Free and conjugated bile acids (BA's) cannot cross cell membranes; therefore, a particular transport system is required by the cell. Members of the family of ABC (ATP-binding proteins) transporters transfer bile acids in and out of the cell, preventing their accumulation. High intracellular concentrations of bile acids, such as those observed in cholestasis, have been related to oxidative stress and apoptosis, which in many cases are the leading causes of hepatocyte damage. MRP3 and MRP4 (multidrug resistance-associated protein 3 and 4) proteins belong to the ABC subfamily C, and are transporters of the hepatocyte's basolateral membrane with a compensatory role. Both transporters’ increased expression constitutes an essential role in the protective and adaptive responses of bile acid overload, such as cholestasis. This work aimed to analyze both transporters’ mRNA and protein expression in an in vitro model of cholestasis using HepG2 cell line treated with main bile acids.
MethodsThe expression of transporters was investigated through confocal microscopy immunofluorescence, Western Blot, and RT-qPCR after the main bile acids in HepG2 line cells.
ResultsThe results showed the relation between confluence and expression of both transporters in the plasma membrane. MRP3 showed atypical and heterogeneous distribution in this cell line. CDCA (chenodeoxycholic acid) at low concentrations induced the expression of mRNA of both transporters. In contrast, protein expression was induced by CA (cholic acid) at high concentrations.
ConclusionPrimary bile acids (CDCA and CA) induce overexpression of the MRP4 and MRP3 transporters in the HepG2 cell line.
ATP-binding cassette
ATP-binding protein subfamily C
ATP-binding protein subfamily C member 3-4
cholic acid
chenodeoxycholic acid
deoxycholic acid
glycocholic acid
glycochenodeoxycholic acid
lithocholic acid
multidrug resistance-associated protein 3-4
3-4, 4,5-dimethyl thiazolyl-2)-2,5 diphenyltetrazolium
reactive oxygen species
retro transcriptase-quantitative polymerase chain reaction
The bile acids (BA's) are the main components of bile and are produced exclusively by hepatocytes. They are known for their amphipathic property to participate in the solubilization and emulsion of fatty acids, xenobiotics, and fat-soluble vitamins. BA's also stimulate lipid bile secretion and have the ability to produce mixed micelles with bile phospholipids to solubilize cholesterol and other lipid compounds; therefore, these micelles emulsify fats and vitamins from the diet, participate in calcium absorption [1], and modulate the release of pancreatic secretion and cholecystokinin [2].
During cholestatic diseases, the increase in BA's levels induces oxidative stress and apoptosis, leading to liver parenchymal damage [1]. High concentrations of BA's in feces (secondary, see below), blood and bile, have been associated with other diseases such as cholestasis, Barret's esophagus, liver and colon cancers [3], liver cirrhosis, diabetes mellitus[4,5] and other diseases related to genetic alterations in synthesis, biotransformation, and transport, conducting to hepatic alterations [1].
Although having basic chemical structures, there are different BA's, since they exhibit various physical properties and biological effects [1]. Their biological activity is related to the chemical properties, such as the number and orientation of hydroxyl groups or their conjugation with aminoacids, affecting directly their hydrophobicity, which is a predictor of toxicity and lithogenicity that in turn influences the cellular signaling dependent on bile acid, regulating the interaction of BA's with their receptors. They are primary BA's, which have been synthesized directly by cholesterol from pericentral hepatocytes. The most abundant of these are cholic acid and chenodeoxycholic acid. When subsequently conjugated mainly with glycine or taurine, they are called conjugated BA's, such as glycocholic acid and glycochenodeoxycholic acid. Finally, they are secreted to the gallbladder and reserved in this organ. After passing through the large intestine, they become secondary BA's by the process of oxidation produced by bacterial enzymes, and the most abundant are deoxycholic acid and lithocholic acid, which are derived from the above.
