Introduction. Detection of hepatitis C virus (HCV) has been reported in extrahepatic sites such as peripheral blood mononuclear cells and platelets. Quantitation of HCV-RNA in platelets from patients under antiviral therapy has not been reported.
Material and methods. HCV-RNA levels in paired serum and platelet samples of 17 chronically HCV-infected patients were determined at baseline, week 12, end-of-treatment, and 24 weeks after completion of treatment with pegylated interferon plus ribavirin. Quantitation of HCVRNA load was performed using COBAS® TaqMan® HCV Test v 2.0 (lower limit of detection, 25 IU/mL). The cohort predominantly consisted of female (59%) with a mean age of 50.7 ± 10.0 years.
Results. Measurements of HCV-RNA in relation to different timepoints of therapy revealed baseline viral load was most frequently detected in higher levels in serum than in platelets (5.6 × 104 lU/mL vs. 379.0 lU/mL; p = 0.0002), a trend also demonstrated in most samples throughout the study. HCV-RNA was also found at low levels (< 25.0-314.0 UI/mL) persistently in platelets of three patients who have lost detectable HCV-RNA in serum during antiviral therapy, resulting in virological relapse.
Conclusion. HCV-RNA levels are most frequently detected in higher levels in serum than in platelets, independent of timepoint of antiviral therapy. Further studies with an increase in size of the samples are needed to better evaluate whether or not patients who presented HCV-RNA at low levels in platelets after having lost detectable HCV-RNA in serum during antiviral therapy are at increased risk of relapse of HCV infection during follow-up evaluation.
Detection of hepatitis C virus (HCV) has been reported in extrahepatic sites, particularly in different sub-populations of the peripheral blood mononuclear cells (PBMCs).1 Regarding peripheral blood platelets, the presence of HCV in this extrahepatic compartment has been demonstrated in a limited number of full text publications.2–5 In this way, platelets may serve as important reservoirs of HCV and sequester HCV virions from immune recognition.6
The current standard for diagnosing and monitoring of HCV infection is based on the detection of HCV-RNA in plasma or serum samples.7,8 However, in the last decade, occult HCV infection was recognized as a novel entity, characterized by the detection of HCV in liver samples and PBMCs in the absence of serum anti-HCV and HCV-RNA in patients with persistently abnormal values of liver enzymes.9,10 Subsequently, patients who achieved sustained virological response (SVR), but with persistence of HCV in the liver and in PBMCs, have been reported.11 In this way, an effective therapy should be capable of inducing clearance of extrahepatic (platelet) HCV-RNA.12 Even though low serum HCV-RNA levels are among the predictors of beneficial response to antiviral therapy for chronic hepatitis C,13 the influence of HCV-RNA levels in extrahepatic sites on antiviral therapy is an issue to be explored. In this study, we aim to determine HCV-RNA levels in paired serum and platelet samples of chronically HCV-infected patients at baseline and during antiviral therapy with pegylated interferon (PEG-IFN) plus ribavirin (RBV).
Material and MethodsDesignThis open-label, prospective pilot study was approved by the Ethical Committee of the Gaffrée & Guinle University Hospital/Federal University of the State of Rio de Janeiro (UNIRIO), on March 12, 2009 (no 026/2009) and conforms to the ethical guidelines of the 1975 Declaration of Helsinki.
PatientsPatients monoinfected with HCV-genotype 1 were prospectively evaluated on an outpatient basis. The diagnosis of chronic HCV infection was made on the basis of the presence of anti-HCV antibodies (ORTHO HCV 3.0 ELISA, Ortho Clinical Diagnostics, Raritan, NJ, USA) and HCV-RNA in serum samples of patients with infection of more than 6 month’s duration. All patients tested negative for hepatitis B surface antigen, antibodies to human immunodeficiency virus (HIV-1/2), and antibodies to human T-cell leukemia virus type 1/2 (HTLV-1/2).
