Circular RNA (circRNA) has been demonstrated as a critical regulator in human cancer, including hepatocellular carcinoma (HCC). Nevertheless, the role of circ-PRMT5 in HCC remains largely unknown.
Patients or materials and methodsThe real-time quantitative polymerase chain reaction (RT-qPCR) was performed to assess the expression levels of circ-PRMT5, miR-188-5p and anti-Hexokinase II (HK2) in HCC tissues and cells. The cell proliferation, migration and glycolysis were determined by 3-(4, 5-dimethylthiazol-2-yl)-2, 5-diphenyl-2H-tetrazol-3-ium bromide (MTT), transwell migration assay, and indicated kits, respectively. The interaction relationship between miR-188-5p and circ-PRMT5 or HK2 was analyzed by the bioinformatics database, dual-luciferase reporter assay, and RNA immunoprecipitation (RIP) assay. The western blot assay was used to analyze the expression level of HK2. The functional role of circ-PRMT5 in vivo was assessed by a xenograft experiment.
ResultsCirc-PRMT5 was elevated in HCC tissues and cells than matched control groups. Furthermore, loss-of-functional experiments revealed that the silencing of circ-PRMT5 could repress proliferation, migration, glycolysis in vitro and tumor growth in vivo. Moreover, we also confirmed that overexpression of circ-PRMT5 abolished the effects on HCC cells induced by upregulating miR-188-5p. In addition, overexpression of miR-188-5p could repress the development of HCC. More importantly, HK2 was a target gene of miR-188-5p, and miR-188-5p regulated proliferation, migration, glycolysis of HCC cells by specifically binding to HK2. Mechanistically, circ-PRMT5 could act as a sponge of miR-188-5p to regulate the expression of HK2.
ConclusionIn summary, circ-PRMT5 might play a key role in proliferation, migration, glycolysis of HCC cells via miR-188-5p/HK2 axis, which indicated that circ-PRMT5 might be a potential therapeutic target for HCC treatment.
Hepatocellular carcinoma (HCC) is a primary clinical tissue subtype of liver cancer, comprising 75–85% of cases in all liver cancer types [1]. Infection with hepatitis B/C virus, smoking, obesity, aflatoxin-exposed, and alcoholism were considered to be the main risk factors for HCC occurrence [2]. Notably, clinical prognosis and long-term survival rate of HCC patients were far from satisfaction, although technological means have made progress [3,4]. In addition, HCC was a continual and complex process that involved numerous factors and stages of evolution, which hampered the development of new treatments for HCC [5].
Currently, circular RNA (circRNA), a kind of evolutionary conservation and non-coding RNA, has emerged as a potential and promising biomarker for cancer diagnosis and treatment [6]. For instance, Su et al. reported that circRNA Cdr1as acted as a well-known biomarker for HCC [7]. Zhu et al. confirmed that hsa_circ_0027089 was a reliable diagnostic biomarker for hepatitis B virus-related HCC [8]. Here, circ-PRMT5 (hsa_circ_0031242) was derived from the PRMT5 gene and located on chr14 (23,389,732–23,392,044). Chen et al. revealed that circ-PRMT5 promoted bladder cancer development by regulating epithelial-mesenchymal transition via sponging miR-30c [9]. Analogous tumor promoter role by circ-PRMT5 was also confirmed in lung cancer [10]. However, the investigation about the mechanism of circ-PRMT5 regulating cell proliferation, migration and glycolysis was still insufficient.
MiRNA had regulatory functions on gene expression with 21–25 nucleotides in length [11]. Although without protein-coding potential, miRNA has the important functions in a wide range of biological processes, including development, cell differentiation and regulation of cell cycle and apoptosis, thereby participating in tumorigenesis [12]. Mechanically, miRNA can modulate specific gene expression through binding to the 3′untranslated regions (3′UTR) of target mRNAs [13]. MiR-188-5p has been described to participate in development of malignant tumors, including colorectal cancer [14], gastric cancer [15], and prostate cancer [16]. Furthermore, it has been verified that miR-188-5p was tightly connected with the proliferation and migration of HCC cells [17]. Here we aimed to investigate the function of miR-188-5p regulating proliferation, migration and glycolysis of HCC cells.
Furthermore, it has been reported that anti-Hexokinase II (HK2) was a well-characterized rate-limiting enzyme in glycolysis [18]. HK2, a pivotal transcription factor which played a key role in the regulation of cancer cell glycolysis, has been documented to be related to tumorigenesis [19]. In addition, upregulation of HK2 was usually found in malignant diseases, including HCC [20]. In this research, circ-PRMT5 expression and its function were analyzed in HCC. Furthermore, we also investigated how circ-PRMT5 affected miR-188-5p and HK2 to regulate proliferation, migration and glycolysis in HCC cells.
2Materials and methods2.1Patient specimensTwenty HCC tissues (obtained from patients with HCC) and paired neighboring normal tissues (away from HCC tissues at least 3cm) were received from the Xiangyang NO.1 People's Hospital, Affiliated Hospital of Hubei University of Medicine. The clinic pathologic features of HCC patients are summarized in Table 1. All tissue samples were surgically resected from patients with the guidance of pathologists and snap-frozen in liquid nitrogen, then stored at −80°C refrigerator. Every patient in the study had received no other therapies before the operation and signed the written informed consent. All the procedures got permission by the Ethics Committee of Xiangyang NO.1 People's Hospital, Affiliated Hospital of Hubei University of Medicine.
Clinical characteristics of 20 HCC patients.
| Variable | Number (%) |
|---|---|
| Age | |
| ≤50 | 7 (35) |
| >50 | 13 (65) |
| Gender | |
| Male | 15 (75) |
| Female | 5 (25) |
| Tumor size (cm) | |
| ≤5 | 11 (55) |
| >5 | 9 (45) |
| Liver cirrhosis | |
| Absent | 9 (45) |
| Present | 11 (55) |
| Cirrhosis etiology | |
| Viral (HCV, HBV) | 5 (45.5) |
| ASH | 3 (27.3) |
| NASH | 2 (18.2) |
| Other | 1 (9.1) |
| TNM stage | |
| I–II | 9 (45) |
| III–IV | 11 (55) |
The human liver cell line (THLE-2) and HCC cell line (SNU-387) were gained from the American Type Culture Collection (Rockville, MD, USA). Human HCC cell line (HCCLM3; with high metastatic potential) was purchased from KeyGen (Nanjing, China). Dulbecco's modified Eagle medium (Wisent, Shanghai, China), which supplemented with 10% (v/v) fetal bovine serum (FBS; Wisent) and 1% penicillin/streptomycin (Wisent), was used to culture above cells under standard culture conditions (5% CO2, 37°C).
2.3RNA isolation, RNase R treatment and real-time quantitative polymerase chain reaction (RT-qPCR)Total RNA was isolated by Trizol reagent (Solarbio, Beijing, China) in accordance with the manufacturer's protocols. Furthermore, to attain the cytoplasmic and nuclear RNAs, the NE-PER Reagent (Thermo Fisher Scientific, Carlsbad, CA, USA) was utilized based on the specification. Subsequently, RNase R treatment was carried out, and partial RNA was treated with 3U/mg RNase R (Epicenter Technologies, Madison, WI, USA) for 15min at 37°C. 5μg of the RNA as template was converted to complementary DNA using High-capacity cDNA Reverse Transcription kit (Applied Biosystems, Foster City, CA, USA) either random or oligo(dT)18 primers. MiRNA was detected using Hairpin-it TM miRNAs qPCR Quantitation Kit (Genepharma, Shanghai, China) and was standardized to endogenous small nuclear RNA U3. RT-qPCR assay was performed using SYBR Green Master Mix kit (Takara, Dalian, China) on AB7300 thermo-recycler (Applied Biosystems). The transcript levels of circ-PRMT5, miR-188-5p and HK2 were assessed based on the 2−ΔΔCt method. The endogenous small nuclear RNA U3 and glyceraldehyde-3-phosphate dehydrogenase (GAPDH) were used as internal controls for miRNA and circRNA/mRNA detection, respectively.
