The prevalence of HCV infection is very diversified according to geographical areas and ranges from 1% in the Northern regions of the world to more than 20% as we move South. Due to the presence of HCV-associated liver diseases and the development of effective treatments, the diagnosis of HCV infection is a growing medical need. Several tests are available, from simple screening to identify the presence of anti-HCV antibodies to the more sophisticated quantification of viral load and genotyping. However, these tests are to be used in a logical, consequential and cost-effective manner. This review article will report on the protocol in use in the North-Eastern part of Italy for the screening and diagnosis of HCV infection. The protocol is based on a consensus among several experts and may be the basis for a more rational approach in this rapidly growing field.
In recent years, considerable advances have been made in diagnostic testing for hepatitis C virus (HCV). Tests for antibodies to HCV (anti-HCV) have improved in sensitivity and specificity, providing rapid and inexpensive means to identify the subjects who have been infected. Well standardized qualitative and quantitative tests for HCV RNA are available. Qualitative tests reveal the presence of viremia and have become the gold standard for monitoring a successful antiviral therapy. The World Health Organization has recently established an international standard for HCV RNA quantification and all commercial HCV RNA quantitative assays now use the IU, which should be preferred to the old units in reporting results. Quantitative assays and HCV genotyping are useful to tailor treatment to individual patient and to determine its effectiveness. An enzyme immunoassay has been recently developed for the detection and the quantitation of core antigen as an alternative to qualitative and quantitative RNA detection, at least in particular conditions.
This article reviews the currently available laboratory tests for diagnosis and management of HCV infected patients and suggests their better use in clinical practice. This is the result of the work of a panel of experts of Friuli-Venezia Giulia Region (North-East of Italy) that produced a consensus document to be adopted by the regional sanitary Authorities and followed in the clinical setting. A correct use and interpretation of sero-virological assays avoids unnecessary tests, thus reducing the cost for the diagnosis and management of HCV-infected patients and improving the clinical outcome.
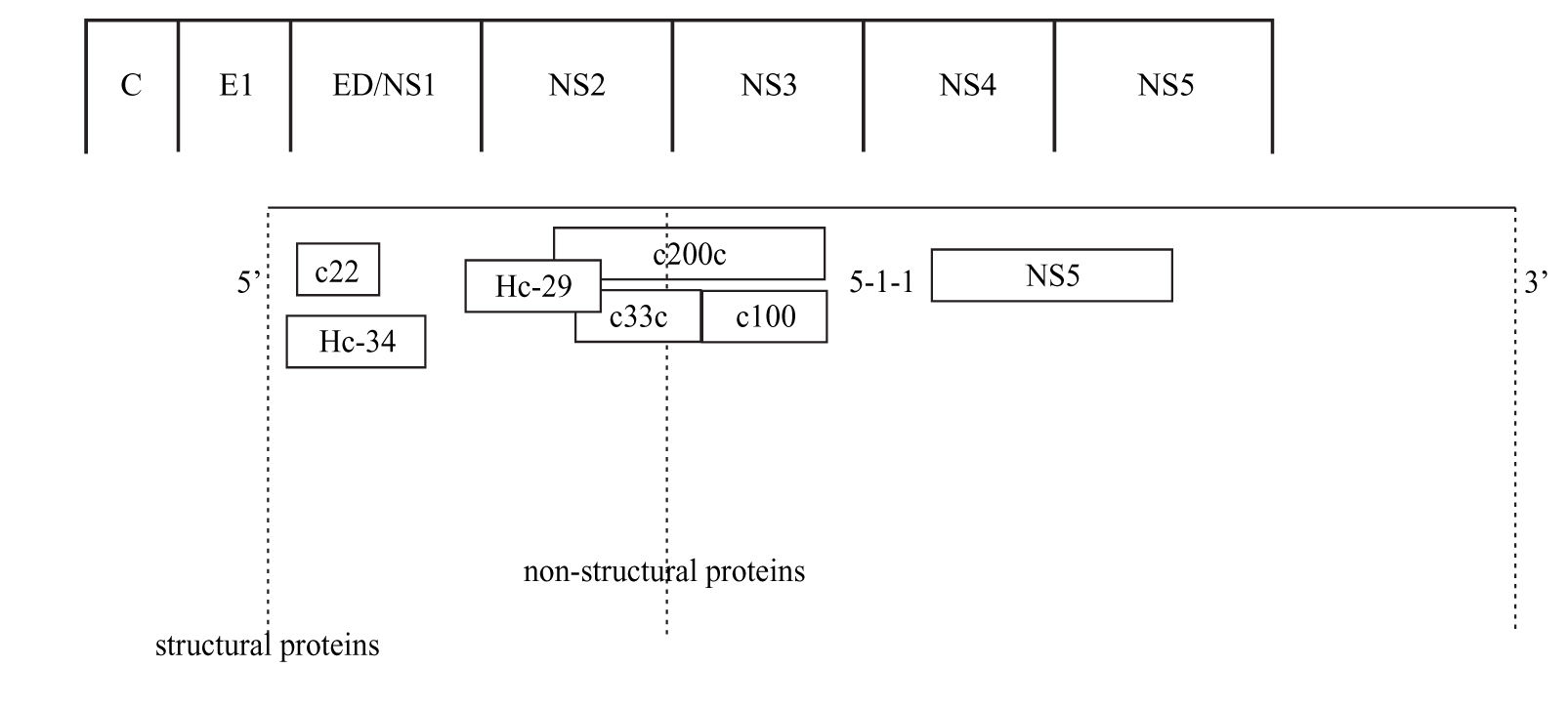
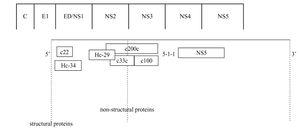
Anti-HCV antibody detectionScreening assayThe detection of anti HCV antibodies in plasma or serum is based on the use of enzyme immunoassays (EIAs). Advantages of this technique include automation ease, highly reproducible results, and low costs. The first-generation EIAs contained a single recombinant antigen (c100-3) from nonstructural 4 region (NS4) (Figure 1). Even though these assays were an important step in clarifying the diagnosis of most patients with non-A, non-B hepatitis and in blood donors screening, it became soon apparent that the method had to be improved in terms of sensitivity and specificity.
