Atopic dermatitis is an inflammatory skin disease in which both genetic and environmental factors interact to determine the susceptibility and severity of the disease.
ObjectiveThe aim of this study was to determine the association between atopic dermatitis and IL-10 and TGF-β1 gene polymorphisms.
MethodsThe allele and genotype frequencies of genes encoding for IL-10 and TGF-β1 were investigated in 89 patients with atopic dermatitis in comparison with 138 in the control group using the PCR-SSP method.
ResultsA significant increase was found in the frequency of the TGF-β1 codon 10/C allele among patients (p<0.001, OR=6.77), whereas a significant decrease was observed in the frequency of the T allele at the same position (p<0.001, OR=0.14). The frequency of the TGF-β1 codon 25/G allele in the control group was significantly higher than among patients (p<0.001, OR=0.08). A significant positive correlation was seen between CC (p<0.001, OR=15.10) and CG (p<0.001) genotypes and AD at codons 10 and 25, respectively. The most frequent haplotypes among patients was TGF-β1 CG which was significantly higher than in the control subjects (50% in patients vs. 39.9% in controls, p=0.042). A significant increase was found in the frequency of TGF-β CC (36% in patients vs. 7.6% in controls, p<0.001) and TC (14% in patients vs. 0% in controls, p<0.001) haplotypes among patients compared to controls. By contrast, the TGF-β1 TG haplotype was significantly lower in patients than controls (0% in patients vs. 52.5% in controls, p<0.001). There were no significant differences in the frequency of alleles, genotypes and haplotypes of the IL-10 gene.
ConclusionsWe found a strong association between the polymorphisms of the TGF-β1 gene at codon 10 and codon 25 positions and atopic dermatitis.
Atopic dermatitis (AD), the inflammatory chronic relapsing condition of the skin, has a strong genetic background with interference of environmental factors. The concordance rate for AD is 77% and 15% in monozygotic and dizygotic twins, respectively. It has been hypothesized that AD may be attributed to the immune dysregulations which occur within the skin. The imbalance between cytokines of T helper (Th)-1 and Th-2 cells has an important role in the pathophysiology of AD. Genes encoding these cytokines have been identified, using candidate gene studies. Several studies established a linkage between single nucleotide polymorphisms (SNPs) within cytokine genes and the pathogenesis of AD.1
IL-10 and TGF-β1 are cytokines which act as regulators in the different aspects of immune functions. They inhibit both Th1 and Th2 cytokines. It has been implicated that polymorphisms in the TGF-β1 and IL-10 genes have been associated with differences in in vitro cytokine production. Several polymorphic sites within the promoter region of the IL-10 gene have been identified, associated with the development of cancers, inflammatory and infectious diseases, including AD. Previous studies demonstrated a significant association between IL-10, TGF-β1 gene polymorphisms and a number of immunological diseases.2–14 SNPs of TGF-β1 gene have also been described to affect TGF-β1 production in AD patients.15
This study was performed to investigate the SNPs within the TGF-β1 and IL-10 genes among Iranian patients with AD. Such investigations have been done among patients from different ethnic backgrounds, with conflicting results.
Patients and methodsSubjectsEighty-nine children with AD who were referred to the Immunology Clinic of the Children's Medical Center, affiliated to Tehran University of Medical Sciences (Tehran, Iran) were included in the study. The diagnosis of AD was made based on the standard criteria of Hanifin and Rajka.16 One hundred and thirty-eight, unrelated, healthy subjects with no history of atopy were also selected as the control group. This project was approved by the ethical committee of Tehran University of Medical Sciences. Written informed consent was obtained from all participants prior to blood sampling.
GenotypingCytokine genotyping was performed on DNA samples using polymerase chain reaction with sequence-specific primers (PCR-SSP assay Kit, Heidelberg University, Germany). Briefly, gene amplification was performed using Tedane Flexigene thermal cycler (Rosche, Cambridge, UK). The frequencies of alleles, genotypes and haplotypes of IL-10 (−1092 A/G, −592 A/C and −819 T/C) and TGF-β1 genes (codon (cdn) 10 C/T and cdn 25 G/C) were counted.
Statistical analysisSPSS statistical software was used for data analysis. Allele frequencies were determined by direct gene counting. Frequencies of alleles, genotypes and haplotypes were compared between patients and controls, using chi-square test. The odds ratios were calculated with 95% confidence intervals. A probability of less than 0.05 was considered statistically significant.
