneumatoceles represent other HIES clinical hallmarks. In 2007, genetic association of the autosomal dominant form of HIES (AD-HIES) with heterozygous dominant-negative mutations in the signal transducer and activator of transcription 3 (STAT3) gene were reported.1 Most of the clinical characteristics of AD-HIES confirm the widespread role of STAT3 in immunity since mutations in this gene affect mainly Th17 cell differentiation leading to a failure of the IL-17 and IL-22 secretion, associated with reduced antimicrobial peptides and chemokines production.2 However, STAT3 has pleiotropic functions in the innate and adaptive immunity.3–7 Specifically in HIES, Giacomelli et al. investigated the dendritic cells (DC) differentiation, maturation and function in the presence of IL-10 and lipopolysaccharide (LPS) in seven patients with STAT3 mutations and healthy controls. A time-dependent higher production of TNF-alpha, IL-6 and IL-12 in the HIES DC was observed.8 Yaganeh et al. also found increased TNF-alpha and IL-8 production in HIES peripheral blood mononuclear cells (PBMC) after stimulation with several concentrations of LPS and Staphylococcus aureus suspension.9 Another study demonstrated a failure in the generation of tolerogenic primary or monocyte-derived DCs in STAT3-deficient HIES patients after stimulation with IL-10.10 These patients exhibited a marked increase in the expression of the costimulatory molecules CD80 and CD86 in monocyte-derived DCs after LPS stimulation, a finding that could not be inhibited with IL-10 pre-treatment. Therefore, it is plausible to hypothesise that STAT3-deficient HIES patients would have defects in the numbers and function of innate immune cells that may partially explain susceptibility to infection. Moreover, other innate immune cells such as invariant natural killer T (iNKT) cells which have important roles in innate defence against microbes have not been previously investigated in HIES. iNKT cells are considered CD1-restricted specialised T cells that directly act as effector cells at mucosal surfaces recognising lipid antigens. More importantly, iNKT cells often participate in the pathogenesis of several inflammatory disorders.11,12
On the other hand, it is possible that single nucleotide polymorphisms (SNP) in Toll-like receptor (TLR) genes may explain the clinical heterogeneity in AD-HIES. SNPs of the gene encoding TLR2 have an impact on susceptibility to infections by modifying cell response to lipoteichoic acid (LTA) and other stimuli.13,14 Within the gene encoding TLR4, the cosegregating SNPs Asp299Gly and Thr399Ile, which affect the extracellular domain of the molecule conferring hyporesponse to LPS stimulation, positively correlate with several infectious diseases.15 Moreover, a number of rare non-synonymous SNPs within the coding region of TLR9 have also been associated with pathogen defence in ocular toxoplasmosis and other diseases.16–19
In the present study, we evaluate the frequency and functional response of innate immune cells from peripheral blood (PB) in patients with HIES and healthy controls upon activation with agonist of TLR2, TLR4 and TLR9. In addition, several SNPs within these TLR genes were examined.
Materials and methodsStudy populationSeven unrelated patients (age range: 12–28 years) were enrolled in this study. All the patients have a score >40 according to the NIH scoring system that is suggestive of AD-HIES.20 The control group included six healthy volunteers (age range: 12–46 years). All the individuals evaluated in this study were from the Metropolitan Area of Medellin, Colombia and gave their informed consent. The study obtained approval of the Institutional Review Board from the University of Antioquia.
Antibodies and reagentsFluorochrome labelled monoclonal antibodies (mAbs) against human molecules CD3, CD11c, CD14, CD16, CD19, CD56, CD69, CD80, CD86, CD123, HLA-DR, the invariant CDR3 loop of human canonical Vα24Jα18 TCR α chain (for the identification of iNKT cells, clone 6B11), lineage markers (Lin, a mixture of anti-CD3, CD14, CD16, CD19, CD20 and CD56) and the corresponding isotype control antibodies were purchased from Becton Dickinson (San Jose, CA, USA). FITC-labelled anti-Vβ11 and PE-labelled anti-Vα24 were obtained from Beckman Coulter Immunotech (Marseille, France).
LTA purified from S. aureus was obtained from InvivoGen (San Diego, CA, USA), and LPS from Escherichia coli 0127:B8 was purchased from Sigma-Aldrich (Saint Louis, MO, USA). The A-Class CpG ODN 2216 (sequence 5′-ggGGGACGATCGTCgggggG-3′, lower case, phosphorothioate linkage; upper case, phosphodiester linkage 3′ of the base) with no detectable endotoxin levels was kindly provided by Coley Pharmaceutical Group (Wellesley, MA, USA).