The sequences of primers were listed:
circ-PRMT5 (F, 5′-TACCATTGGCCTCTAGCCCT-3′; R, 5′-CAAGGGGAATCACAGCCCAT-3′);
mRNA-PRMT5 (F, 5′-CTGTCTTCCATCCGCGTTTCA-3′; R, 5′-GCAGTAGGTCTGATCGTGTCTG-3′);
miR-188-5p (F, 5′-GCCGAGCATCCCTTGCATG-3′; R, 5′-CTCAACTGGTGTCGTGGA-3′);
HK2 (F, 5′-GAGCCACCACTCACCCTACT-3′; R, 5′-CCAGGCATTCGGCAATGTG-3′);
GAPDH (F, 5′-TCCCATCACCATCTTCCAGG-3′; R, 5′-GATGACCCTTTTGGCTCCC-3′);
U3 (F, 5′-AGAGGTAGCGTTTTCTCCTGAGCG-3′; R, 5′-ACCACTCAGACCGCGTTCTC-3′).
2.4Transfection assayTo generate stable circ-PRMT5-asbsence HCC cell lines, specific short hairpin RNA (shRNA) targeting circ-PRMT5 (sh-circ-PRMT5) were synthesized and inserted into lentiviral vector (GenePharma), followed by injecting into SNU-387 and HCCLM3 cells by Lipofectamine 2000 (Thermo Fisher Scientific) in compliance with the producer's direction. Similarly, HK2-asbsence HCC cell lines were established. MiR-188-5p mimic (miR-188-5p) and its negative control (miR-NC), and miR-188-5p inhibitor (anti-miR-188-5p) and its negative control (anti-NC) were generated by GenePharma. Circ-PRMT5-overexpression vector (circ-PRMT5), and its negative control (Vector) were purchased from RiboBio Corporation (Guangzhou, China).
2.5Cell proliferation assayFor cell proliferation assay, SNU-387 and HCCLM3 cells were implanted in 96 wells with a density of 3000cells per well. After 48h, HCC cells were reacted with 20μL of MTT (Promega, Madison, WI, USA) at 37°C for 4h. The medium was replaced with 150μL of dimethyl sulfoxide and then oscillation reaction lasted for 10min. Absorbance at 490nm of each well was examined using a microplate reader (Bio-Rad, Hercules, CA, USA).
2.6Migration assayThe 24-well transwell chamber was used for migration assay. In brief, SNU-387 or HCCLM3 cells (about 1×104cells) were added into the upper chamber with 200μL of serum-free medium, and the bottom chamber was filled with the complete medium. After incubation 48h, the cells on the lower surface were fixed with 95% ethanol, and then stained with 0.1% crystal violet. The five randomly fields were photographed and analyzed using a microscope (Olympus, Tokyo, Japan) and Image J software (National Institutes of Health, Bethesda, MD, USA), respectively.
2.7Glucose consumption and lactate production and ATP level assaysGlucose consumption, lactate production and ATP level could reflect the intracellular glycolysis of cells. Transfected SNU-387 or HCCLM3 cells were seeded into 12-well plates at 1×105cells/well and cultured overnight. The cell supernatant was collected to measure glucose and lactate concentration by Glucose Assay Kit (Nanjing Jiancheng Bioengineering Institute, Nanjing, China) and Lactic Acid assay kit (Nanjing Jiancheng Bioengineering Institute), individually. Moreover, cells were lysed to measure ATP level with ATP assay kit (Nanjing Jiancheng Bioengineering Institute) according to the manufacturer's instructions. The equal amount of un-transfected cells were used as control.
2.8Dual-luciferase reporter assayWe predicted the miR-188-5p binding sites in circ-PRMT5 or 3′UTR of HK2 using the bioinformatics databases starbase3.0 (http://starbase.sysu.edu.cn/). The sequences contained the presumed binding sites of miR-188-5p were designed from the circ-PRMT5 or 3′UTR of HK2 and then inserted pGL3 vectors (Promega), named as wt-circ-PRMT5 or wt-HK2, respectively. The binding sites that interacted with the miR-188-5p were mutated using site-directed mutation primers to generate mut-circ-PRMT5 or mut-HK2, respectively. For the dual-luciferase reporter assay, SNU-387 or HCCLM3 cells were planted into 6-well plates and co-transfected with indicated luciferase reporter vectors according to the experiment designed and miR-188-5p or miR-NC. After transfection 48h, luciferase activity in SNU-387 or HCCLM3 cells was detected by the dual-luciferase reporter assay system (Promega) and normalized to Renilla luciferase.
2.9RNA immunoprecipitation (RIP) assayThe RIP-assay was carried out by using Imprint® RNA immunoprecipitation kit (Sigma, St. Louis, MO, USA) according to manufacturer's instruction. Firstly, SNU-387 or HCCLM3 cells were lysed by RIP-buffer combined with a protease inhibitor cocktail and RNase inhibitors. After centrifugation, 50μl of the supernatant was retained as input and the remaining part was incubated with magnetic beads pre-covered with Argonaute-2 (Ago2, Millipore, Billerica, MA, USA) or IgG (Millipore) antibodies at 4°C overnight. After washing off unbound material, immunoprecipitated RNA was purified by proteinase K buffer and the abundance of RNA was detected by RT-qPCR.
2.10Western blot assayBriefly, total protein of HCC tissues or cells was isolated using Radio-Immunoprecipitation assay (RIPA) buffer (Cell Signaling Technology, Danvers, MA, USA) according to the operation manual. 30μg of protein for each sample was isolated by 12% sodium dodecyl sulfate polyacrylamide gel electrophoresis and shifted onto polyvinylidene fluoride membranes (GE Healthcare, Piscataway, NJ, USA). After the incubation with a high affinity antibody anti-HK2 (ab209847; 1:1000 dilution; Abcam, Cambridge, MA, USA) at 4°C overnight, with anti-GAPDH (ab181602; 1:1000 dilution; Abcam) as negative control, then the membranes were incubated with a secondary antibody (1:2000 dilution, Boster, Wuhan, China). After being washed, signals were detected using a chemiluminescence system (Bio-Rad) and quantified using Image LabTM Software (Bio-Rad).
2.10.1In vivo experimentThe experiment in nude mice was licensed by the Institutional Animal Care and Use Committee of Xiangyang NO.1 People's Hospital, Affiliated Hospital of Hubei University of Medicine. 12 male BALB/c nude mice (females, 4 weeks of age; Shanghai Experimental Animal Center, Shanghai, China) were randomly divided into two groups (n=6). SNU-387cell stably transfected with lentiviral vector contained sh-circ-PRMT5 were subcutaneously inoculated into the right flank near the forelimb, with sh-NC as control. Tumor growth was monitored starting from 7 days post-inoculation using V=1/2×ab2 method (length (a) and width (b) length of the tumor). 28d after injection, mice were sacrificed by cervical dislocation and tumors were harvested for further determination.
2.11Statistical analysisAll data were expressed as mean±standard deviation and P value less than 0.05 meant significant difference. Comparison between any two groups or among multiple groups was analyzed by using Student's t-test or one-way analysis of variance, respectively. All analyses were conducted using the SPSS 21.0 software (IBM, Somers, NY, USA). Pearson's correlation coefficient analysis was used to analyze the correlations between circ-PRMT5 and miR-188-5p.