ResultsPatients’ characteristicsA total of 89 patients with AD (52 males and 37 females) were enrolled. 58.4% and 41.6% of subjects had moderate and severe disease, respectively. All subjects were older than six months of age. Almost 90% of patients had a family history of atopy. Skin prick skin test was positive in 67.4% of patients (to at least one allergen). Mean eosinophil count was 269/mm3, and median total serum IgE was 33IU/mL.
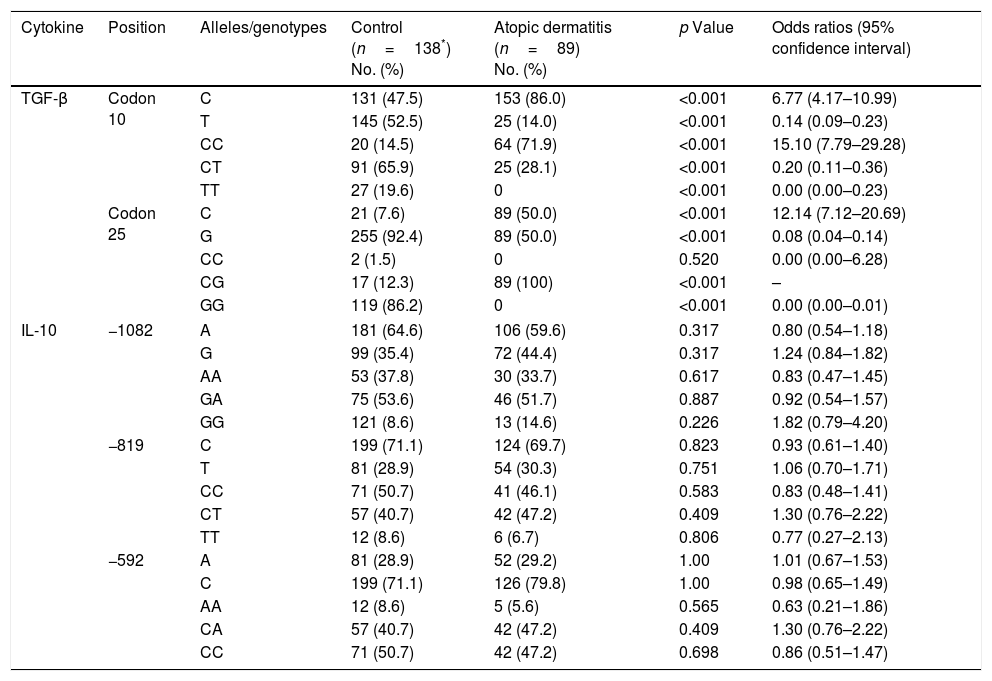
Alleles and genotypes frequenciesThe results of allele and genotype frequencies are presented in Table 1. A significant increase was found in the frequency of the TGF-β1 cdn 10/C allele among patients (p<0.001, OR=6.77), whereas a significant decrease was observed in the frequency of the T allele at the same position (p<0.001, OR=0.14). The frequency of the TGF-β1 cdn 25/G allele in the control group was significantly higher than among patients (p<0.001, OR=0.08). A significant positive correlation was seen between CC (p<0.001, OR=15.10) and CG (p<0.001) genotypes and AD at cdn 10 and 25, respectively. There were no statistically significant differences in the frequency of the alleles and genotypes of IL-10 gene.
IL-10 and TGF-β1 gene polymorphisms in patients with atopic dermatitis and controls.