Free and conjugated bile acids do not cross the cell membrane by theirself, they require transporters of the ABC family, which mediate the transport of various substrates through the cell membranes at the expense of ATP hydrolysis and are responsible for the efficient in and out moving in the cell's membranes [6,7], these transporters protect the cells from the detergent properties of the BA'S in humans [8]. There are nine members of the ABCC subfamily, also referred to as MRP's [9]. One member of this subfamily, MRP4 (ABCC4), is a carrier with a broad substrate specificity, expressed in diverse human tissues, including basolateral and apical plasma membranes from liver and kidneys, respectively [10–12]. It works as an outflow pump for bile acids together with glutathione offering a possible route for its elimination [13,14]. Another transporter of this subfamily is MRP3 (ABCC3), which similarly mediates the transport of anionic conjugates, preferably glucuronides of endogenous lipophilic substances, xenobiotics [15], and sulfated bile salts [16]. The location of MRP3 is controversially discussed; in humans, MRP3 is located at the basolateral membrane of hepatocytes [17,18] and in orthologue rats in the canalicular membrane [19].
This work aimed to evaluate the expression and localization of MRP3 and MRP4 transporters in a cholestasis in vitro model using HepG2 cell line treated with the main human bile acids (primary: cholic acid and chenodeoxycholic acid, their respective conjugates: glycocholic acid and glycochenodeoxycholic acid, and respective secondary bile acids: deoxycholic acid and lithocholic acid). This study was conducted as an approximation to know what happens in cholestasis.
2Material and methodsBile salts: cholic Acid (CA), glycocholic acid (GCA), deoxycholic acid (DCA), chenodeoxycholic acid (CDCA), glycochenodeoxycholic acid (GCDCA), lithocholic acid (LCA) all provided by Sigma & Aldrich, (CA, USA). The bile salts were dissolved in dimethyl sulfoxide (DMSO), Sigma & Aldrich, CA, USA). HepG2 cells were cultured in DMEM advanced medium supplemented with 5% fetal bovine serum, 4mM glutamine and 100U/ml penicillin/streptomycin (all provided by Gibco, Thermo Fisher Scientific, Waltham, MA, USA), at 37°C and 5% CO2, harvested with 0.1% trypsin-EDTA. HepG2 cells are a human hepatoma cell line derived from a liver biopsy of a 15 years old Caucasian male with hepatocellular carcinoma. This cell line has high proliferation rates, epithelial-like morphology polarization, and it can form bile canaliculi-like structures between adjacent cells. However, HepG2 cells express poorly many transporter proteins and metabolize enzymes, making them more sensitive than a primary hepatocyte [20].
2.1Viability assayViability assay was performed with the MTT reagent (4,5-dimethyl thiazolyl-2)-2,5 diphenyltetrazolium) (Roche, Basel, Switzerland), following the manufacturer's instructions. 8×104 cells seeded in culture medium were allowed to grow overnight, then bile acids were given separately at different concentrations and incubated 4h at 37°C and 5% CO2. The medium was removed, the cells were washed with PBS, and then the MTT reagent (1μg/ml) was added and incubated for 3h at 37°C and 5% CO2. After this time, the cells were washed, the formazan solubilized with cold isopropanol (Sigma & Aldrich, (CA, USA), and the absorbance was obtained at OD570.
2.2Subcellular fractionationCells incubated with the bile acids were collected and washed 2× with cold PBS and treated with lysis buffer (10mM HEPES pH 7.9, 1.5mM MgCl2, 10mM KCl, 0.5mM DTT, IP-3X, NP-40 1%) for 25min at 4°C. The cell extract was then centrifuged at 14,000rpm 25min, and the supernatant containing the cytoplasmic fraction was recovered and stored at -20°C until use.
2.3Western Blot and immunofluorescenceCytoplasmic fraction was separated by 12% SDS-PAGE and transferred to 0.45μm nitrocellulose membrane (Bio-RAD, Marnes-la-Coquette, France). It was incubated with mouse anti-MRP4 antibody (Santa Cruz sc-59614/1:200, Santa Cruz, CA, USA), then, membranes were incubated with peroxidase-conjugated goat anti-rat IgG antibody (ab6845/1:1000, Abcam, Cambridge, MA, USA). Finally, the antibody-reactive protein complexes were developed with the luminol substrate (Immobilon, Millipore, Saint-Quentin en Yvelines, France) and acquiring the image in the software iS (image Studio, LI-COR, Biosciences, Nebraska, USA). Densitometric analysis was performed with ImageJ software.