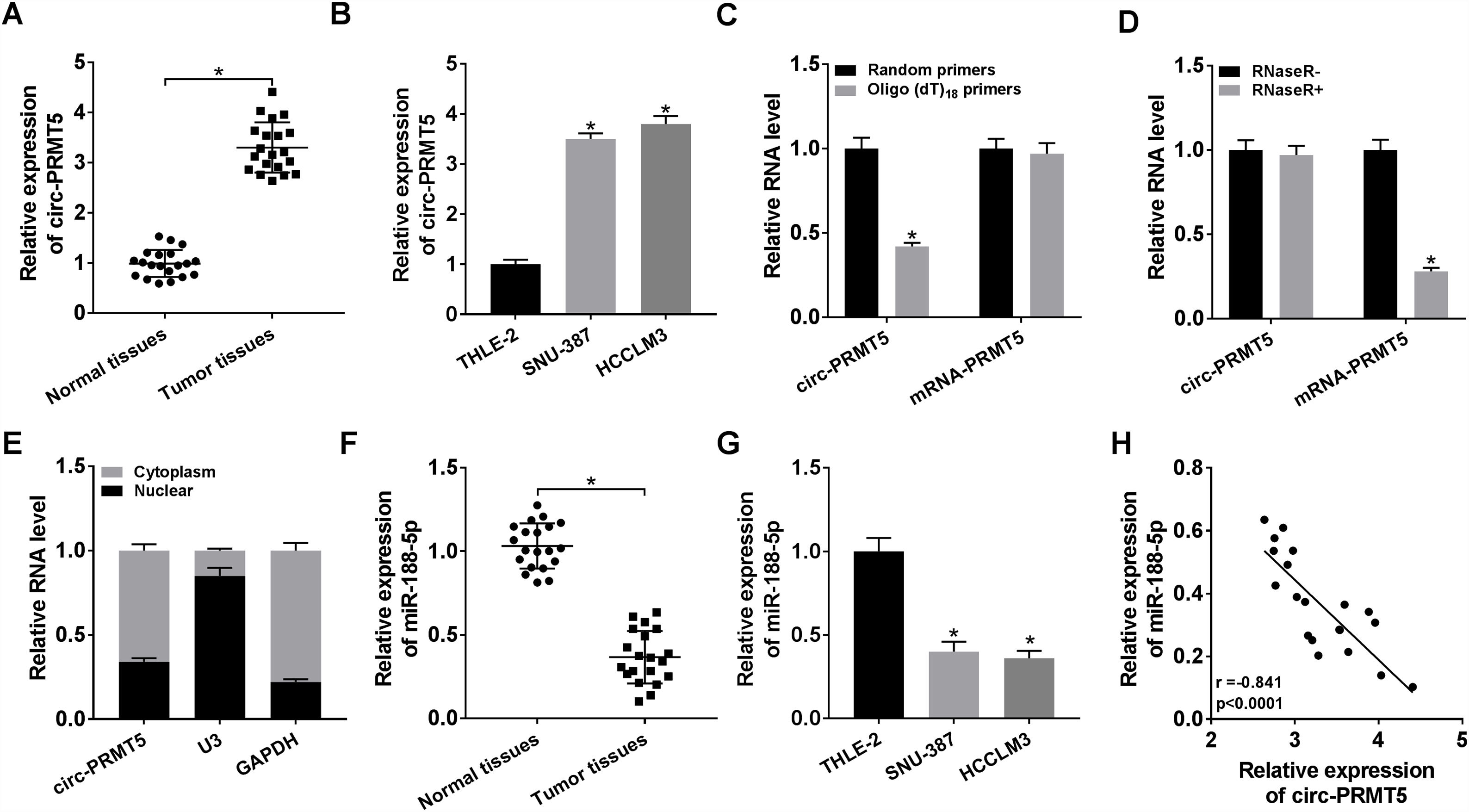
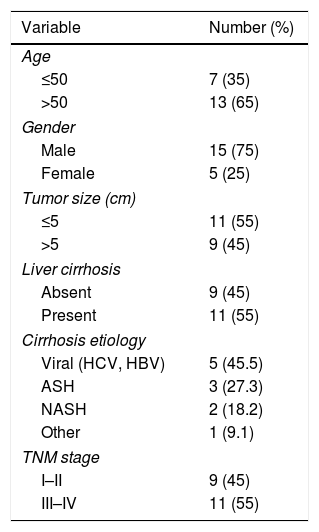
3Results3.1Circ-PRMT5 was overexpressed while miR-188-5p was declined in HCC tissues and cellsFirstly, the expression level of circ-PRMT5 in 20 HCC tissues and neighboring normal tissues was assessed by RT-qPCR analysis, and the data suggested that circ-PRMT5 was evidently upregulated in HCC tissues compared with control group (Fig. 1A). Also, circ-PRMT5 was apparently increased in HCC cells (SNU-387 and HCCLM3) compared with THLE-2cells (Fig. 1B). As described in Fig. 1C and D, circ-PRMT5 had no poly-A tail and was resistant to RNase R, suggesting that circ-PRMT5 was circular RNA. In addition, RT-qPCR assay further established the cytoplasmic localization of circ-PRMT5 (Fig. 1E). As shown in Fig. 1F and G, miR-188-5p was observed to be dramatically decreased in HCC tissues and cells compared with matched control groups. By measuring the expression of circ-PRMT5 and miR-188-5p in the HCC tissues using RT-qPCR assay, we found that there was existed a negative linear correlation between circ-PRMT5 and miR-188-5p (Fig. 1H). These data implied that circ-PRMT5 was upregulated and negatively correlated with miR-188-5p expression in HCC.
The expression levels of circ-PRMT5 and miR-188-5p in hepatocellular carcinoma tissues and cells. (A and B) The relative expression level of circ-PRMT5 in hepatocellular carcinoma tissues and neighboring normal tissues, as well as in THLE-2, SNU-387 and HCCLM3 cells was measured by RT-qPCR. (C) Random or oligo (dT)18 primers were used for reverse transcription experiments and the expression levels of circ-PRMT5 and PRMT5 mRNA were analyzed by RT-qPCR. (D) The relative levels of circ-PRMT5 and PRMT5 mRNA were analyzed by RT-qPCR after treatment with RNase R. (E) RT-qPCR was conducted for analysis of nuclear and cytoplasmic circ-PRMT5 levels in SNU-387 and HCCLM3. (F and G) RT-qPCR was carried out to test miR-188-5p level in hepatocellular carcinoma tissues and cells as well as matched control group. (H) Correlation analysis between circ-PRMT5 and miR-188-5p was performed by Pearson's correlation coefficient analysis. *P<0.05.
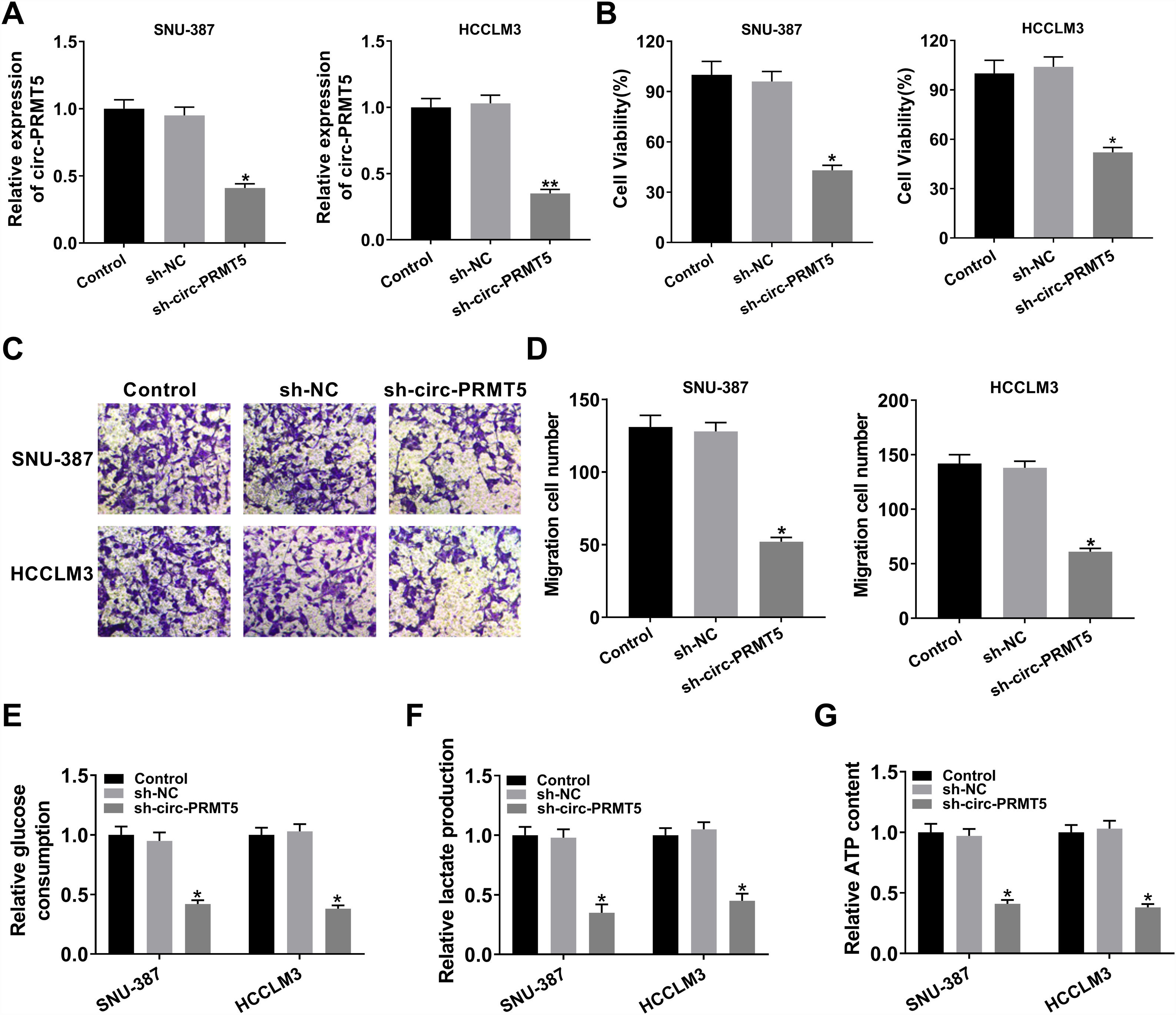
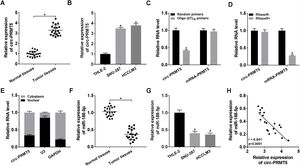
To further assess the function of circ-PRMT5 in tumorigenesis of HCC, SNU-387 and HCCLM3 cells were transfected with sh-circ-PRMT5 or sh-NC. As presented in Fig. 2A, the expression level of circ-PRMT5 was declined in SNU-387 and HCCLM3 cells infected with sh-circ-PRMT5 than cells transfected with sh-NC. Furthermore, downregulation of circ-PRMT5 inhibited cell proliferation, as shown by the MTT assay (Fig. 2B). Transwell migration analysis also revealed that silencing of circ-PRMT5 led to reduced numbers of migration cells than the sh-NC transfected group or un-transfected group (Fig. 2C and D). Additionally, glucose consumption, lactate production, and ATP level were observed to be decreased by transfection with sh-circ-PRMT5 in the SNU-387 and HCCLM3 cells (Fig. 2E–G). These data strongly implied that circ-PRMT5 may promote cell proliferation, migration and glycolysis in HCC cells.