Expression of IL-17PBMC isolated from heparinised blood were washed with phosphate-buffered saline and resuspended in RPMI 1640 medium supplemented with 10% heat-inactivated foetal calf serum, 100U/mL penicillin G and 100μg/mL streptomycin. Cells were then stimulated with 10ng/mL of phorbol 12-myristate 13-acetate (PMA) and 1μmol/L ionomycin (Sigma) for 2h at 37°C. Thereafter, brefeldin A 10μg/mL was added for 4h (Sigma) in the same conditions. Intracellular staining was performed with the BD Cytofix/Cytoperm™ Fixation/Permeabilization Kit. The following surface and intracellular marker were used for staining: CD3 Quantum-dot 705 (Invitrogen) and IL-17 PE (eBioscience). PBMC were then acquired in a FACS-canto II instrument (BD Biosciences) and analysed using FlowJo software (TreeStar, Ashland, OR, USA).
Quantification of innate immune cellsThe absolute number of PB eosinophils, neutrophils, lymphocytes and monocytes was calculated on the basis of total and differential blood cell counts using Wright staining of blood smears and conventional light microscopy. Frequency and phenotype of PB myeloid dendritic cells (mDC, Lin−/CD11c+/HLA-DR+), plasmacytoid dendritic cells (pDC, Lin−/CD123+/HLA-DR+), monocytes (CD14+/CD3−), NK cells (CD3−/CD16+/CD56+), iNKT cells (6B11+/CD3+) and NK T cells (CD3+/CD56+) were determined by multi-parametric flow cytometry. Briefly, PBMC were incubated with the corresponding mAbs for 20min/RT in the dark. Non-specific mAb binding was controlled by blocking FcγR with 20μL of blocking reagent for each 1×107cells (20min/4°C) (Miltenyi Biotech, Bergisch Gladbach, Germany). Erythrocytes were lysed by incubation of the cell suspensions with 1× FACS lysing solution (Becton Dickinson) following the manufacturer's instructions. Finally, cells were fixed with 250μL of 2% formaldehyde. Appropriate isotype-matched control antibodies were also included. Flow cytometry was performed using the Becton Dickinson FACSORT and analysed using the CellQuest software (Becton Dickinson).
Isolation and culture of mononuclear cellsPBMC were obtained from heparinised blood samples by density gradient centrifugation using lymphocyte separation medium (BioWhittaker, Walkersville, MD, USA). Viability was determined by trypan blue exclusion. For cell culture, PBMC (1×106mL−1) were suspended in complete culture media (RPMI 1640 supplemented with 10% of heat-inactivated foetal calf serum, 100U/mL penicillin, 100μg/mL streptomycin and 2mM of l-glutamine) and stimulated with/without 15μg/mL of LTA, 2μg/mL of LPS or 4μg/mL of A-Class CpG ODN 2216 for 24h at 37°C in 5% CO2. The CD80 and CD86 expression were analysed by flow cytometry in the different cell types using specific mAbs for plasmacytoid dendritic cells (pDC, Lin−/CD123+), myeloid dendritic cells (mDC, Lin−/CD11c+) and monocytes (CD14+). Culture supernatants were collected and stored at −70°C for measurement of cytokine concentration.
Measurement of cytokinesConcentration of IL-12p70, TNF-α, IL-10, IL-6, and IL-1β in PBMC cultures supernatants was determined by flow cytometry using the BD® Cytometric Bead Array following the manufacturer's instructions (CBA, Human Inflammatory Kit, BD Biosciences Pharmingen, San Diego, CA, USA) after TLR-ligand stimulation using the stimulus conditions described before. The assay sensitivities were 1.9, 3.7, 3.3, 2.5 and 7.2pg/mL, respectively. IFN-α was measured using a commercial ELISA kit (PBL Biomedical Laboratories, Piscataway, NJ, USA) following the manufacturer's instructions (detection limit: 3.5pg/mL).
Detection of polymorphisms in TLR genesGenomic DNA was isolated from blood samples using the DNA Isolation Kit provided by Puregene™ (Gentra Systems, Minneapolis, MN, USA) following the manufacturer's instructions. For TLR-2, two SNPs located at position 17,023 (C/T, Arg677Trp, rs121917864) and 17,252 (G/A, Arg753Gln, rs5743708) of the gene were studied. Specific primers flanking both SNPs in a single PCR product (approx. 370bp) were used. For the TLR-4 gene, two SNPs located at position 8719 (A/G, Asp299Gly, rs4986790) and 9019 (C/T, Thr399Ile, rs4986791) were also analysed. Specific primers flanking both SNPs in a single PCR product (approx. 509bp) were used as well. Additionally, the TLR-9 gene (from position +474 to +3765) was sequenced by primer walking. We further confirmed these findings by amplifying and sequencing of three different PCR products using specific primers targeting the regions +745 to +1290, +1291 to +1837 and +3324 to +3837 of the TLR-9 gene. Seven SNPs have been reported within this gene, two of which (G1174A, rs352139 and G2848A, rs352140) have been found at high frequencies in the Hispanic population.16 Primer design and synthesis as well as PCR product amplification and sequencing were performed by Macrogen Corporation (Seoul, Korea). Sequences were analysed and edited using Chromas Lite version 2.1.1. Homology search against the human genome database at NCBI was performed using nucleotide Blast (Blast-n version 2.2). Multiple alignments to map the SNPs were performed using the programme ClustalW version 1.83 (http://www.ebi.ac.uk/clustalw/index.html). Some sequences were submitted to the GenBank and the following accession numbers were assigned: GQ228031 and GQ228033 for the TLR-2 gene sequences; GQ903698, GQ903699, GQ903700 and GQ903702 for the TLR-4 gene sequences and FJ946863, FJ946864, FJ946865, FJ946866, FJ946867 and FJ946872 for TLR-9 gene sequences. Genotypes counts were also compared with those of the Medellin, Colombia subpopulation obtained from http://browser.1000genomes.org.