The second-generation EIAs were implemented by the introduction of another non-structural protein (NS3) and the core protein. These tests were more sensitive and specific than the first-generation EIAs, further reducing the risk of post-transfusion hepatitis C from one hand, and false-positive results among blood donors from the other.1 These tests also proved to be quite effective in the screening of HCV infection, particularly in high-risk subjects. Approximately 92-95% of patients who allegedly have HCV infection can be detected by using the second-generation EIAs.1 The tests shorten the window period between HCV infection and the detection of specific anti-bodies to approximately 10 weeks, compared with an average of 16 weeks with the first-generation EIAs.1
The third-generation EIAs that were subsequently introduced, contained also antibodies to the NS5 protein. The increase in sensitivity is ascribed to a reconfiguration of core and NS3 antigens rather than to the addition of NS5 antigen, responsible of frequent false-positive results in low prevalence populations.2 The sensitivity of these assays was estimated to range from 98.8% to 100%3,4 in immunocompetent subjects, showing that most immuno-competent subjects with active or past HCV infection can be identified by EIAs. In hemodialysis and in immunocompromised subjects the sensitivity of anti-HCV EIAs is lower, ranging from 50 to 95%, according to the depth of immunosuppression.5-9 False HCV EIAs negative results have also been reported in patients with HCV-associated mixed cryoglobulinemia, probably related to the concentration of anti-HCV antibodies within cryoglobulin complex.10 The third-generation EIAs have shortened the seroconversion time by 2-3 weeks11 and are now the most widely used screening test for HCV.12,13 They are suitable for screening at-risk populations and are recommended as initial test for patients with liver disease.14 The very high sensitivity and specificity prevent the need for confirmatory immunoblot assay in the diagnosis for patients with liver disease,15 while a negative EIA test is sufficient to exclude a chronic infection in immune-competent patients.14
Supplementary or confirmatory assaysSupplemental tests have been developed to establish the “true positivity” of anti-HCV EIA tests results. The most commonly used are the recombinant immunoblot assay (RIBA, Chiron Corporation, Emeryville, CA) and the line immunoassay (LIA, Innogenetics, Ghent, Belgium) which are modifications of Western blot technique. These tests use the same antigens contained in EIA tests in an immunoblot format and allow to identify antibodies against individual antigens. The result of the tests may be positive, indeterminate or negative, depending on type and the version of the assay and the criteria defined by the manufacturer. Although the specificity is higher, the sensitivity is lower than that of EIAs.1,16 The rational in accepting a test with a lower sensitivity to confirm another test with higher sensitivity was object of controversy. Accordingly, “supplemental” rather than “confirmatory” may be a better term.