MethodsPaired samples of serum and platelets were collected from 17 patients at baseline and at longitudinal timepoints of antiviral treatment (at week 12, end-of-treatment, and 24 weeks after completion of treatment) between March 2010 and August 2011. Only one serum and two platelet samples correspondent to week 12 were not available for testing. All paired samples of each patient were tested in the same assay. All patients were assigned to receive PEG IFN-a2b/RBV for 48 weeks. Viral RNA was extracted from paired serum and platelet pellets with QIAamp Viral RNA and RNeasy Mini Kit (QIAGEN, Hilden, Germany), respectively. Platelet pellets were obtained from 500 μL of platelet-rich plasma obtained after appropriate centrifugation of whole blood. Platelet pellets (10 μL) were washed seven times with Tyrode’s solution before viral RNA extraction and the final washing solution was used as negative control. Platelet-rich plasma and platelet pellets leukocyte contamination was excluded by cytomorpho-logical analysis (May-Grünwald-Giemsa staining) and counting (Neubauer chamber and automated cell counter) assays, as previously described.3 Quantitation of HCV-RNA load in platelets samples were determined after performing vortexing of platelets pellets over a minute and then resuspended in 500 μL of anti-HCV/HCV-RNA negative human serum. After this platelet processing, quantitation of HCV-RNA load in paired platelets and serum samples was performed by using COBAS® TaqMan® HCV Test, v 2.0 (Roche Molecular Systems Inc., Branchburg, NJ, USA), which has a dynamic range of detection between 25-391,000,000 IU/mL.14 HCV genotypes was determined in serum samples by using direct nucleotide sequencing of the PCR products from part of HCV NS5A gene. The sequencing protocol was utilized as described by Otto, et al.15
Statistical analysisData are expressed as frequencies, and mean ± standard deviation (SD) if quantitative variables were normally distributed. For continuous variables which did not pass normality test (Kolmogorov-Smirnov test), we used median and ranges, and nonparametric (Wilcoxon test for paired samples) statistics to compare medians. A p value < 0.05 was considered to be statistically significant. Statistical analysis was performed using MedCalc for Windows, version 7.6.0.0 (MedCalc Software, Mariakerke, Belgium).
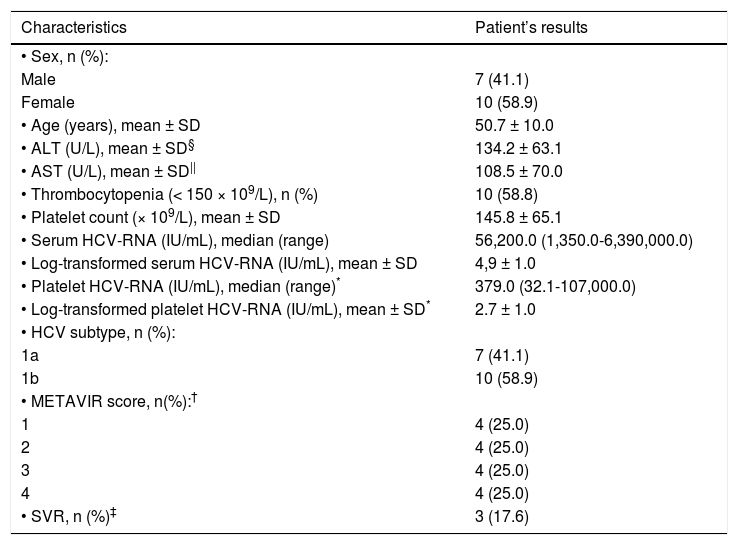
ResultsThe baseline clinical and laboratory characteristics of the 17 patients included in the study are presented in table 1. Our cohort predominantly consisted of female (59%) with a mean age of 50.7 ± 10.0 years. Mean ALT and AST values were as follows: 134.2 ± 63.1 (18.0-257.0 U/L) and 108.5 ± 70.0 (50.8-276.0 U/L), respectively. Mean platelet count was 145.8 ± 65.1 (66.0-306.0 x 109/L) and 10/17 patients presented thrombocytopenia (platelets < 150 × 109/L). Only 3/17 (17.6%) had high baseline HCVRNA viral load ( > 600.000 IU/mL) in serum. Only one patient was found to have undetectable baseline HCV-RNA in platelets whereas three patients had platelet levels lower than 25 IU/mL. HCV subtype 1b was the most prevalent (58.9%). Based on the METAVIR Cooperative Study Group, there were not differences in the prevalence of hepatic fibrosis score in the patients included in the study.