Statistical analysisResults are presented as mean±standard deviation (SD). Statistical comparison between two different groups was performed using the Mann–Whitney U test, with a confidence level of 95%. Statistical comparisons among three or more different groups were performed using the non-parametric Kruskal–Wallis test, with a confidence level of 95%. We also applied the Shapiro Wilk test to evaluate normality (Statistical version 5) and the Bartlett's test to evaluate equality of variances (GraphPad Prism 5.0 Software). A p value <0.05 was considered significant.
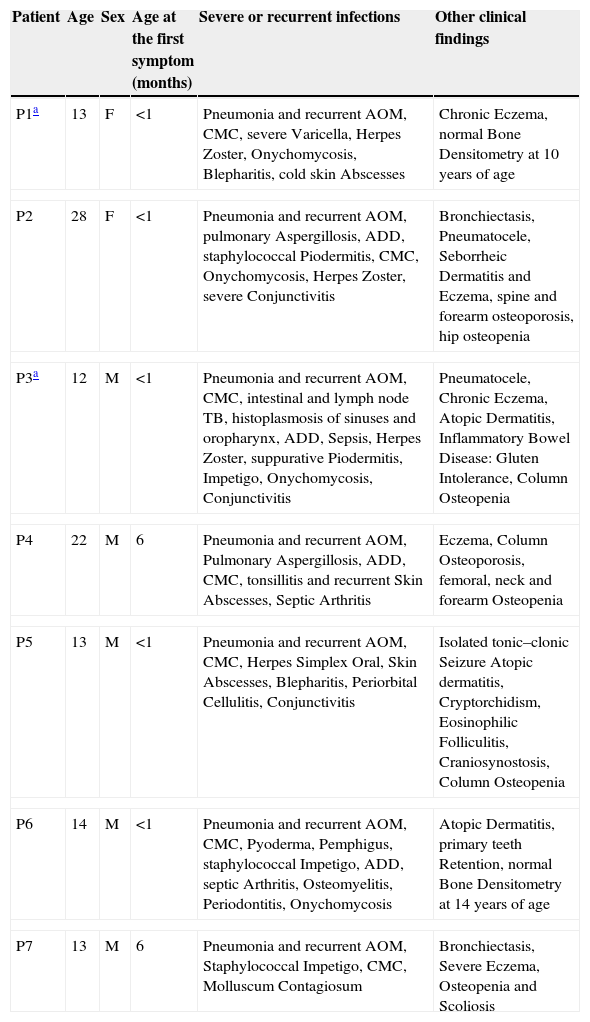
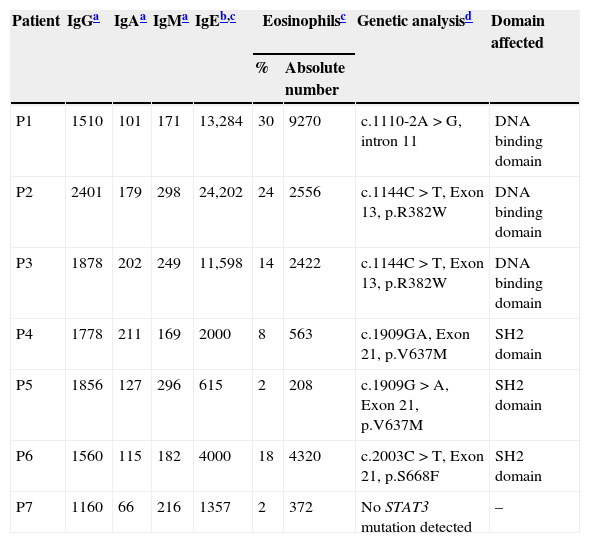
ResultsClinical features of the HIES patients and STAT3 genetic analysisClinical and immunological characteristics of our cohort are shown in Tables 1 and 2. Genetic analysis within the STAT3 gene is listed in Table 2. Family history of pneumonias, dermatitis, bone fractures and S. aureus infections was found only in P1. Evidence of consanguinity (a second cousins marriage) was observed only in P2. All patients demonstrated an early onset disease and presented with pneumonia and recurrent acute otitis media. All but one of them manifested chronic mucocutaneous candidiasis. More importantly, eczema and osteopenia were reported in 71% of the cases. Infections such as S. aureus and Herpes disease, onychomycosis, acute diarrhoeal disease as well as skin abscesses were also very frequent in our cohort. Chronic and opportunistic infections, including aspergillosis and hystoplasmosis were also observed in three of our patients. Mutations at the STAT3 gene affecting either the DNA-binding or the SH2 domains were detected in six patients. Although mutations were not found in the coding region and adjacent splice sites of the STAT3 gene in the genomic DNA from P7, clinical features and the NIH score were also compatible with the disease.