There are two interpretative problems related with the use of confirmatory tests. First, an indeterminate or negative result after a sensitive EIA positive test: in this case, to discriminate the false positivity a molecular HCV RNA test and/or follow-up of the patients are necessary. Second, the predictivity of a positive or indeterminate RIBA test to HCV viremia in subjects with high or low prevalence of HCV infection, respectively. About 50% of the RIBA-3 positive blood donors are HCV RNA positive by RT-PCR assay,1 while a small number of indeterminate samples are found to be HCV RNA positive. The 85% of immunoblot positive samples and the 20-50% of indeterminate samples with core or NS3 reactivity are found to be HCV RNA positive in high HCV prevalence population.15 On the other hand, confirmed anti-HCV results are not a true indicator of an active HCV infection, since cleared patients may remain anti-HCV positive for years.17 For all these reasons, the use of a HCV RNA qualitative test to confirm the presence of an active HCV infection is more effective than any supplemental assay. Immunoblot tests could still be useful in blood screening for donation, a setting in which positive EIA results are poorly predictive of true HCV infection.18 Even their utility in blood donor screening decreased with systematic molecular testing for HCV RNA in the European Union and in the United States.19
Anti-HCV IgM assayThe significance of the presence of anti-HCV IgM antibodies in patients with HCV infection is still unclear. Anti-HCV IgM are found in 50%-93% of patients with acute hepatitis C, but also in 50-70% of patients with chronic hepatitis C.20-22 Therefore, anti-HCV IgM cannot be used as a reliable marker of acute HCV infection. In the evaluation of HCV vertical transmission, IgM positivity in the mother was found to be a prognostic factor of neonatal infection.23
HCV RNA detectionHepatitis C virus replicates at relatively low levels and viral genomes may be present in small amounts so that HCV RNA cannot be detected by classical hybridization-based techniques. As a result, a preliminary amplification step is necessary, which can be carried out using a molecular biology-based technique, namely target amplification. The purpose is to synthesize a large number of copies of viral genome (amplicons) in a cyclic enzymatic reaction, such as polymerase chain reaction (PCR) and transcription-mediated amplification (TMA).
Many variations in the qualitative HCV PCR assay have been described and standardization of in-house assays has been difficult.24-26 Several factors contribute to reverse-transcription PCR assay variability such as specimen handling and storage conditions,27 the presence of inhibitors, the design of the primers, the DNA product contamination and the efficiency in the detection of the amplification products. Initial PCR tests were found to be of a very low accuracy.24,26 Further experience and standardization of tests increased the number of laboratories obtaining accurate results and in a recent evaluation, a concordance of more that 90% was reported.1,28 The use of non-standardized “home-made” assays in clinical setting should be avoided and commercial assays, such as Amplicor HCV v2.0 (Roche Molecular System, Pleasanton, CA or its automated version Cobas Amplicor HCV v2.0; Roche Molecular System, Pleasanton, CA) have to be preferred both for the accuracy and comparative purposes.