Effects of circ-PRMT5 silencing on proliferation, migration and glycolysis of hepatocellular carcinoma cells. (A–G) SNU-387 and HCCLM3 cells were transfected with sh-circ-PRMT5 or sh-NC, un-transfected cells were regarded as the control group. (A) RT-qPCR was conducted to confirm the knockdown efficiency of circ-PRMT5 in SNU-387 and HCCLM3 cells. (B) The cell viability of SNU-387 and HCCLM3 cells was measured using MTT assay. (C and D) Transwell migration assay was used to measure the migration ability of SNU-387 and HCCLM3 cells and representative images were shown. (E–G) Glucose consumption, lactate production, and ATP level in SNU-387 and HCCLM3 cells were measured by matched assay kits. *P<0.05.
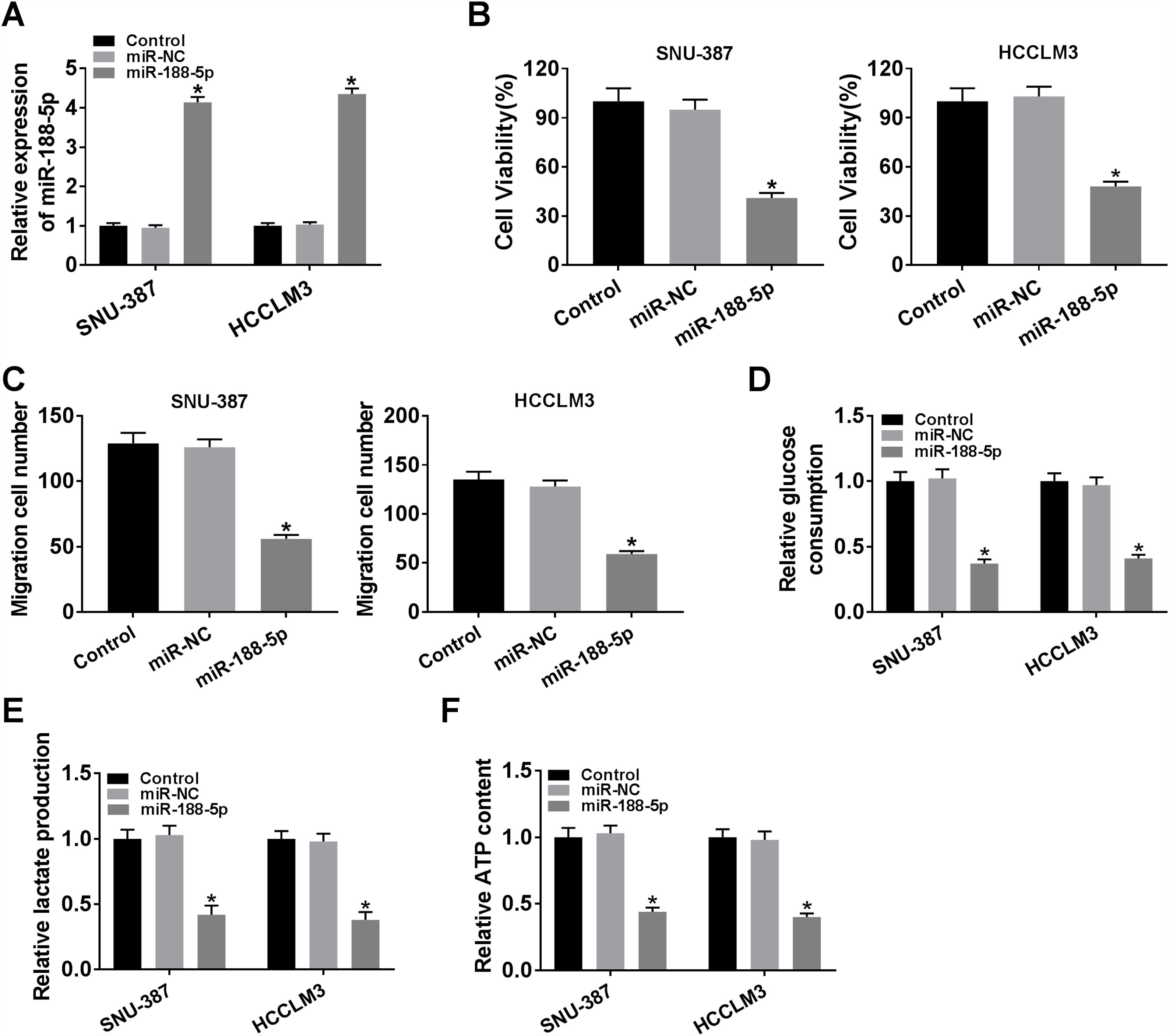
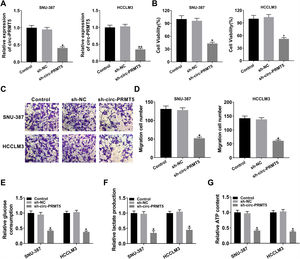
The expression level of miR-188-5p was overly strengthened in SNU-387 and HCCLM3 cells transfected with miR-188-5p compared with miR-NC-transfected cells (Fig. 3A). MTT assay results indicated that upregulation of miR-188-5p blocked cell proliferation (Fig. 3B). Besides, the results of the transwell migration assay suggested that the migration ability of SNU-387 and HCCLM3 cells transfected with miR-188-5p was declined compared to control or miR-NC group (Fig. 3C). Importantly, overexpression of miR-188-5p led to reduced glucose consumption, lactate production, and ATP content in SNU-387 and HCCLM3 cells (Fig. 3D–F). Thus, the above data indicated that miR-188-5p may be a tumor-suppressor gene in HCC.
Influences of miR-188-5p overexpression on proliferation, migration and glycolysis of hepatocellular carcinoma cells. (A–F) SNU-387 and HCCLM3 cells were divided into three groups: Control, miR-NC, and miR-188-5p groups. (A) The relative expression level of miR-188-5p in SNU-387 and HCCLM3 cells was determined by RT-qPCR. (B) The cell viability of SNU-387 and HCCLM3 cells was evaluated by MTT assay. (C) The migration ability of SNU-387 and HCCLM3 cells was assessed by transwell migration assay. (D–F) Glucose consumption, lactate production, and ATP content in SNU-387 and HCCLM3 cells were shown and normalized to the value detected in the Control group. *P<0.05.