Clinical and immunological parameters of the HIES patients included in this study.
| Patient | Age | Sex | Age at the first symptom (months) | Severe or recurrent infections | Other clinical findings |
|---|---|---|---|---|---|
| P1a | 13 | F | <1 | Pneumonia and recurrent AOM, CMC, severe Varicella, Herpes Zoster, Onychomycosis, Blepharitis, cold skin Abscesses | Chronic Eczema, normal Bone Densitometry at 10 years of age |
| P2 | 28 | F | <1 | Pneumonia and recurrent AOM, pulmonary Aspergillosis, ADD, staphylococcal Piodermitis, CMC, Onychomycosis, Herpes Zoster, severe Conjunctivitis | Bronchiectasis, Pneumatocele, Seborrheic Dermatitis and Eczema, spine and forearm osteoporosis, hip osteopenia |
| P3a | 12 | M | <1 | Pneumonia and recurrent AOM, CMC, intestinal and lymph node TB, histoplasmosis of sinuses and oropharynx, ADD, Sepsis, Herpes Zoster, suppurative Piodermitis, Impetigo, Onychomycosis, Conjunctivitis | Pneumatocele, Chronic Eczema, Atopic Dermatitis, Inflammatory Bowel Disease: Gluten Intolerance, Column Osteopenia |
| P4 | 22 | M | 6 | Pneumonia and recurrent AOM, Pulmonary Aspergillosis, ADD, CMC, tonsillitis and recurrent Skin Abscesses, Septic Arthritis | Eczema, Column Osteoporosis, femoral, neck and forearm Osteopenia |
| P5 | 13 | M | <1 | Pneumonia and recurrent AOM, CMC, Herpes Simplex Oral, Skin Abscesses, Blepharitis, Periorbital Cellulitis, Conjunctivitis | Isolated tonic–clonic Seizure Atopic dermatitis, Cryptorchidism, Eosinophilic Folliculitis, Craniosynostosis, Column Osteopenia |
| P6 | 14 | M | <1 | Pneumonia and recurrent AOM, CMC, Pyoderma, Pemphigus, staphylococcal Impetigo, ADD, septic Arthritis, Osteomyelitis, Periodontitis, Onychomycosis | Atopic Dermatitis, primary teeth Retention, normal Bone Densitometry at 14 years of age |
| P7 | 13 | M | 6 | Pneumonia and recurrent AOM, Staphylococcal Impetigo, CMC, Molluscum Contagiosum | Bronchiectasis, Severe Eczema, Osteopenia and Scoliosis |
F: female, M: male, ADD: acute diarrhoeal disease, AOM: acute otitis media, TB: tuberculosis, CMC: chronic mucocutaneous candidiasis.
Laboratory findings of the HIES patients included in this study.