Patient’s characteristics during pretreatment evaluation (n = 17).
| Characteristics | Patient’s results |
|---|---|
| • Sex, n (%): | |
| Male | 7 (41.1) |
| Female | 10 (58.9) |
| • Age (years), mean ± SD | 50.7 ± 10.0 |
| • ALT (U/L), mean ± SD§ | 134.2 ± 63.1 |
| • AST (U/L), mean ± SD|| | 108.5 ± 70.0 |
| • Thrombocytopenia (< 150 × 109/L), n (%) | 10 (58.8) |
| • Platelet count (× 109/L), mean ± SD | 145.8 ± 65.1 |
| • Serum HCV-RNA (IU/mL), median (range) | 56,200.0 (1,350.0-6,390,000.0) |
| • Log-transformed serum HCV-RNA (IU/mL), mean ± SD | 4,9 ± 1.0 |
| • Platelet HCV-RNA (IU/mL), median (range)* | 379.0 (32.1-107,000.0) |
| • Log-transformed platelet HCV-RNA (IU/mL), mean ± SD* | 2.7 ± 1.0 |
| • HCV subtype, n (%): | |
| 1a | 7 (41.1) |
| 1b | 10 (58.9) |
| • METAVIR score, n(%):† | |
| 1 | 4 (25.0) |
| 2 | 4 (25.0) |
| 3 | 4 (25.0) |
| 4 | 4 (25.0) |
| • SVR, n (%)‡ | 3 (17.6) |
Excluded one patient with undetectable HCV-RNA and three patients with values < 25 IU/mL in platelets.
According to the METAVIR Cooperative Study Group (Bedossa & Poynard, 1996).16 Excluded one patient who did not undergo hepatic biopsy.
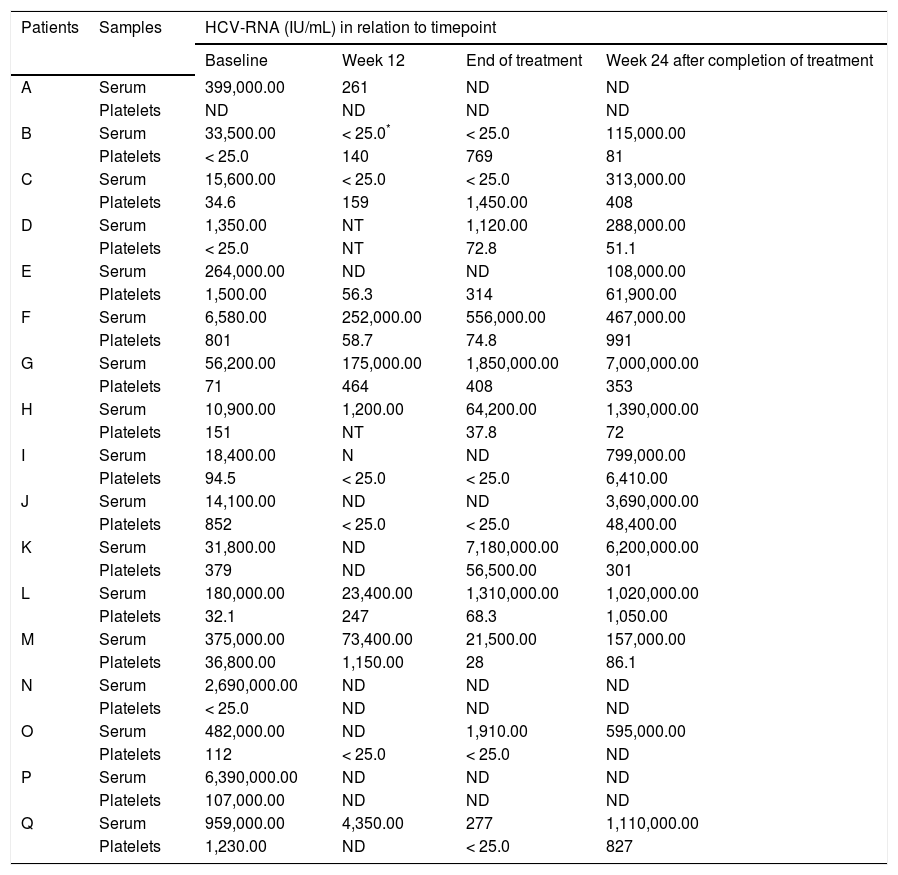
Measurements of HCV-RNA in relation to different timepoints of antiviral therapy are presented in table 2. Baseline HCV-RNA levels in platelets were found to be significantly lower than levels observed in serum (5.6 × 104 IU/mL vs. 379.0 IU/mL; p = 0.0002). This trend was also demonstrated in most samples throughout the study period. HCV-RNA was quantified at low levels (< 25.0-314.0 UI/mL) persistently in platelets during all therapy period in three patients (E, I, and J), who had lost detectable HCV-RNA in serum samples collected at the same periods. However, after 24 weeks of follow-up they relapsed virologically in serum as demonstrated by detectable viremia (108,000.0-3,690,000.0 IU/mL). Another patient (O) presented low levels of HCVRNA (< 25.0 IU/mL) at week 12 in platelets but undetectable in serum, and, at the end of therapy, this patient had also detectable HCV-RNA (1,910.0 IU/ mL) in serum.
HCV viral loads measured longitudinally in a cohort of 17 patients receiving antiviral therapy with pegylated interferon plus ribavirin.
| Patients | Samples | HCV-RNA (IU/mL) in relation to timepoint | |||
|---|---|---|---|---|---|
| Baseline | Week 12 | End of treatment | Week 24 after completion of treatment | ||
| A | Serum | 399,000.00 | 261 | ND | ND |
| Platelets | ND | ND | ND | ND | |
| B | Serum | 33,500.00 | < 25.0* | < 25.0 | 115,000.00 |
| Platelets | < 25.0 | 140 | 769 | 81 | |
| C | Serum | 15,600.00 | < 25.0 | < 25.0 | 313,000.00 |
| Platelets | 34.6 | 159 | 1,450.00 | 408 | |
| D | Serum | 1,350.00 | NT | 1,120.00 | 288,000.00 |
| Platelets | < 25.0 | NT | 72.8 | 51.1 | |
| E | Serum | 264,000.00 | ND | ND | 108,000.00 |
| Platelets | 1,500.00 | 56.3 | 314 | 61,900.00 | |
| F | Serum | 6,580.00 | 252,000.00 | 556,000.00 | 467,000.00 |
| Platelets | 801 | 58.7 | 74.8 | 991 | |
| G | Serum | 56,200.00 | 175,000.00 | 1,850,000.00 | 7,000,000.00 |
| Platelets | 71 | 464 | 408 | 353 | |
| H | Serum | 10,900.00 | 1,200.00 | 64,200.00 | 1,390,000.00 |