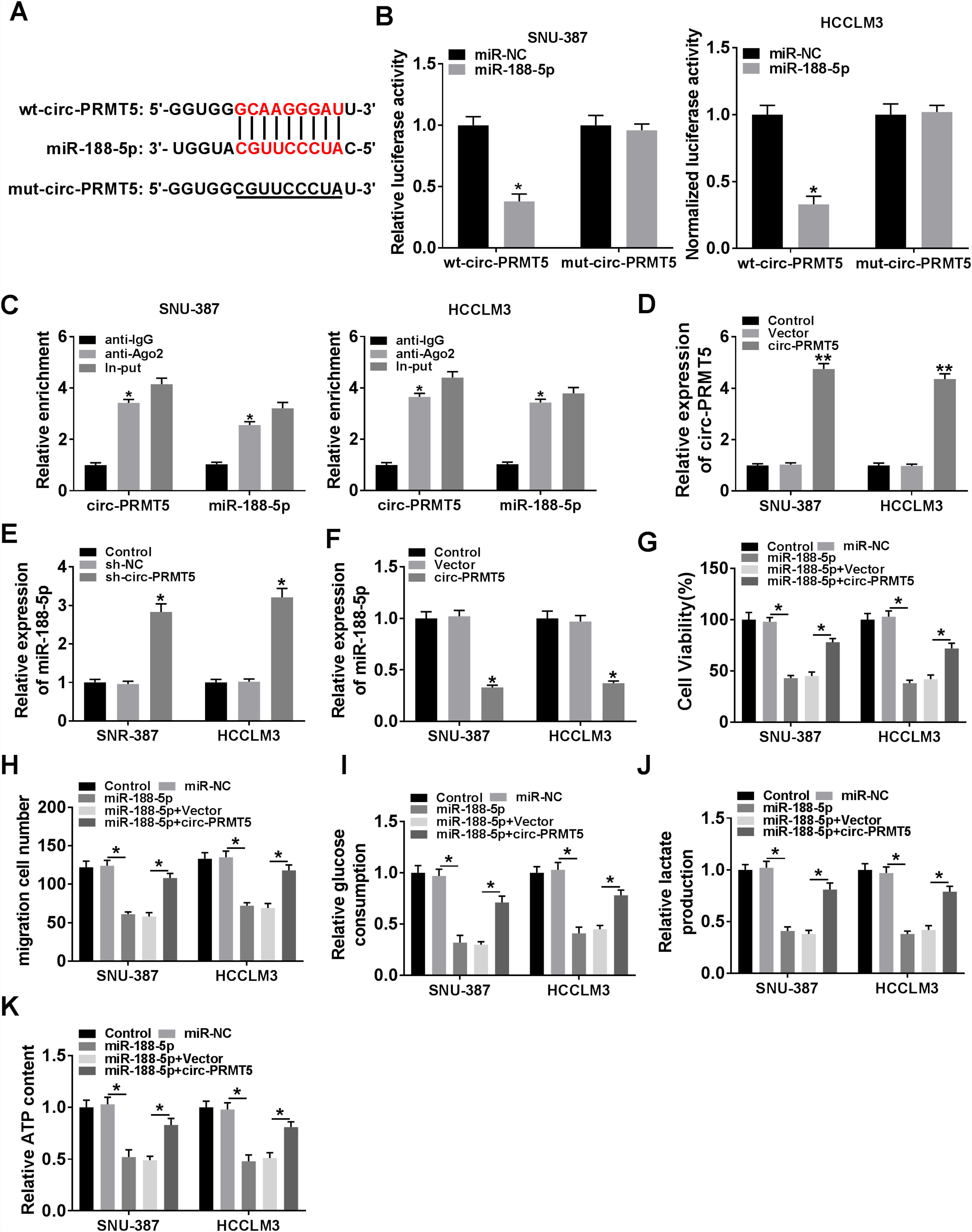
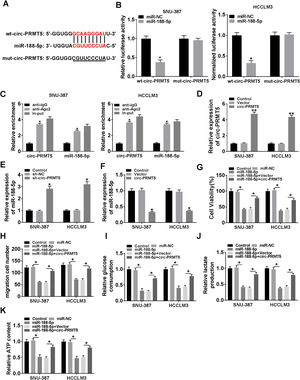
Bioinformatics assay suggested that there was one putative miR-188-5p binding site in the circ-PRMT5 (Fig. 4A). To figure out whether miR-188-5p was targeted by circ-PRMT5, dual-luciferase reporter and RIP assays were conducted. We found that luciferase activity of wt-circ-PRMT5 in HCC cells transfected with miR-188-5p was obviously inhibited compared with the control group, whereas mut-circ-PRMT5 blocked the miR-188-5p-induced repression effects on luciferase activity (Fig. 4B). RIP assay revealed that a prominent enrichment of circ-PRMT5 and miR-188-5p in Ago2-immunoprecipitated complex when compared to the anti-IgG group (Fig. 4C). Additionally, circ-PRMT5 was distinctly increased in the cells transfected with circ-PRMT5 than those transfected with empty vector or control group (Fig. 4D). The expression level of miR-188-5p was elevated in SNU-387 and HCCLM3 cells transfected with sh-circ-PRMT5, while opposite results were found in SNU-387 and HCCLM3 cells transfected with circ-PRMT5, suggesting that circ-PRMT5 can negatively regulate the miR-188-5p expression (Fig. 4E and F). Meanwhile, upregulation of miR-188-5p can impair proliferation ability of HCC cells, however, repression effects on cell growth ability can be partly rescued by simultaneously overexpressing of circ-PRMT5 in SNU-387 and HCCLM3 cells (Fig. 4G). Analogously, upregulation of circ-PRMT5 markedly counteracted suppression effect on cell migration in SNU-387 and HCCLM3 cells caused by overexpression of miR-188-5p (Fig. 4H). Furthermore, upregulation of circ-PRMT5 rescued the decreased glucose consumption, lactate production, and ATP level in SNU-387 and HCCLM3 cells caused by overexpression of miR-188-5p (Fig. 4I–K). All the data revealed that circ-PRMT5 regulated proliferation, migration and glycolysis of HCC cells by targeting miR-188-5p.
MiR-188-5p was a direct target of circ-PRMT5 and upregulation of miR-188-5p-mediated effects on proliferation, migration and glycolysis of hepatocellular carcinoma cells could be abolished by overexpression of circ-PRMT5. (A) Binding region between miR-188-5p and circ-PRMT5, as well as matched mutant sites were shown. (B) The relative luciferase activity was examined in SNU-387 and HCCLM3 cells co-transfected with miR-188-5p mimic or miR-NC and luciferase reporter vectors wt-circ-PRMT5 or mut-circ-PRMT5. (C) RIP assay was performed in SNU-387 and HCCLM3 cells using Ago2 antibody, then the enrichment of miR-188-5p and circ-PRMT5 was detected. (D) The expression level of circ-PRMT5 was examined by RT-qPCR assay in SNU-387 and HCCLM3 cells transfected with circ-PRMT5 or Vector, with un-transfected cells as Control group. (E and F) RT-qPCR assay was conducted to evaluate the expression level of miR-188-5p in SNU-387 and HCCLM3 cells transfected with circ-PRMT5, Vector, sh-NC, or sh-circ-PRMT5 with un-transfected cells as Control group. (G-K) SNU-387 and HCCLM3 cells were transfected with miR-NC, miR-188-5p, miR-188-5p+Vector, or miR-188-5p+circ-PRMT5. (G) MTT assay was introduced to assess the cell viability of SNU-387 and HCCLM3. (H) The migration ability of SNU-387 and HCCLM3 cells was evaluated using transwell migration assay. (I–K) Glucose consumption, lactate production, and ATP content in SNU-387 and HCCLM3 cells were shown. *P<0.05.