| Patient | IgGa | IgAa | IgMa | IgEb,c | Eosinophilsc | Genetic analysisd | Domain affected | |
|---|---|---|---|---|---|---|---|---|
| % | Absolute number | |||||||
| P1 | 1510 | 101 | 171 | 13,284 | 30 | 9270 | c.1110-2A>G, intron 11 | DNA binding domain |
| P2 | 2401 | 179 | 298 | 24,202 | 24 | 2556 | c.1144C>T, Exon 13, p.R382W | DNA binding domain |
| P3 | 1878 | 202 | 249 | 11,598 | 14 | 2422 | c.1144C>T, Exon 13, p.R382W | DNA binding domain |
| P4 | 1778 | 211 | 169 | 2000 | 8 | 563 | c.1909GA, Exon 21, p.V637M | SH2 domain |
| P5 | 1856 | 127 | 296 | 615 | 2 | 208 | c.1909G>A, Exon 21, p.V637M | SH2 domain |
| P6 | 1560 | 115 | 182 | 4000 | 18 | 4320 | c.2003C>T, Exon 21, p.S668F | SH2 domain |
| P7 | 1160 | 66 | 216 | 1357 | 2 | 372 | No STAT3 mutation detected | – |
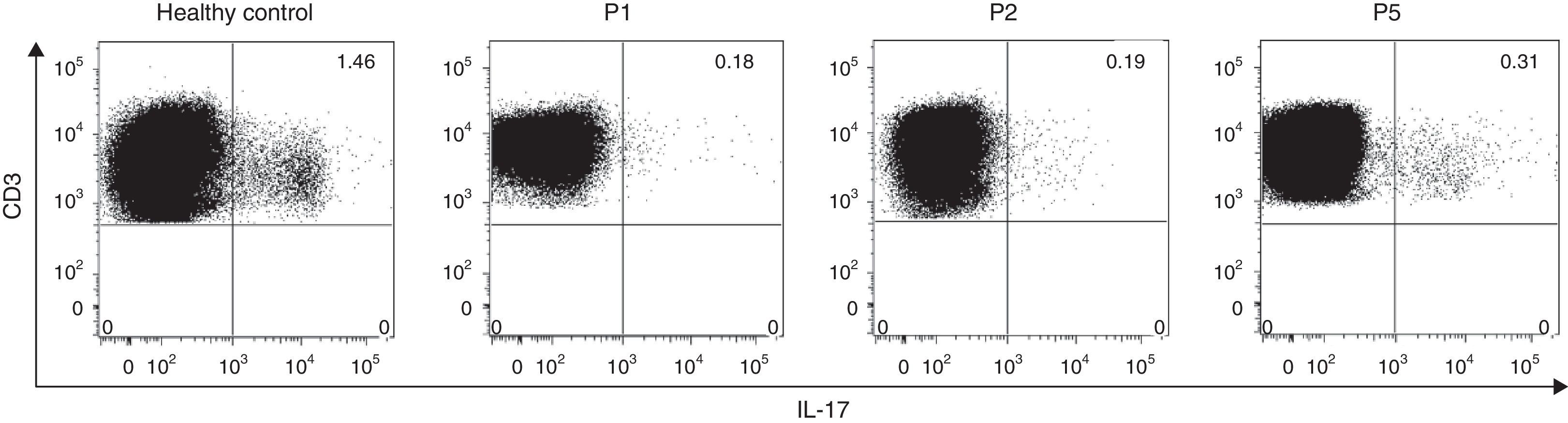
Since several studies have described that the Th17-mediated cytokine response is regulated by STAT3 and in addition, mutations in this gene impair the IL-17A, IL-17F and IL-22 production, we aimed to evaluate the intracelullar production of IL-17 post-PMA/ionomycin stimulation in three HIES patients and a healthy control. Flow cytometry analysis revealed decreased IL-17 producing-lymphocytes regardless of the type of STAT3 mutations in all HIES patients evaluated (Fig. 1). Interestingly, P5, who presented with mutations in the STAT3 SH2-domain exhibited 1.6 times more IL-17-producing cells approximately, than P1 and P2, with mutations in the DNA-binding domain (Fig. 1). These finding confirmed the defective STAT3 signalling pathways from our patients.
IL-17 intracellular expression in CD3+ T cells. The intracellular expression of IL-17 in CD3+ cells was evaluated by flow cytometry in a healthy control and three HIES patients from our cohort. PBMC were stimulated with PMA+ionomycin in the presence of the protein transport inhibitor Brefeldin A as indicated in materials and methods. Numbers at the upper right quadrant represent the percentages of IL-17-producing CD3+ cells.
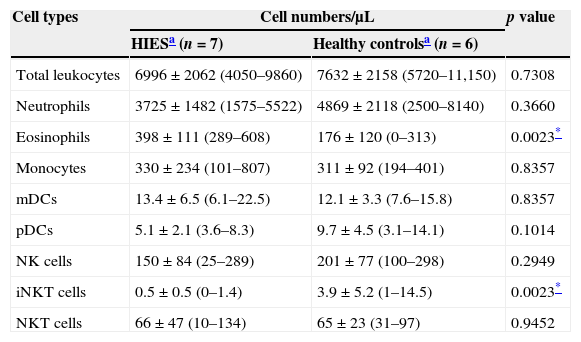
The absolute number and frequency of innate immune cells was evaluated in PB of HIES patients and healthy controls. No differences were observed in the absolute number of total leucocytes, neutrophils, monocytes, mDCs, pDCs, NK and NK T cells in PB from HIES patients and healthy controls (Table 3). As expected, a significant increase in the number of circulating eosinophils was observed in HIES in comparison with healthy controls, although three out of the seven patients included exhibited normal blood eosinophil counts (<350cells/μL; P4, P5 and P7)21 and the other four demonstrated only mild blood eosinophilia (351–1500cells/μL) at the time of sample collection. For the last four patients, blood eosinophil counts at this time point were much lower than the maximum numbers reported in their clinical records (Table 2). All HIES patients exhibited also a significant decrease in the blood iNKT cell numbers in comparison to those observed in healthy controls.