| Cytokine | Position | Alleles/genotypes | Control (n=138*) No. (%) | Atopic dermatitis (n=89) No. (%) | p Value | Odds ratios (95% confidence interval) |
|---|---|---|---|---|---|---|
| TGF-β | Codon 10 | C | 131 (47.5) | 153 (86.0) | <0.001 | 6.77 (4.17–10.99) |
| T | 145 (52.5) | 25 (14.0) | <0.001 | 0.14 (0.09–0.23) | ||
| CC | 20 (14.5) | 64 (71.9) | <0.001 | 15.10 (7.79–29.28) | ||
| CT | 91 (65.9) | 25 (28.1) | <0.001 | 0.20 (0.11–0.36) | ||
| TT | 27 (19.6) | 0 | <0.001 | 0.00 (0.00–0.23) | ||
| Codon 25 | C | 21 (7.6) | 89 (50.0) | <0.001 | 12.14 (7.12–20.69) | |
| G | 255 (92.4) | 89 (50.0) | <0.001 | 0.08 (0.04–0.14) | ||
| CC | 2 (1.5) | 0 | 0.520 | 0.00 (0.00–6.28) | ||
| CG | 17 (12.3) | 89 (100) | <0.001 | – | ||
| GG | 119 (86.2) | 0 | <0.001 | 0.00 (0.00–0.01) | ||
| IL-10 | −1082 | A | 181 (64.6) | 106 (59.6) | 0.317 | 0.80 (0.54–1.18) |
| G | 99 (35.4) | 72 (44.4) | 0.317 | 1.24 (0.84–1.82) | ||
| AA | 53 (37.8) | 30 (33.7) | 0.617 | 0.83 (0.47–1.45) | ||
| GA | 75 (53.6) | 46 (51.7) | 0.887 | 0.92 (0.54–1.57) | ||
| GG | 121 (8.6) | 13 (14.6) | 0.226 | 1.82 (0.79–4.20) | ||
| −819 | C | 199 (71.1) | 124 (69.7) | 0.823 | 0.93 (0.61–1.40) | |
| T | 81 (28.9) | 54 (30.3) | 0.751 | 1.06 (0.70–1.71) | ||
| CC | 71 (50.7) | 41 (46.1) | 0.583 | 0.83 (0.48–1.41) | ||
| CT | 57 (40.7) | 42 (47.2) | 0.409 | 1.30 (0.76–2.22) | ||
| TT | 12 (8.6) | 6 (6.7) | 0.806 | 0.77 (0.27–2.13) | ||
| −592 | A | 81 (28.9) | 52 (29.2) | 1.00 | 1.01 (0.67–1.53) | |
| C | 199 (71.1) | 126 (79.8) | 1.00 | 0.98 (0.65–1.49) | ||
| AA | 12 (8.6) | 5 (5.6) | 0.565 | 0.63 (0.21–1.86) | ||
| CA | 57 (40.7) | 42 (47.2) | 0.409 | 1.30 (0.76–2.22) | ||
| CC | 71 (50.7) | 42 (47.2) | 0.698 | 0.86 (0.51–1.47) | ||
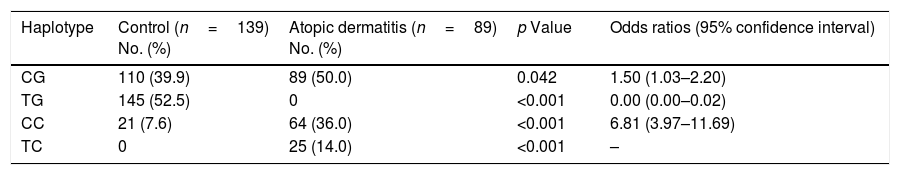
Haplotype frequencies are shown in Table 2. The most frequent haplotype in our patients was TGF-β1 CG, which was significantly higher than in the control subjects (50% in patients vs. 39.9% in controls, p=0.042). Moreover, we found a significant increase in the frequency of TGF-β1 CC (36% in patients vs. 7.6% in controls, p<0.001) and TC (14% in patients vs. 0% in controls, p<0.001) haplotypes in patients compared to controls. By contrast, the TGF-β1 TG haplotype was significantly lower in patients than controls (0% in patients vs. 52.5% in controls, p<0.001). In the case of IL-10, no significant differences were found in the frequency of haplotypes (data not shown).
TGF-β1 haplotype polymorphisms in patients with atopic dermatitis and controls.
| Haplotype | Control (n=139) No. (%) | Atopic dermatitis (n=89) No. (%) | p Value | Odds ratios (95% confidence interval) |
|---|---|---|---|---|
| CG | 110 (39.9) | 89 (50.0) | 0.042 | 1.50 (1.03–2.20) |
| TG | 145 (52.5) | 0 | <0.001 | 0.00 (0.00–0.02) |
| CC | 21 (7.6) | 64 (36.0) | <0.001 | 6.81 (3.97–11.69) |
| TC | 0 | 25 (14.0) | <0.001 | – |
TGF-β1 is a pleiotropic cytokine, produced by most cell types, which suppresses the production of pro-inflammatory cytokines and is an inhibitor of in vitro and in vivo T cell-mediated immune responses. In an animal model of asthma, TGF-β1 reduced airway inflammation.17 Lee et al.18 demonstrated that peripheral blood leukocytes of patients with AD express reduced levels of TGF-β1 mRNA. It has been shown that abnormal Smad (signal transducers of TGF-β1) 3 signaling causes reduced inflammation in the murine model of AD.19 Gambichler et al.20 observed significantly higher Smad 3/4 in healthy controls than in AD patients, while no significant difference was found regarding TGF-β1 mRNA expression. Moreover, Peng et al.21 showed that monocytes from AD patients expressed low response to TGF-β1 in an experimental study.