| Platelets | 151 | NT | 37.8 | 72 | |
| I | Serum | 18,400.00 | N | ND | 799,000.00 |
| Platelets | 94.5 | < 25.0 | < 25.0 | 6,410.00 | |
| J | Serum | 14,100.00 | ND | ND | 3,690,000.00 |
| Platelets | 852 | < 25.0 | < 25.0 | 48,400.00 | |
| K | Serum | 31,800.00 | ND | 7,180,000.00 | 6,200,000.00 |
| Platelets | 379 | ND | 56,500.00 | 301 | |
| L | Serum | 180,000.00 | 23,400.00 | 1,310,000.00 | 1,020,000.00 |
| Platelets | 32.1 | 247 | 68.3 | 1,050.00 | |
| M | Serum | 375,000.00 | 73,400.00 | 21,500.00 | 157,000.00 |
| Platelets | 36,800.00 | 1,150.00 | 28 | 86.1 | |
| N | Serum | 2,690,000.00 | ND | ND | ND |
| Platelets | < 25.0 | ND | ND | ND | |
| O | Serum | 482,000.00 | ND | 1,910.00 | 595,000.00 |
| Platelets | 112 | < 25.0 | < 25.0 | ND | |
| P | Serum | 6,390,000.00 | ND | ND | ND |
| Platelets | 107,000.00 | ND | ND | ND | |
| Q | Serum | 959,000.00 | 4,350.00 | 277 | 1,110,000.00 |
| Platelets | 1,230.00 | ND | < 25.0 | 827 | |
A number of studies have demonstrated the presence of HCV in various extrahepatic human tissues by providing evidence of HCV genome detection in different cells other than hepatocytes, such as B lymphocytes, T lymphocytes, monocytes, dendritic cells, and platelets. Nevertheless, its influence on antiviral therapy is still controversial.17 The present work explored the serial quantitative detection of HCV-RNA in peripheral blood platelets of HCV-monoinfected patients under antiviral therapy. According to our results we could demonstrate that most patients with chronic HCV infection harbor HCVRNA sequences in their platelets before, during, and after completion of therapy. Considering that platelet pellets were washed seven times with a platelet-washing buffer before viral RNA extraction and the final washing solution of platelets tested negative for HCV-RNA in all patients, and that HCV-RNA was detected in platelet but not in serum samples in a few patients throughout the study period, we can assume that HCV-RNA in platelets was not due to contamination from circulating viral particles.
It is known that during the course of chronic HCV infection, viremia level can vary from phases of high viral load to phases when the viral load is detected at a very low or undetectable level. However, there is little information on viral kinetics in extrahepatic compartments during therapy for chronic HCV infection.18,19 In the present study, baseline HCV-RNA levels in platelets were found to be significantly lower than those levels observed in serum. This trend was also demonstrated for a number of samples throughout the study period. Besides, antiviral therapy contributed to reduce the number of patients with HCV-RNA detected in serum (100 to 50%) and platelets (94 to 73%) at week 12. Chary, et al. have also obtained median plasma HCV viral load higher than in PBMCs at baseline (2.6 × 106 IU/mL vs. 485.0 IU/mL).20 In addition, these authors reported a patient initiating IFN-based HCV therapy who presented an increase in PBMCs viral load at week 36 after successfully diminished the HCV-RNA in PBMCs during the first 24 weeks of therapy.20 Whether or not antiviral therapy with PEG-IFN plus RBV may affect only a certain HCV subtype in platelets is not known and remains to be clarified. To the best of our knowledge, this is the first report on studying viral kinetics in platelets during antiviral therapy.