Quantitative evaluation of innate immune cells in peripheral blood from patients with HIES and healthy controls.
| Cell types | Cell numbers/μL | p value | |
|---|---|---|---|
| HIESa (n=7) | Healthy controlsa (n=6) | ||
| Total leukocytes | 6996±2062 (4050–9860) | 7632±2158 (5720–11,150) | 0.7308 |
| Neutrophils | 3725±1482 (1575–5522) | 4869±2118 (2500–8140) | 0.3660 |
| Eosinophils | 398±111 (289–608) | 176±120 (0–313) | 0.0023* |
| Monocytes | 330±234 (101–807) | 311±92 (194–401) | 0.8357 |
| mDCs | 13.4±6.5 (6.1–22.5) | 12.1±3.3 (7.6–15.8) | 0.8357 |
| pDCs | 5.1±2.1 (3.6–8.3) | 9.7±4.5 (3.1–14.1) | 0.1014 |
| NK cells | 150±84 (25–289) | 201±77 (100–298) | 0.2949 |
| iNKT cells | 0.5±0.5 (0–1.4) | 3.9±5.2 (1–14.5) | 0.0023* |
| NKT cells | 66±47 (10–134) | 65±23 (31–97) | 0.9452 |
mDCs, myeloid dendritic cells; pDCs, plasmacytoid dendritic cells; NK, natural killer cells; iNKT, invariant natural killer T cells.
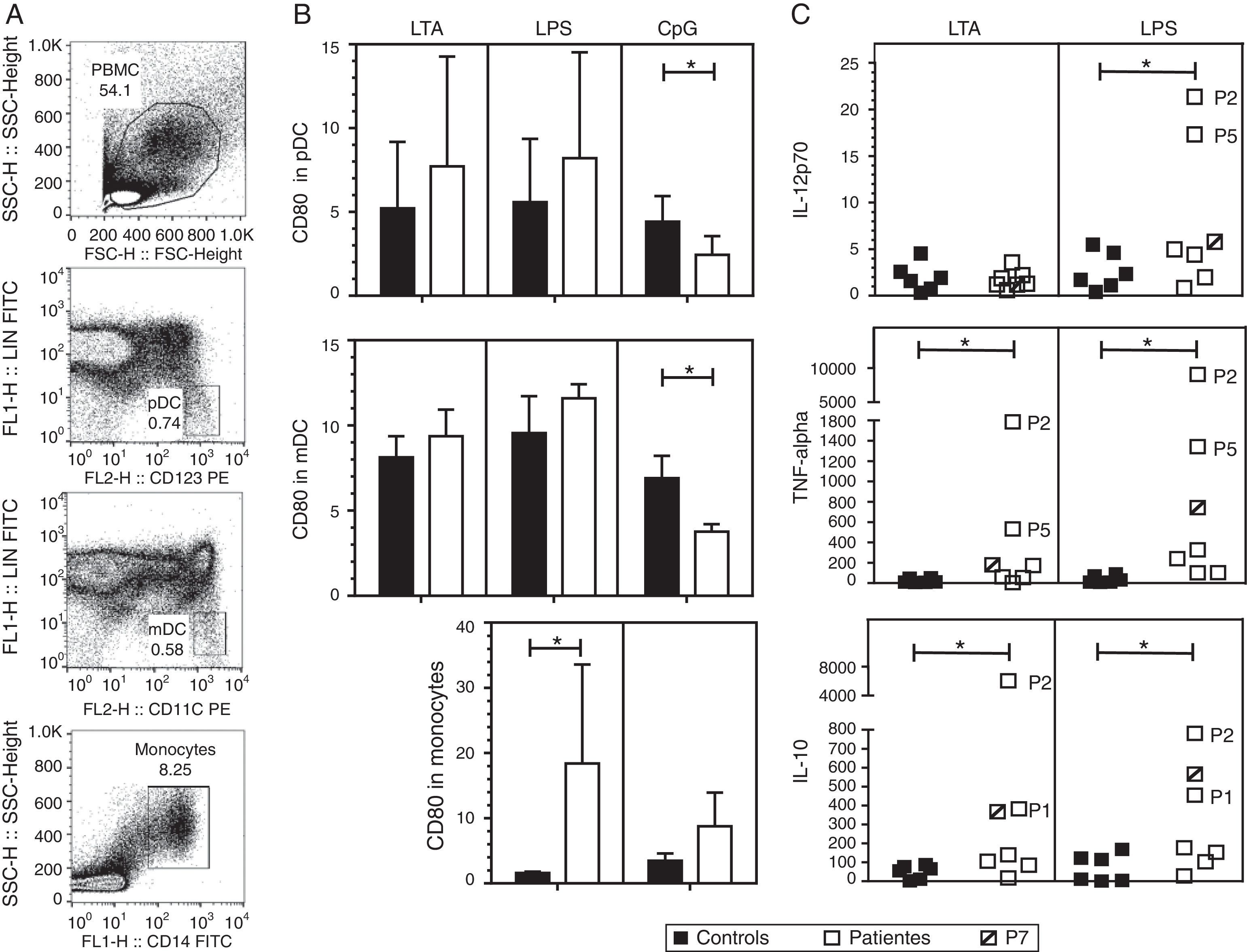
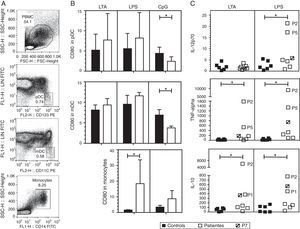
We investigated the effect of different TLR agonists in the expression of CD80 and CD86 in pDC, mDC and monocytes after PBMC stimulation with LTA, LPS or CpG. All these stimuli induced an increase in the expression of CD80 and CD86 on the surface of pDCs and mDCs from both patients and controls. Surface CD86 expression did not exhibit a significant difference in cells from HIES patients as compared with healthy controls (data not shown). As shown in Fig. 2B, after LTA and LPS stimulation, the expression levels of CD80 on pDC were similar in HIES patients and controls but a significantly lower increase in CpG-stimulated pDC from HIES PBMC was observed. Similar results were observed in PBMC-derived mDC from HIES patients (Fig. 2B).