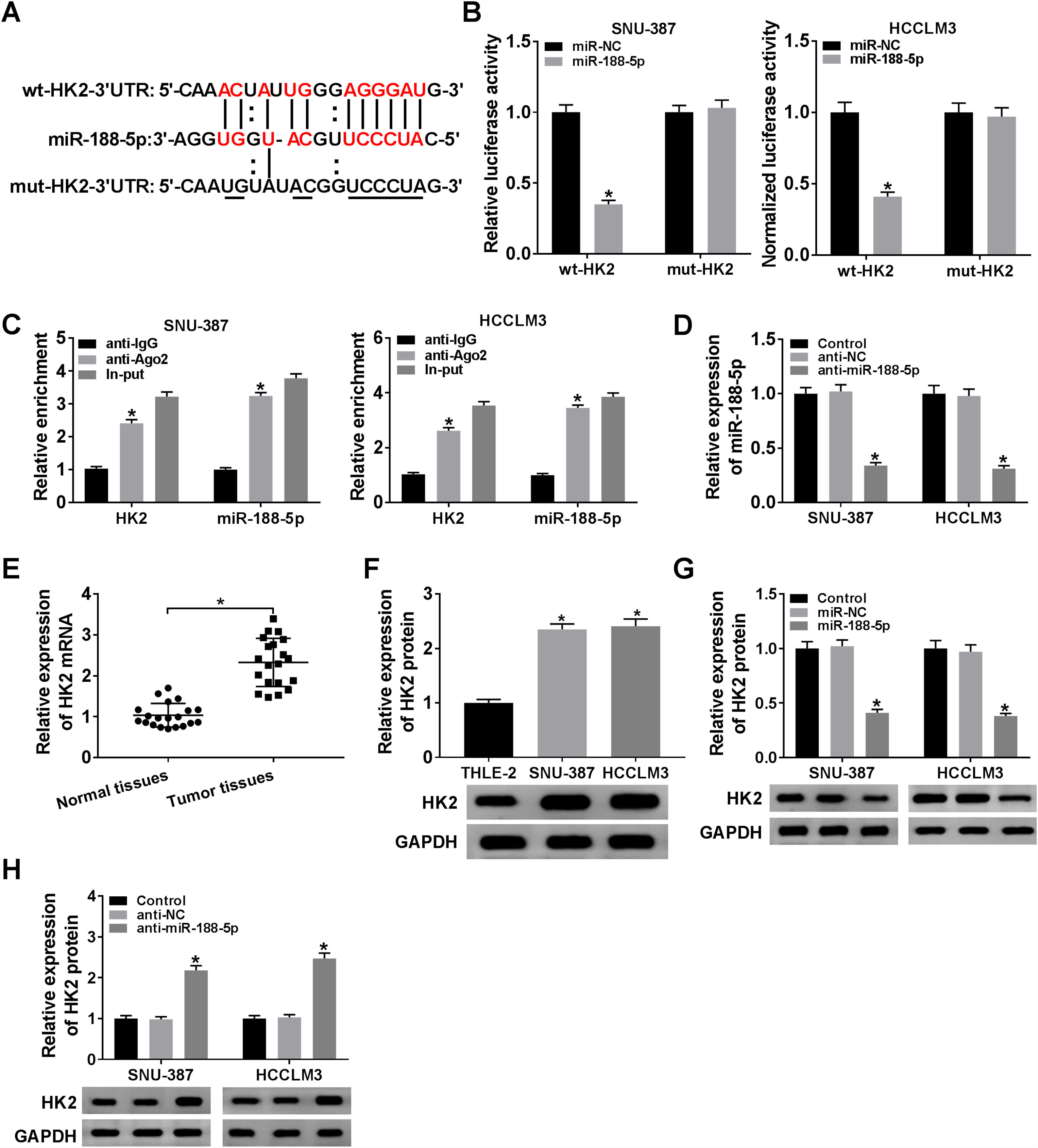
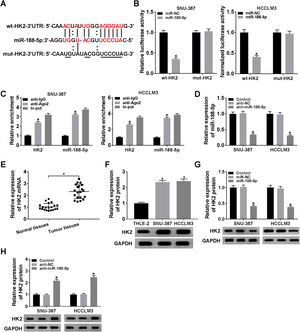
The miR-188-5p binding sites in the 3′UTR of the HK2 mRNA and matched mutated sites are shown in Fig. 5A. We also observed that the miR-188-5p mimic declined the luciferase activity of the wt-HK2, while luciferase activity of the mut-HK2 was not changed by miR-188-5p mimic (Fig. 5B). Moreover, HK2 was enriched in RIP-Ago2 group compared with that in RIP-IgG group (Fig. 5C). The expression level of miR-188-5p was estimated by RT-qPCR assay, and the results suggested that miR-188-5p was decreased in SNU-387 and HCCLM3 cells infected with anti-miR-188-5p than control (Fig. 5D). In addition, the mRNA and protein expression levels of HK2 in HCC were notably reinforced than matched controls by performing RT-qPCR and western blot assays (Fig. 5E and F). More importantly, transfection miR-188-5p into SNU-387 and HCCLM3 cells resulted in a decrease of protein expression level of HK2, while anti-miR-188-5p triggered the opposite results (Fig. 5G and H). In summary, HK2 was negatively regulated by miR-188-5p in HCC cells.
HK2 was regulated by miR-188-5p. (A) The complementary sequences between HK2 and miR-188-5p we as well as mutated nucleotides of HK2 3′UTR were shown. (B) Dual-luciferase reporter assay was used to confirm the interaction between HK2 and miR-188-5p. (C) After performing RIP assay in SNU-387 and HCCLM3 cells, the enrichment of HK2 and miR-188-5p were detected. (D) The interference efficiency of anti-miR-188-5p was checked RT-qPCR analysis. (E–H) The mRNA and protein expression levels of HK2 in hepatocellular carcinoma tissues and cells as wells matched controls, or SNU-387 and HCCLM3 cells infected with miR-NC, miR-188-5p, anti-NC, or anti-miR-188-5p were assessed by RT-qPCR and western blot assays, respectively. *P<0.05.
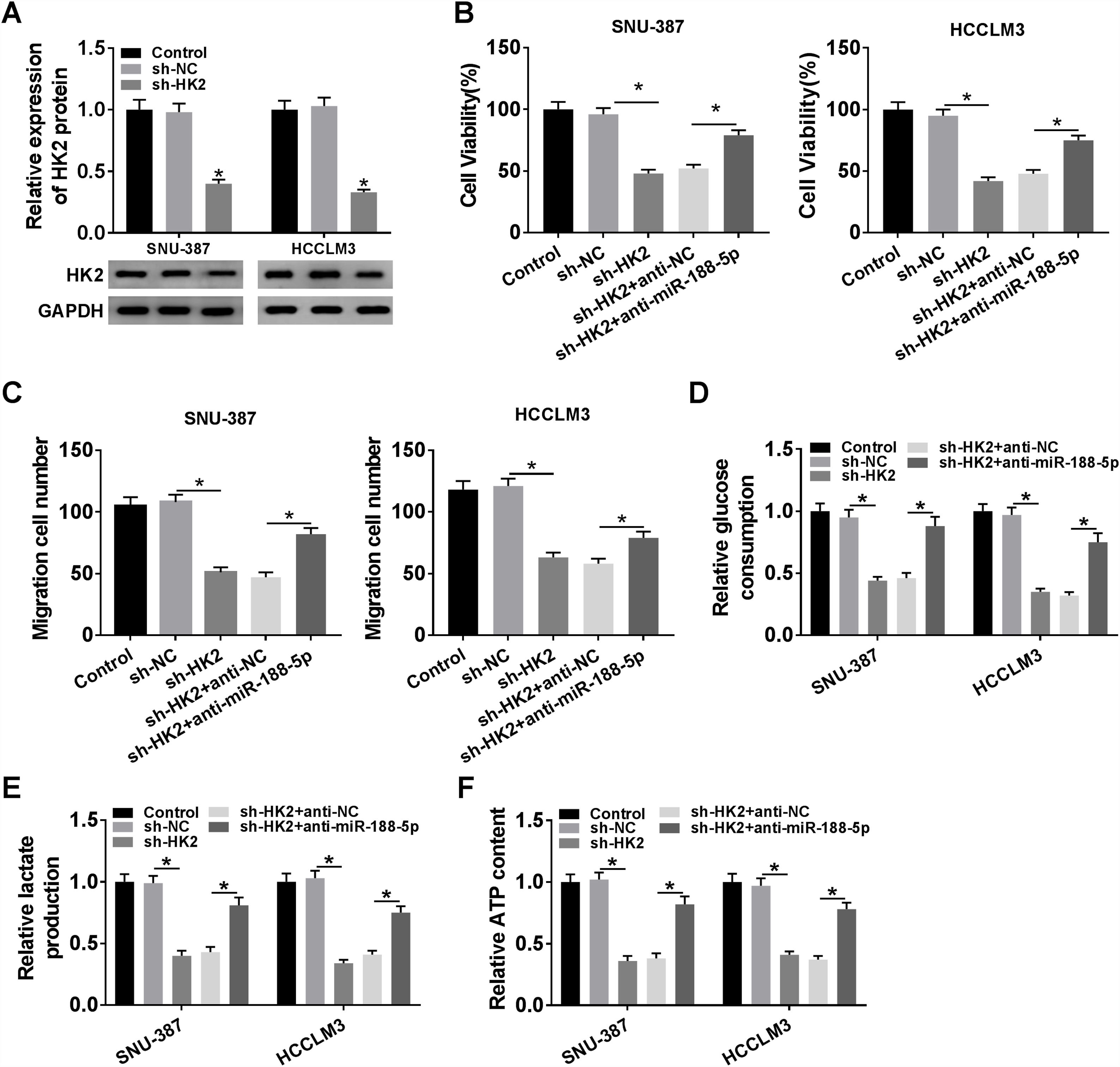
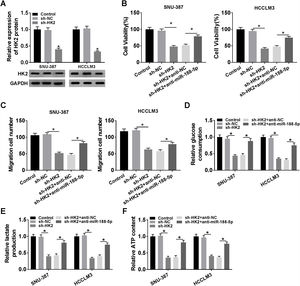
Transfection with sh-HK2 into SNU-387 and HCCLM3 cells could impede HK2 expression, as demonstrated in Fig. 6A. Furthermore, cell proliferation capacity was restored in SNU-387 and HCCLM3 cells co-transfected with sh-HK2 and anti-miR-188-5p compared with those cells alone transfected with sh-HK2 (Fig. 6B). The suppression of cell migration triggered by sh-HK2 was partly abolished by transfection of miR-188-5p inhibitor (Fig. 6C). The sh-HK2 exerted a suppressor function on glycolysis by limiting glucose consumption and generating lactate and ATP, which was reversed by silencing miR-188-5p (Fig. 6D–F). These findings revealed that inhibition of miR-188-5p expression could reverse the effects of HK2 silencing on proliferation, migration and glycolysis of HCC cells.