The first generation Amplicor HCV had a manufacturer’s stated cut-off of 1,000 copies/mL, but the assay appeared to be slightly less sensitive for HCV genotype 2 or 3 than for genotype 1. In the second generation of the assay, the detection cut-off is of 50 international units (IU) of HCV RNA per mL and has an equal sensitivity for the detection of all genotypes. The specificity of the Amplicor HCV v2.0 appears to be of 97-99%.1 The commercial TMA-based assay is currently available by Bayer Corporation (Versant HCV RNA Qualitative Assay, Bayer Corporation, Diagnostic Division, Tarrytown, NY). It is fully manual at present and it has a lower detection limit of 10 UI/mL for all of the major HCV genotypes.29 The specificity of this assay exceeds 98%.14
Whichever test is used, while a single qualitative positive assay for HCV RNA confirms active viral replication, a single negative test does not exclude viremia and may reflect only a viral load below the detection limit of the assay. Therefore, a follow-up qualitative HCV RNA is required to exclude an active HCV replication. Once HCV infection is confirmed, the repeating qualitative assay does not help in managing untreated patients, except for determining whether an acute infection has resolved.14
Qualitative HCV RNA assays must still be used to assess the virological response to therapy, owing to its sensitivity. A sensitive qualitative HCV RNA assay is necessary at 24 weeks in patients infected with genotype 1 with indication of treatment for 48 weeks, since the probability of sustained virological response (SVR) is extremely low when HCV RNA is still detectable at week 24.30-32 HCV RNA negativity for all genotypes 24 weeks after stopping the treatment indicates a sustained virological response.
HCV RNA quantificationThe HCV RNA level can be quantified by means of target amplification technique (PCR) or signal amplification technique (“branched DNA” assay). In the target amplification techniques, the quantification is based on the competitive amplification of viral genome with a known amount of synthetic standard added to each reaction tube. The relative amount of viral template and standard amplicons are measured at the end of the procedure. The results can be read in a standard curve established in parallel. In the signal amplification techniques, the HCV RNA is captured in a microtiter well by hybridization to synthetic oligonucleotide probes, complementary in sequence to the 5’-non coding region and core of the HCV genome. Additional target probes bind the HCV RNA to branched DNA (bDNA) molecules, which are then amplified and labelled with a chemiluminescent probe.33 The quantification is based on a standard curve generated simultaneously using known standards.
Two commercial standardized assays were developed and widely employed in the recent years. Amplicor HCV Monitor v2.0 (Roche Molecular System) is a quantitative reverse-transcriptase PCR-based assay with a stated cutoff of 1,000 copies/mL. Versant HCV RNA 2.0 Assay (Bayer Corporation) is a bDNA-based signal amplification with a stated cut-off of 200,000 genome equivalents/mL. The measures as “copy” or “genome equivalent” do not represent the same amount of HCV RNA, because they were defined independently using quantified standards of different natures, lengths and sequences.34 The World Health Organization has defined an international standard for HCV RNA quantification35,36 and all commercial assays now use the IU. HCV RNA levels obtained with previous assays can be transformed in IU standards by means of conversion factors.37 The lower detection cut-off of the current assays ranges from 30 IU/mL (SuperQuant, National Institute, Los Angeles, CA) to 615 IU/mL (Versant HCV RNA 3.0 Assay, Bayer Corporation). The upper end of the linear range stretches from less than 500,000 IU/mL (Amplicor HCV Monitor v2.0 and Cobas Amplicor HCV Monitor, Roche Molecular System) to 7,700,000 IU/mL (Versant HCV RNA 3.0 Assay). The samples with a viral level higher than the upper limit must be re-tested after 1:10 or 1:100 dilutions for accurate quantification.