CD80 expression and secretion of cytokines by TLR agonist-stimulated PBMC. PBMC from healthy controls (black bars and shapes, n=6) and HIES patients (white bars and shapes, n=7) included in this study were incubated w/o LTA, LPS or A-class CpG ODN 2216 for 24h. Thereafter, cells were collected and stained with specific fluorescent antibodies to evaluate the expression of CD80 on pDC, mDC and monocytes by flow cytometry. Also cytokine levels were evaluated in the supernatants by BD® CBA. Shown is the gating strategy of the different cell types (A), the fold increase in the CD80 MFI±SD (B) and in the cytokine levels (C) when comparing stimulated and non-stimulated cells in picograms/mL. The patient with no STAT3 mutation is denoted. *p<0.05.
In LPS-stimulated monocytes, CD80 expression was higher in HIES patients in comparison with healthy controls. The same effect to a lesser extent was observed in CpG-stimulated monocytes; however, these differences were not statistically significant.
Increased secretion of IL-12p70, TNF-alpha and IL-10 by TLR agonist-stimulated PBMC from HIES patientsThe capacity of PBMC from HIES patients to secrete different inflammatory cytokines following stimulation with TLR agonist was evaluated. After stimulation with LTA and LPS, measurable levels of IL-12, TNF-alpha, IL-10, IL-6, IL-8, IL-1beta and IFN-alpha were obtained; however, significant differences between patients and controls were observed only for the cytokines depicted in Fig. 2C. Additionally, as CpG was used to stimulate PBMC, secretion of the different cytokines evaluated was induced in supernatants, however no differences were observed in the levels from patients and controls cells (data not shown).
As mentioned, albeit LTA induced IL-12 production in PBMC, no differences were observed in controls and HIES patients’ cells. However, a significant increase in IL-12 secretion was observed in cells from HIES patients as compared to healthy controls after LPS stimulation. Likewise, TNF-alpha and IL-10 secretion was significantly higher in LTA and LPS-stimulated cells from HIES patients when compared to healthy controls. Notably, these differences were mainly due to the higher secretion of cytokines shown by P2 and P5 cells (Fig. 2C) but this was related neither with the STAT3 mutation detected in these patients nor with the clinical phenotype documented (see Tables 1 and 2).
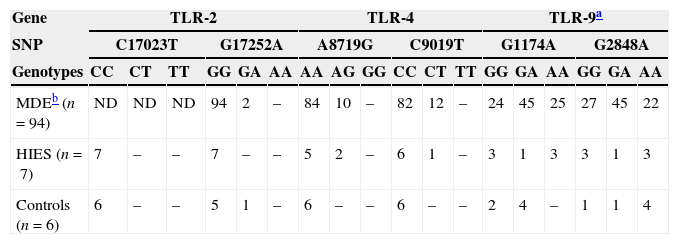
Analysis of polymorphisms in TLR2, TLR4, and TLR9 genesWe also investigated the presence of several previously reported SNPs in the TLR2, TLR4, and TLR9 genes in HIES patients and healthy controls (Table 4). Within the TLR2 gene, we did not observe the presence of the C17023T SNP in any of the individuals studied. A heterozygous individual for G17252A was found in the healthy control group. Regarding the TLR4 gene, two heterozygous HIES patients for A8719G and one for C9019T were found. Of note, these two SNPs were not present in our healthy controls and they were shown to alter the LPS responsiveness in vitro also at the heterozygous state.15 Finally, our work showed that the two TLR9 SNPs were also highly frequent in our study groups. Of the healthy controls analysed, four were heterozygous for G1174A and one for the G2848A SNP. In addition, we found four controls homozygous for G2848A. Within the HIES group, we found one individual heterozygous and three homozygous for G1174A. The same genotype counts were obtained for the G2848A SNP. All these data agree with the genotype counts reported for the Medellin, Colombian subpopulation at the 1000 genomes project (http://browser.1000genomes.org).