The subcellular localization of MRP3 and MRP4 was achieved by confocal microscopy. Cultured cells on coverslips were washed with PBS and fixed with cold 95% methanol for 10min at −20°C. Then, fixed cells were incubated with 10% FBS (fetal bovine serum) for 1h at 37°C, washed with PBS, and incubated with either anti-MRP4 antibody (Santa Cruz, sc-59614/1:50, Santa Cruz, CA, USA) or anti-MRP3 antibody(Santa Cruz, sc-59612/1:25, Santa Cruz, CA, USA). Washed cells were incubated with a secondary antibody coupled to FITC (goat anti-rat, ZyMax™ 81-9511/1:100, Invitrogen, Cergy Pontoise, France) or goat anti-mouse IgG coupled to FITC (Abcam, ab6785/1:2500, Cambridge, MA, USA). The nuclei were counterstained with 4′,6-diamidino-2-phenylindole (DAPI) in VectaShield (Vector Laboratories Inc., Burlingame, CA, USA). All samples were analyzed, and images were acquired with a Carl Zeiss LSM700 confocal microscope, and the Zeiss Zen black and blue editions software was used for analysis (Oberkochen, Germany).
2.4RT-qPCR assayTotal RNA was isolated from the cells with the TRIzol™ Reagent (Invitrogen, Cergy Pontoise, France), following the manufacturer's instructions and verifying its integrity in 1% agarose gels stained with ethidium bromide. Then, after treatment with DNase I (Invitrogen, Cergy Pontoise, France), cDNA was synthesized from 5μg of RNA by using SuperScript II reverse transcriptase (Invitrogen, Cergy Pontoise, France) according to the manufacturer's instructions. Real time-qPCR was performed using NZY qPCR Green Master Mix (2×), ROX (NZYTech, Lisboa, Portugal). Each sample was measured in triplicates in two independent experiments using as control the GAPDH expression. Primer pairs were 5′-TGATGAGCCGTATGTTTTGC-3′ and 5′-CTTCGGAACGGACTTGACAT-3′ for MRP4; 5′-AAAAGCAGACGGCACGACA-3′ and 5′-GCAGGCACTGATGAGGAAGC-3′ for MRP3; 5′-CTTTGGTATCGTGGAAGGACTC-3′ and 5′-GTAGAGGCAGGGATGATGTTCT-3′ for GAPDH. Thermal cycling conditions were 95°C for 2min, followed by 40 cycles of 95°C for 2s, 60°C for 30s. The data were obtained with the 7500 Applied Biosystem thermocycler (CA, USA). Relative expression levels were calculated by the Pfaffl modification of the ΔΔCt method [21].