The role of the measurement of the HCV RNA levels for therapy varies according to different genotypes. Baseline HCV RNA quantification is not necessary in patients with genotype 2 or 3. Measurement of HCV RNA before treatment and again after 12 weeks of treatment is useful to monitor patients with genotype 114,38-41 and, according to the present knowledge, also those with genotype 4, 5 and 6.42 A 2-log drop at least or undetectable HCV RNA at week 12, is defined as early virological response (EVR)38 which is now believed to have a poor positive predictive value but an excellent negative predictive value to sustained virological response (SVR). In other words, in the absence of EVR, a patient has a minimal chance of a sustained virological response. These results would allow taking the decision to either stop or continue the treatment as early as 12 weeks after the start of therapy. As indicated above, the clinical reliability of serial HCV viral load testing in a patient is dependent on the use of the same quantitative assay.
More recently, “real-time” PCR techniques have been developed. The principle is to detect amplicon synthesis and to assess the viral load during rather than at the end of the PCR.43 These methods are theoretically more sensitive than the classical target amplification techniques and are not prone to a carryover contamination. The dynamic range is substantially wide, making them particularly useful for quantifying the full range of viral loads in untreated and treated patients.44-46 Unfortunately, no commercial standardized assay is currently available.
HCV genotypingHepatitis C is a heterogeneous virus with at least 6 genotypes and numerous subtypes identified around the world.47,48 Although considerable disagreement exists on the natural history of the disease in patients infected with different genotypes, there is a general consensus on the fact that the HCV genotype is one of the most important predictors to antiviral therapy response.14,42,49
The gold standard for genotyping is the direct sequencing of the NS5B or E1 region, followed by the sequence alignment with reference sequences and phylogenetic analysis.50 In clinical practice, HCV can be geno-typed with several methods such as: 1) restriction fragment length polymorphism (RFLP) analysis of the highly conserved 5’ non coding region;51 2) nested PCR analysis of the HCV core region using genotype-specific primers;52 3) reverse hybridization analysis using genotype-specific probes of 5’ non coding region sequences;53 and 4) direct sequencing of 5’ non coding region and sequence comparison with reference database.54 The first method is rapid and inexpensive, but lacks standardization while the second is labour-intensive and yields to high rates of cross amplification. On the contrary, standardized commercial kit are available for the other two methods (INNO-LIPA HCV II, Innogenetics and Trugene HCV 5’NC Genotyping kit, Visible Genetics Inc., Toronto, Ontario). Both assays can identify the six HCV types and a large number of subtypes. Typing errors are uncommon, but subtyping errors may occur in about 10% of cases.55,56 Subtyping has no clinical significance, since no therapeutically relevant decision is currently taken on the HCV subtype assessment.
The HCV genotype can also be determined by serological methods, namely the detection of antibodies directed to genotype-specific HCV epitopes. The available commercial assay (Murex HCV Serotyping 1-6 Assay, Murex Diagnostic, Dartford, UK) uses NS4 peptides in a competitive EIA. This test provides interpretable results in approximately 90% of immunocompetent patients with chronic HCV infection.57 Its reliability is obviously lower in hemodialysis and immunocompromised patients.58,59 The assay identifies the type1-6 but not the subtypes of HCV. The concordance with molecular assays is around 95% and is higher for genotype 1 than for the others.57,60 In cases of discrepancy, the sequencing of reference genomic regions, such as NS5B and E1, generally confirms the result of the molecular assay.61 Mixed serologic reactivity is sometimes observed. This test cannot distinguish between true mixed infection and cross-reactivity or recovery from one genotype infection and persistence of viremia with another.