Although the magnitude of the extrahepatic viral contribution to HCV viremia is difficult to be estimated,21 HCV replicative activity, at best limited, in extrahepatic compartments such as platelets can not be rule out. Platelets are small anucleate cell fragments (cytoplasts) derived from megakaryocytes and HCV particles could be transferred from megakaryocytes to platelets during proplatelet formation. Despite its short lifespan (7–10 days), mature platelets are capable of de novo synthesis of proteins.22–24 Detection of both strands (positive and replicative) of HCV in megakaryocytes of patients with HCV-related cryoglobulinemia and thrombocytopenia using in situ hybridization was reported in previous studies.25,26 Moreover, HCV replication was demonstrated in human megakaryoblastic leukaemia cell line.27 A study suggested that the difference in HCV-RNA levels between serum and PBMCs samples may be associated with a differential regulation of HCV-RNA in each extrahepatic compartment, which could explain, at least in part, its adverse influence on virological response.28
In the context of antiviral therapy, to be considered effective, this intervention should be capable of inducing HCV eradication in extrahepatic compartments, since they could constitute reservoir of HCV, thus contributing to HCV relapse after therapy. Nevertheless, in our series, low levels of HCV-RNA was found in platelets throughout the therapy period in three patients (E, I, and J) who had lost detectable HCV-RNA in serum samples collected at the same periods. These patients, after 24 weeks of follow-up, relapsed virologically in serum as demonstrated by detectable viremia. In other patient (O), HCV-RNA reappearance in serum was demonstrated at the end of therapy, the same patient had lost detectable serum HCV-RNA at week 12, but not in platelets during these timepoints (< 25.0 IU/mL). These findings are interesting because they show that platelets may also be involved in virological relapse after apparent clearance of the virus at the end of antiviral treatment. The study by Torres, et al. demonstrated that the presence of HCV in serum, a major criterion for evaluation of virological response, is not necessarily followed by its disappearance in PBMCs after antiviral therapy.18 Nevertheless, another study reported no samples having detectable HCV-RNA (lower detection limit, 20 IU/mL) in PBMCs in the setting of undetectable HCV-RNA in serum samples during longitudinal quantitation of HCV-RNA in patients receiving IFN-based HCV therapy.20 The importance of detection of HCV-RNA in platelets was previously reported in a study using RT-PCR in-house technique (lower detection limit, 50 IU/ mL).12 In that study, comprising 48 chronically HCV-infected patients, De Almeida, et al. reported two patients with HCV-RNA detectable in platelets and normal ALT levels at the end of therapy