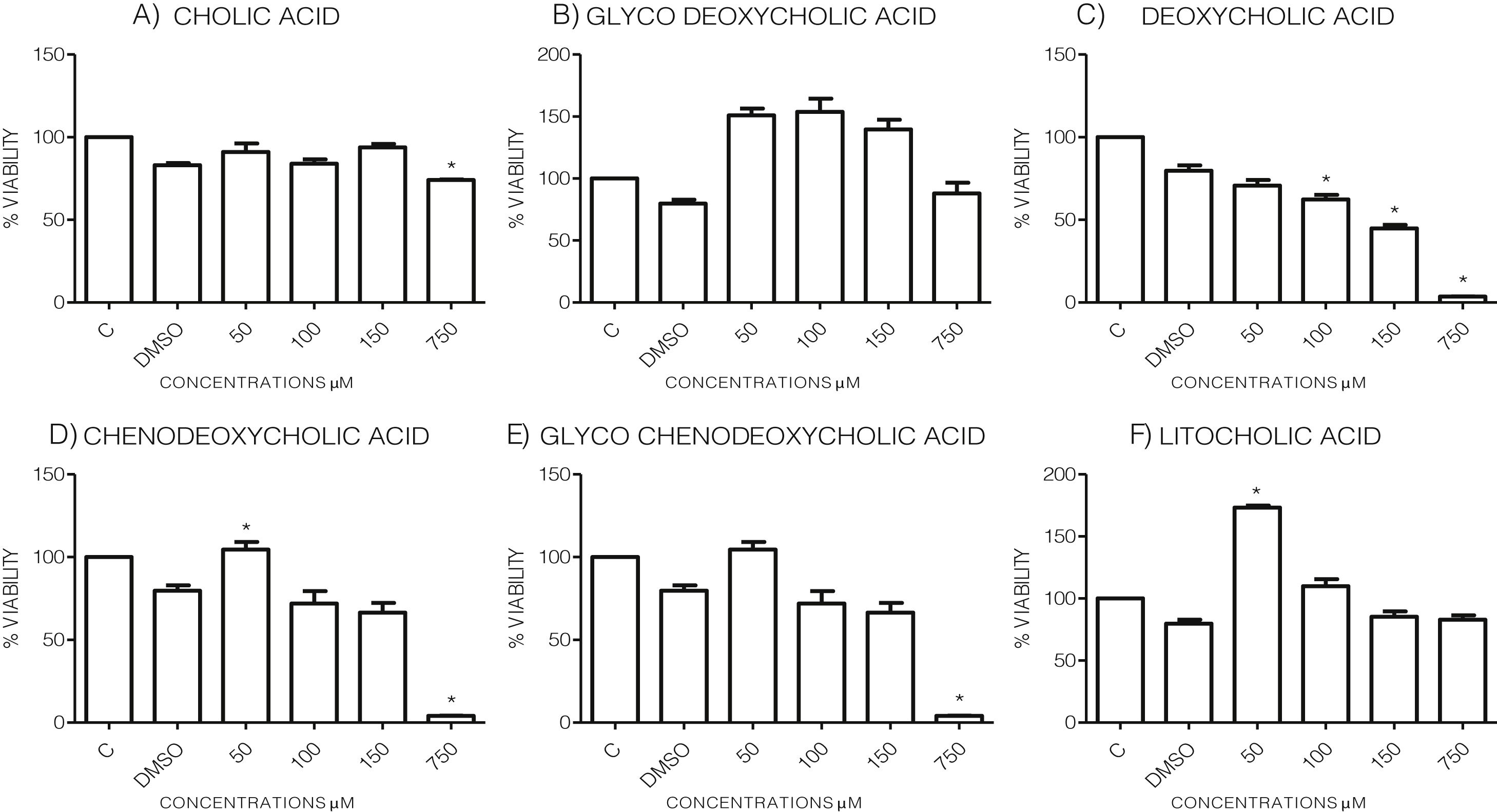
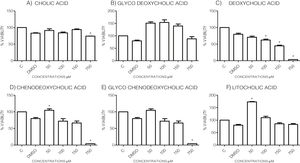
3Results3.1Effect of bile acids on the viability and morphology of HepG2 cellsInitially, we evaluated the effects of various concentrations of BA's on cell viability through MTT tests using the maximum amount of DMSO (3.3%, v/v) as vehicle control. As shown in the graph (Graph 1), each bile acid exerts different effects on the viability and metabolic activity of cells. It can be seen that at the highest concentrations tested (750μM), the treatments DC, CDCA, GCDCA (Graph. 1B, D and E) lysed the culture after 4h of incubation, however CA, GCA, LCA preserved more than 74% the cell viability at this concentration. While low concentrations of GCA, CDCA, GCDCA, and LCA (Graph. 1B and D–F) caused viabilities above 100%, obviously this is not due to an increase in cell population in 4h, we consider that these discrepancies in the MTT assay could be due to effects from treatment in the cellular mitochondria or oxidative stress.
Viability of cells treated with bile salts. Cultured HepG2 cells were incubated with different concentrations of BA's for 4h and tested for viability trough MTT assay. The highest concentrations tested of DC, CDCA, GCDCA (750μM) produces lysis of cell, while CA, GCA, LCA preserved more than 74% the cell viability at same concentration. At medium concentrations GCA, CDCA, LT (150–50μM), was observed an increase of metabolic activity exceeded 100% compared to the untreated control (details of this phenomenon or discrepancy see in discussion). C: Control untreated cells, DMSO: as vehicle control (3.3%, v/v).