HCV genotype should be determined before treatment, as it tailors the therapy to the individual patient.14,37,41,42,49,62
Antigen detection assayA standardized commercial assay using a monoclonal antibody was developed for the qualitative detection of HCV core antigen (Ortho Antibody to Hepatitis C Core Antigen ELISA Test System; Ortho-Clinical Diagnostics, Raritan, NJ). This assay, devoted to screen blood donations, increased safety by significantly reducing the serologic window. Several studies showed that core antigen can be detected 1 to 2 days after HCV RNA positivity during the pre-seroconversion period.63-66 However, this screening assay presents a low sensitivity in HCV antibody-positive subjects. In the new version of the commercial assay (Total HCV core Ag assay; Ortho-Clinical Diagnostics), a preliminary immune-complex dissociation step was introduced to increase the sensitivity. The detection cut-off of this assay is approximately of 2 pg/mL, where 1 pg/mL of total HCV core Ag was estimated to be approximately 8,000 HCV RNA IU/mL.67
When a molecular method is lacking, the total HCV core antigen quantification can be used in the viral load monitoring during therapy, provided the baseline antigen amount is higher than 200 pg/mL.37,67 A new assay with greater sensitivity is currently under development.
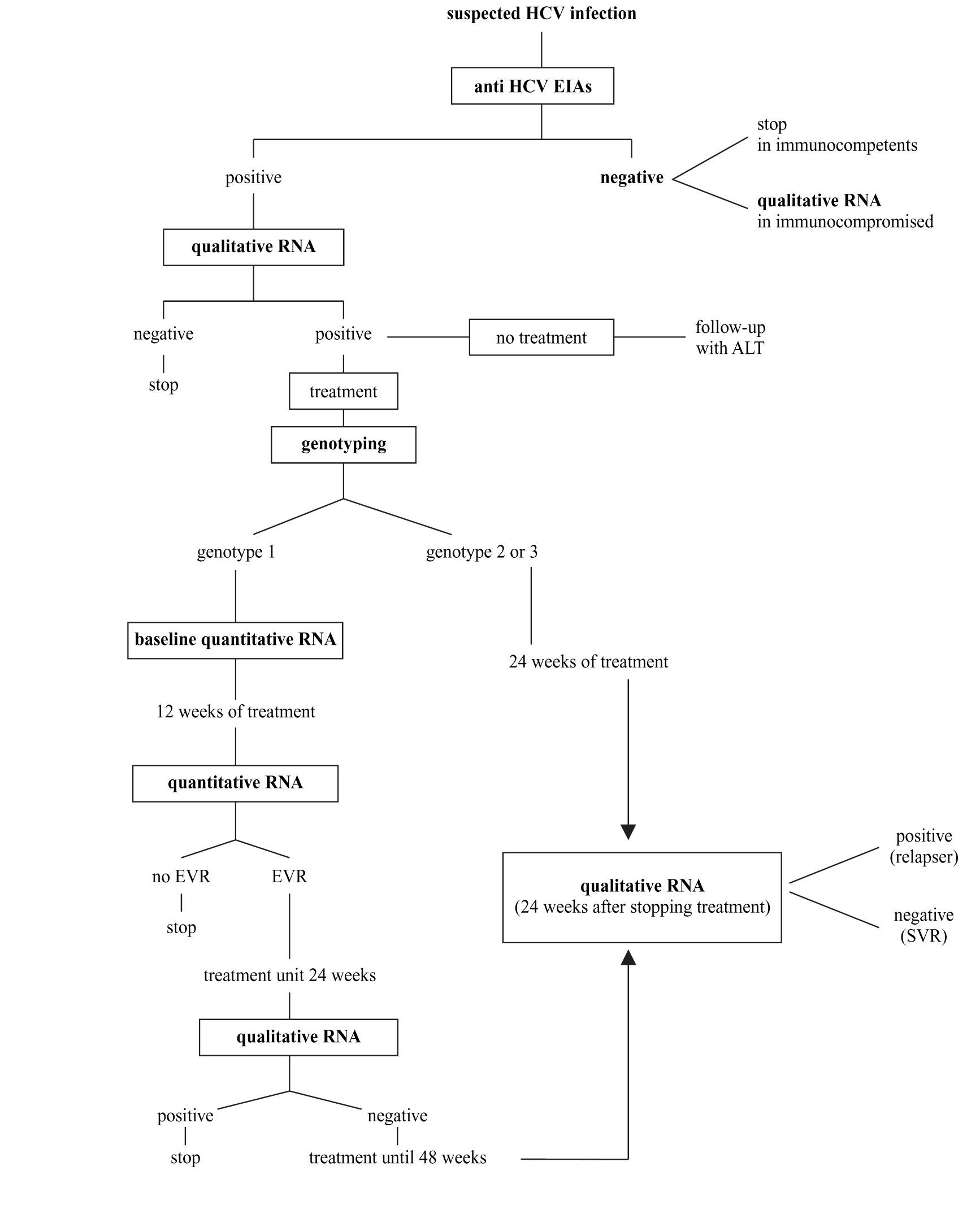
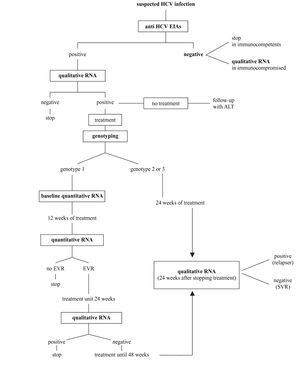
Synopsis of HCV laboratory testingBased on what indicated above and following recent consensus conferences on the best handling of HCV positive subjects,14,41 the diagnostic flowchart shown in figure 2 can be proposed.
A. The screening should be based on a single sample by second-or third-generation EIAs; confirmation of positivity with EIA on a second, different sample might be useful to avoid false-positive results due to sampling or processing errors; no immunoblot-based supplemental assay is needed.
B. HCV RNA detection by PCR or by TMA must be performed whenever the replicative status of HCV needs to be established. The true indications for a qualitative HCV-RNA testing are: 1) seronegative acute hepatitis; 2) seronegative chronic hepatitis in immunocompromised patients; 3) chronic liver disease with several possible causes, including the presence of HCV antibodies; 4) chronic hepatitis C with repeatedly normal ALT; 5) diagnosis of HCV infection in babies born from HCV-infected mothers; and 6) diagnosis after occupational exposure and the therapy monitoring.