Genotype counts of single nucleotide polymorphisms (SNPs) within the TLR2, TLR4, and TLR9 genes in individuals from Medellin, HIES patients and healthy controls.
In the present study, we reported the clinical, immunological features and STAT3 mutation analyses of seven HIES cases. Moreover, we quantified PB innate immune cells in these patients and evaluated the immune response of the patients’ PBMC to different TLR agonists. The presence of several TLR SNPs in genomic DNA from our study group was also investigated. All patients exhibited the clinical features suggestive of AD-HIES and 6/7 reported mutations at the STAT3 gene. A significant decrease in the PB iNKT cells was observed in our HIES group. Also, a decrease in the IL-17 intracellular production was observed in stimulated PBMC from the patients available for these analyses. CpG-stimulated dendritic cells exhibited a lower increase in the CD80 expression and additionally, LTA and LPS-stimulated PBMC's supernatants in the patients showed increased TNF-alpha and IL-10.
All patients included here presented with PB innate cell counts comparable to those observed in healthy controls, except iNKT cells that were significantly decreased. Of interest, PB iNKT cells are reduced in allergic asthma, systemic lupus erythematosus (SLE), rheumatoid arthritis, multiple sclerosis and inflammatory bowel disease.11,22–25 Decreased iNKT cells in PB have been also observed during Mycobacterium tuberculosis and Varicella zoster virus infections.26,27 Moreover, our own work has demonstrated reduced iNKT cells in PB from patients with Common Variable Immunodeficiency.28 However, decreased PB iNKT cells may result from their recruitment to the inflammation sites. Jyonouchi et al. found an association between the reduction of iNKT cells in PB and its increase in oesophageal biopsies from patients with eosinophilic oesophagitis.29 A similar relationship is observed in SLE, since increased iNKT cells infiltration is observed in sites of marked inflammation from patients’ skin biopsies. This result correlates with the lower percentages of circulating CD3+6B11+ PB iNKT cells observed.30
In our study, we also observed a highly variable expression of the costimulatory molecule CD80 after stimulation of innate immune cells with LTA and LPS, especially in pDC and monocytes. The heterogeneity in the TLR response may be due to the small number of patients and controls included. However, no statistical differences were observed among the study groups and these data agree with previous studies that have reported no defective expression of CD80 in immune cells from HIES patients after different stimuli.10,31,32 Of note, stimulation of pDCs and mDCs from PBMC with CpG resulted in a lower increase in the surface expression of CD80 in HIES patients as compared with those levels in healthy controls’ cells. More studies are necessary to elucidate the role of STAT3 in the expression of this costimulatory molecule in DCs, considering that previous evidence indicates that STAT3 participates in the intracellular CpG signalling pathways in B cells.33
Additionally, we revealed an increase in the production of IL-12p70, TNF-alpha and IL-10 in some patients’ PBMC after stimulation with LTA or LPS. These findings agree with several publications that demonstrate higher production of these cytokines by whole blood cells, PBMC and monocytes after stimulation with these TLR ligands.4,8,9,32,34 Interestingly, Giacomelli et al. demonstrate that HIES monocytes are hyporesponsive to the anti-inflammatory effect of IL-10 as they exhibit a failure in the suppression of several pro-inflammatory cytokine production when cultured in the presence of IL-10.8 Saito et al. also reported a deficient IL-10-mediated induction of tolerogenic DC and regulatory T cells in HIES patients.10 Thus, we hypothesise that pro-inflammatory cytokines are elevated in supernatants from our HIES patients’ cells due to a failure in the IL-10 regulation.
More studies are necessary to ascertain if iNKT cells are not only quantitatively but also functionally affected in HIES and to characterise the role of iNKT cells in tissues under inflammatory conditions. The role of STAT3 not only in the iNKT cell generation but also in the different innate cells TLR signalling pathways should also be investigated.
Ethical disclosuresConfidentiality of dataThe authors declare that no patient data appear in this article.
Right to privacy and informed consentThe authors have obtained the informed consent of the patients and/or subjects mentioned in the article. The author for correspondence is in possession of this document.
Protection of human subjects and animals in researchThe authors declare that the procedures followed were in accordance with the regulations of the responsible Clinical Research Ethics Committee and in accordance with those of the World Medical Association and the Helsinki Declaration.
Conflict of interestThe authors have no conflict of interest to declare.
This work was supported by the Colombian Institute for Development of Science and Technology-Colciencias-Grant 111551929012.