Associations between SNPs of the TGF-β1 gene and AD have not been well documented. Our data showed that the TGF-β1 cdn 10/C allele and CC genotype were significantly over-expressed in patients compared to controls, while the frequency of the T allele and TT and CT genotypes were significantly lower in patients than in controls, indicating their protective role against AD. Previous studies have not shown such associations. At TGF-β1 cdn 25, we found a positive association between C allele and AD. Although the frequency of this allele is very low in the normal population, half of our patients had C allele at this position. We also found a protective role of the G allele and GG genotype at cdn 25 position against AD. These results were compatible with Stavric et al.’s study.22 Similarly, Arkwright et al.15 showed that substitution of G to C at this position is accompanied with a higher risk of AD. Given that the presence of C allele is associated with the low producer TGF-β1 genotype,15 we could expect a low level of TGF-β1 in our patients. Regarding Haplotype frequencies, CG, CC and TC haplotypes were shown to be susceptible, whereas TG Haplotype was protective against AD. Consistently, Stavric et al. found positive association for CC and negative association for TG haplotypes.
IL-10 is an anti-inflammatory cytokine produced by different cell types including regulatory T cells, macrophages and dendritic cells.23 It down-regulates anti-microbial peptides (AMPs) expression, increasing the tendency toward bacterial skin infection among AD patient.24 It has been shown that IL-10 enhanced the B-cell proliferation and IgE synthesis under the combined stimuli of IL-4 and CD40.25 Previous studies have found increased levels of IL-10 in peripheral blood mononuclear cells and lesional skin of AD patients; however it has also been shown that the serum level of IL-10 is conversely associated with severity of AD.23 IL-10 also stimulates the mast cell growth by the combination of IL-4 and IL-3.26
Several SNPs within the IL-10 gene have been described to correlate with IL-10 serum levels and differences in nuclear-binding activity.27 Certain haplotypes were also identified which associated with increased levels of IL-10, IgE production and eosinophil count.28 Data regarding the association between SNPs within the IL-10 promoter region and AD were inconsistent. Arkwright et al. and Reich et al. found no correlations.15,29 Sohn et al.30 found genotypic association between AD and IL-10 promoter SNPs. Shin et al.31 showed that haplotype polymorphisms in IL-10 distal promoter (−1082A, −819C, −592C, +117T) were associated with low IgE levels. Moreover, Stavric et al.22 and Kayserova et al.32 observed associations between AD and IL-10 gene polymorphisms. The present study failed to show a significant association between SNPs of IL-10 gene at −1082A/G, −819C/T and −592C/A positions at the level of allele, genotype and Haplotype frequencies and AD. These results indicate that there may be other polymorphisms within the IL-10 gene, interfering in the development of AD. Ethnical differences may also contribute to these inconsistencies. Altogether, this study is the first performed on Iranian patients with AD, proving association between TGF-β1 cdn 10 polymorphisms and AD. Significant differences were seen at TGF-β1 cdn 10 C/T alleles and CC, CT and TT genotypes and TGF-β1 cdn 25 C/G alleles and CG and GG genotypes. In addition, the differences in the frequency of TG, CC and TC haplotypes between AD patients and controls were statistically significant. Nonetheless, further investigations should be conducted to elucidate the association of these SNPs and AD.
Conflict of interestThe authors have no conflict of interest to declare.
Ethical disclosuresConfidentiality of dataThe authors declare that they have followed the protocols of their work center on the publication of patient data and that all the patients included in the study have received sufficient information and have given their informed consent in writing to participate in that study.
Right to privacy and informed consentThe authors have obtained the informed consent of the patients and/or subjects mentioned in the article. The author for correspondence is in possession of this document.
Protection of human subjects and animals in researchThe authors declare that the procedures followed were in accordance with the regulations of the responsible Clinical Research Ethics Committee and in accordance with those of the World Medical Association and the Helsinki Declaration.
This study was supported by a grant from Tehran University of Medical Sciences and Health Services (89-04-80-12136).