Repression of miR-188-5p abolished effects of HK2 silencing on proliferation, migration and glycolysis of hepatocellular carcinoma cells. (A) The knockdown efficiency of sh-HK2 in SNU-387 and HCCLM3 cells was checked by RT-qPCR assay. (B–F) SNU-387 and HCCLM3 cells were transfected with sh-NC, sh-HK2, sh-HK2+anti-NC, or sh-HK2+anti-miR-188-5p. (B) MTT assay was performed for examining the cell viability of SNU-387 and HCCLM3 cells. (C) Cell migration assay was conducted in transfected SNU-387 and HCCLM3 cells. (D-F) Glucose consumption, lactate production and ATP level of transfected SNU-387 and HCCLM3 cells were calculated. *P<0.05.
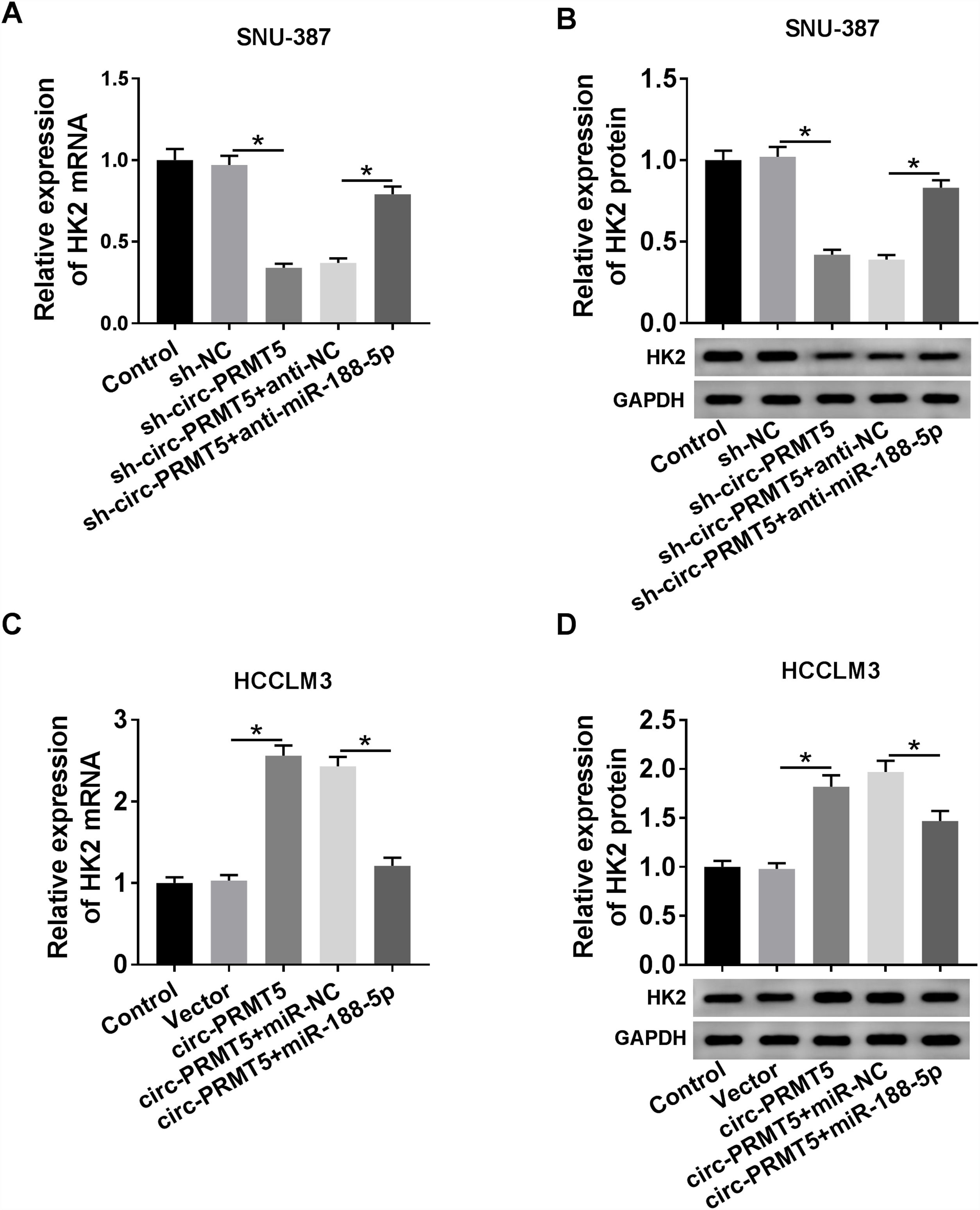
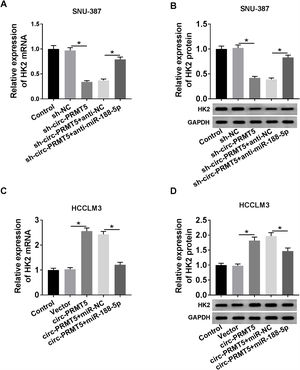
RT-qPCR and western blot assays showed that co-transfection with sh-circ-PRMT5 and anti-miR-188-5p could partly increase the mRNA and protein expression levels of HK2 which were repressed by downregulation of circ-PRMT5 in SNU-387 cells (Fig. 7A and B). We also found that mRNA and protein expression levels of HK2 were enhanced by transfection with circ-PRMT5, which was abrogated by reintroduction with miR-188-5p in HCCLM3 cells (Fig. 7C and D). Collectively, circ-PRMT5 regulated miR-188-5p/HK2 axis in HCC cells.
Circ-PRMT5 regulated HK2 expression by sponging miR-188-5p. (A–D) RT-qPCR and western blot assays were carried out to examine HK2 level in SNU-387 cells transfected with sh-NC, sh-circ-PRMT5, sh-circ-PRMT5+anti-NC, sh-circ-PRMT5+anti-miR-188-5p, or HCCLM3 cells transfected with Vector, circ-PRMT5, circ-PRMT5+miR-NC, or circ-PRMT5+miR-188-5p, un-transfected cells were served as control group. *P<0.05.