C. All patients with chronic hepatitis C must have viral genotyping before treatment to provide prognostic information regarding the SVR as well as to define the length of the treatment and the dose of ribavirin.
D. Patients with genotype 1 must have a quantitative HCV RNA determined by the same method both before and after 12 weeks of treatment. Early virological response is defined as a fall in the HCV RNA level by at least 2 log units or to an undetectable level. Patients with EVR at week 12 should continue the treatment up to 24 weeks when a qualitative HCV test will be performed. Those with negative HCV RNA should be treated for additional 24 weeks, while those where HCV RNA is still detectable must be withdrawn from therapy. The treatment must also be stopped in those patients not showing EVR at 12 weeks.
E. Patients with genotype 2 or 3 should be treated for 24 weeks and do not need to have a 12-week assessment for EVR.
F. HCV RNA must be determined 6 months after the end of the treatment (24 weeks for genotype 2 and 3 and 48 weeks for genotype 1) to assess sustained viral clearance. The assessment of HCV RNA at the end of treatment has the mere role to reassure the patient.
Conclusion and perspectivesDue to the high prevalence of HCV infection in the general population, particularly in subjects older than 40 years,68,69 and the possible associated liver disease, the diagnosis of infection has become a major health problem. The ideal test should be specific, reproducible, reliable and inexpensive. Unfortunately this goal is not yet fully achieved. Most important is, however, that the tests used may give hints to the clinician for a better, more timed treatment of the disease accordingly to our present knowledge. This is the reason why interaction between the virologist and the clinician should be the most stringent and cooperative.
AcknowledgementsThe contribution of Dr. Nora Coppola and Dr. Flora Masutti in gratefully acknowledged. This work was supported by a grant from “Ricerca Corrente” of IRCCS “Burlo Garofolo” and by a grant from C.S.F.