who lost detectable serum HCV-RNA. After 24 weeks of follow-up, HCV-RNA persistence in platelets of both cases was observed in association with the reappearance of HCV in serum, indicating that occult HCV detection in platelets appears to be a rare event during antiviral therapy.12 However, in this longitudinal study quantitation of platelet HCV-RNA was not carried out.12 Indeed, in a study of the prevalence of HCV in paired cervical cytobrush and plasma samples from 85 HCV/HIV coinfected patients, only a woman presented HCV-RNA positivity in cytobrush sample in the setting of plasma HCV-RNA below the limit of quantitation of the assay (6 × 102 IU/mL) used in that study,29 indicating that, as demonstrated in our study, patients can harbor HCV genome in extrahepatic compartment in the absence of detectable viremia.
Evidence of possible distribution of specific HCV variants or quasispecies in PBMCs (compar-tmentalization) was demonstrated in the study by Blackard, et al., after analysis of HCV diversity in the NS5B and envelope 1 genes.30 Furthermore, B-cell tropism of HCV was found to be associated with impaired IFN response.31 However, HCV variants in platelets were not reported in previous studies. Only preliminary results of a recent study aiming to analyse HCV variants or quasispecies (clones from the NS5A region) in serum and platelets obtained from antiviral therapy naïve patients with chronic HCV infection revealed a distinct distribution between serum and platelet compartments.32 Whether or not HCV presents platelet tropism with an interferon-resistant phenotype is not known and deserves to be clarified in appropriate studies.
ConclusionOur data indicated that HCV-RNA levels are most frequently detected in higher levels in serum than in platelets. Further studies with an increase in size of the samples in this population are necessary to better evaluate whether or not patients who presented HCV-RNA at low levels in platelets after having lost detectable HCV-RNA in serum during antiviral therapy are at an increased risk of relapse of HCV infection during follow-up evaluation.
Abbreviations- •
HCV: hepatitis C virus.
- •
PBMCs: peripheral blood mononuclear cells.
- •
SVR: sustained virological response.
- •
PEG-IFN. pegylated interferon.
- •
RBV: ribavirin.
- •
HTLV: Human T-cell leukemia virus.
- •
HIV: human immunodeficiency virus.
- •
PCR: polymerase chain reaction.
- •
NS5A: nonstructural 5A.
- •
SD: standard deviation.
- •
ALT: alanine aminotransferase.
- •
AST: aspartate aminotransferase.
The authors are grateful to the sequencing group of the PDTIS program of Oswaldo Cruz Foundation for performing the DNA sequencing: Conselho Nacional de Desenvolvimento Científico e Tecnológico, CNPq: Coordenaçao de Aperfeiçoamento de Pessoal de Nível Superior, CAPES; and Program Papes V from Oswaldo Cruz Foundation (FIOCRUZ), Rio de Janeiro, Brazil for financial support.
List of Grants and Other Financial SupportConselho Nacional de Desenvolvimento Científico e Tecnológico, CNPq.
Coordenaçao de Aperfeiçoamento de Pessoal de Nivel Superior, CAPES.
Program Papes V from Oswaldo Cruz Foundation (FIOCRUZ).