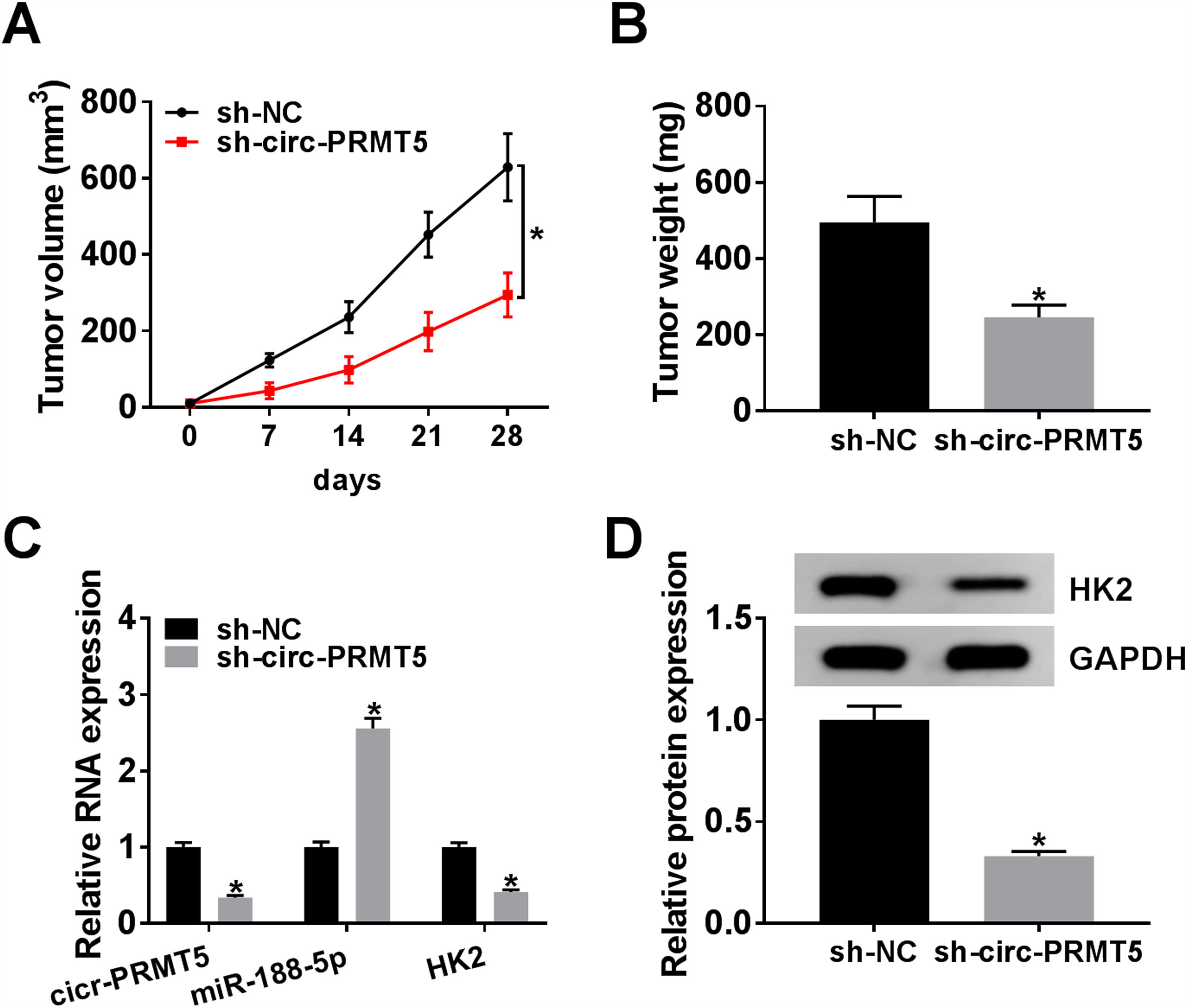
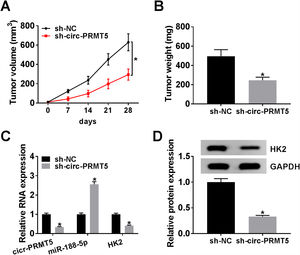
The animal model for in vivo research was established, and our data disclosed that tumors in the sh-circ-PRMT5 group grew more slowly and were smaller with respect to tumors from sh-NC group (Fig. 8A and B). Furthermore, we found that the expression levels of circ-PRMT5 and HK2 were repressed while miR-188-5p was increased in the sh-circ-PRMT5 group compared with the sh-NC group (Fig. 8C). Consistently, the protein expression level of HK2 was declined in the sh-circ-PRMT5 group when compared with sh-NC group (Fig. 8D). Taken together, cell growth was significantly impeded in vivo by knockdown of circ-PRMT5.
Knockdown of circ-PRMT5 impeded tumor growth. (A and B) The growth curves and weight of xenograft tumors were shown. (C) The RNA expression levels of circ-PRMT5, miR-188-5p and HK2 in dissected tumor tissues from different groups were measured with RT-qPCR assay. (D) Western blot assay was used to assess the protein expression level of HK2 in dissected tumor tissues. *P<0.05.
Currently, glycolysis has been reported to be associated with the development of malignant tumors, among which the relevance of oxidative stress, accelerated metabolism, and metastasis [21,22]. The energy source and intermediate macromolecular production of cancer cells were depended on decomposing glucose by glycolytic instead of mitochondrial oxidative phosphorylation [23]. Moreover, the deranged glucose metabolism might contribute to the tumor growth and endue malignant behavior of malignant tumors [24]. Additionally, the abnormal expression of circRNA has received great attention in numerous tumors, including HCC. For example, Su et al. observed that hsa_circ_0070269 impeded the aggressive behavior of HCC by mediation miR-182/NPTX1 pathway [25]. Zhu et al. revealed that circular RNA PVT1/miR-203/homebox D3 axis promoted HCC through regulation growth and migration of HCC cells [26].
Emerging evidence has identified the oncogenic role of circ-PRMT5 in various human cancers, such as bladder cancer and lung cancer [9,10]. Consistently, we also confirmed that circ-PRMT5 was overexpressed in the HCC tissues and cells than conrrols. In addition, circ-PRMT5 was negatively correlated with miR-188-5p expression by Pearson's correlation coefficient analysis. The silencing of circ-PRMT5 repressed cell growth, migration and glycolysis of HCC cells. Furthermore, we revealed that miR-188-5p-induced effects on HCC cells could be alleviated by overexpression of circ-PRMT5, supporting the critical regulatory role in growth, migration and metastasis by the circ-PRMT5/miR-188-5p regulatory axis.
Moreover, upregulation of miR-188-5p led to obvious repression of glucose consumption as well as lactate and ATP production, suggesting that miR-188-5p may inhibit HCC progression by blocking glycolysis. In addition, overexpression of miR-188-5p impeded proliferation and migration of HCC cells. Ma et al. and Cheng et al. also found that miR-188-5p exerted anti-tumor effects in HCC [27,28]. Furthermore, it has been testified in HCC, miR-188-5p can also directly target certain oncogene (fibroblast growth factor 5) [17]. Analogously, we pointed out that miR-188-5p was a critical negative regulator of the HK2 expression by performing functional experiments. Coincidentally, HK2 played a critical role in glycolysis [18], and abnormal expression of HK2 was involved in cancer cell metabolism, suggesting that HK2 may be therapeutically targetable for malignant tumor [29,30].
In this study, we further confirmed that HK2 acted as a novel target of miR-188-5p. Additionally, the silencing of HK2 inhibited cell growth, migration and glycolysis of HCC cells. More interestingly, knockdown of miR-188-5p could abolish the inhibition effects on growth, migration and glycolysis of HCC cells induced by silencing of HK2. Conclusively, our study identified that circ-PRMT5 acted as an endogenous sponge of miR-188-5p to positively regulate HK2 expression. Moreover, as a novel regulatory mechanism, circ-PRMT5/miR-188-5p/HK2 axis may be a therapeutic target in the future treatment of HCC.
5ConclusionCollectively, circ-PRMT5 was upregulated in HCC tissues and cells than that in matched control. In addition, inhibition of circ-PRMT5 can effectively suppress proliferation, migration and glycolysis in HCC cells. The interaction relationship among circ-PRMT5, miR-188-5p, and HK2 was discovered in HCC, importantly, circ-PRMT5/miR-188-5p/HK2 axis might serve as a potential therapeutic target for HCC.AbbreviationscircRNA
circular RNA
HCChepatocellular carcinoma
RIPRNA immunoprecipitation
Authors’ contributionsZhenghua Ding conceptualized the study and wrote the manuscript. Zhongming Deng, Peng Li collected the data and analyzed the data. All the authors revised and approved the final version.
FundingNone.
Conflict of interestThe authors declare that they have no financial conflicts of interest.