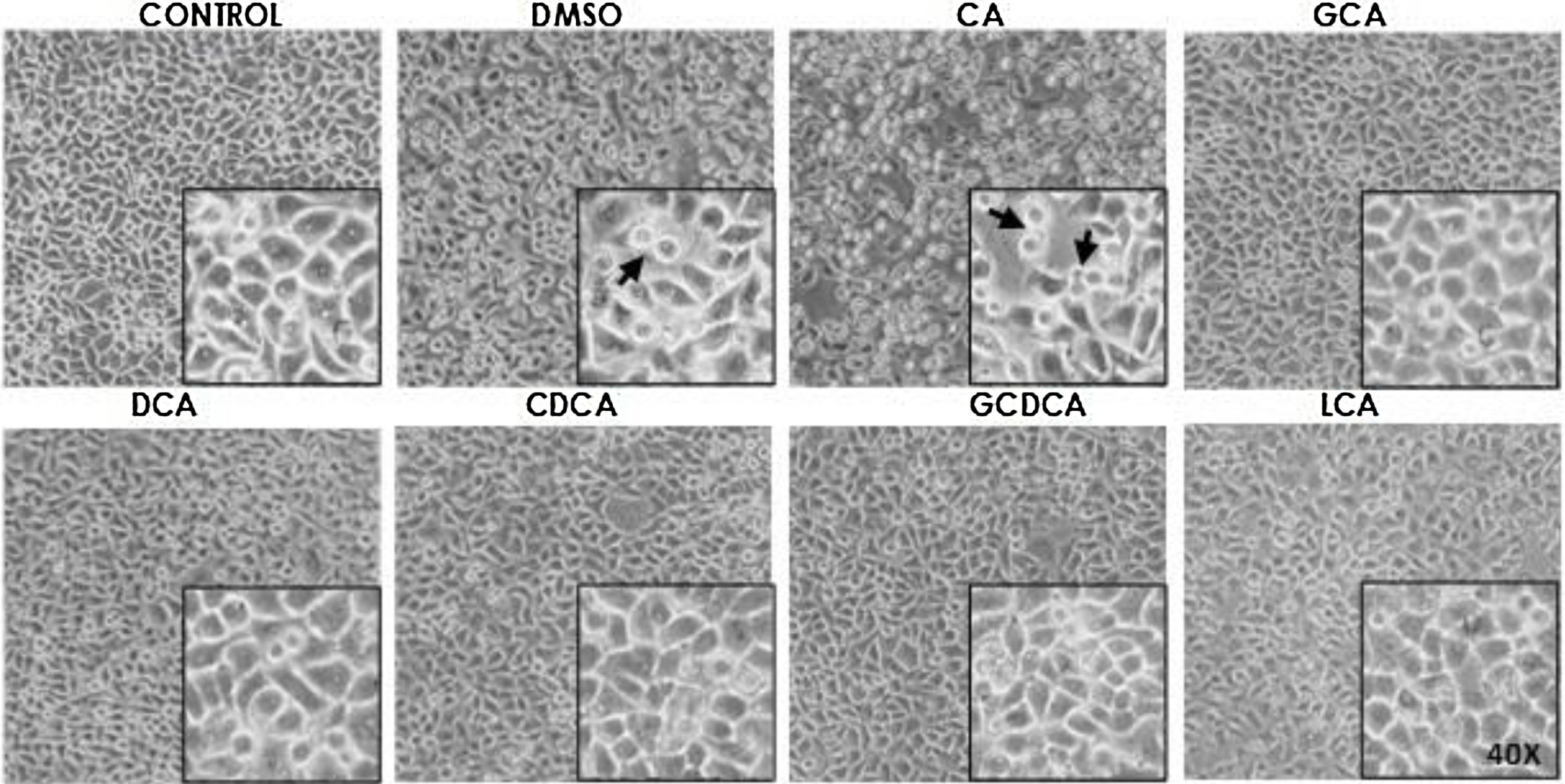
Based on the viability results, in subsequent assays we analyzed the effects of different bile acids on cell morphology. In the case of the treatment with 750μM with CA, rounding of the cells was observed in a similar way to the vehicle (DMSO), while in the other treatments with the concentrations shown in the figures (Fig. 1) they did not show significant morphological changes in the monolayer cell, although they had shown an increase in metabolic activity.
Monolayer of HepG2 cells treated with bile salts. The concentrations of bile salts were selected based on the viability results. No morphological changes were detected in most treatments except in AC, which could be due to the high concentration of this bile acid and vehicle, in these cases displaying round cells with cytoplasm content loss (arrows). Control: untreated cells; DMSO: vehicle control (3.3%, v/v); CA750μM; GCA100μM; DCA:50μM; CDCA150μM; GCDCA100μM; LCA50μM (Inset at 40×).
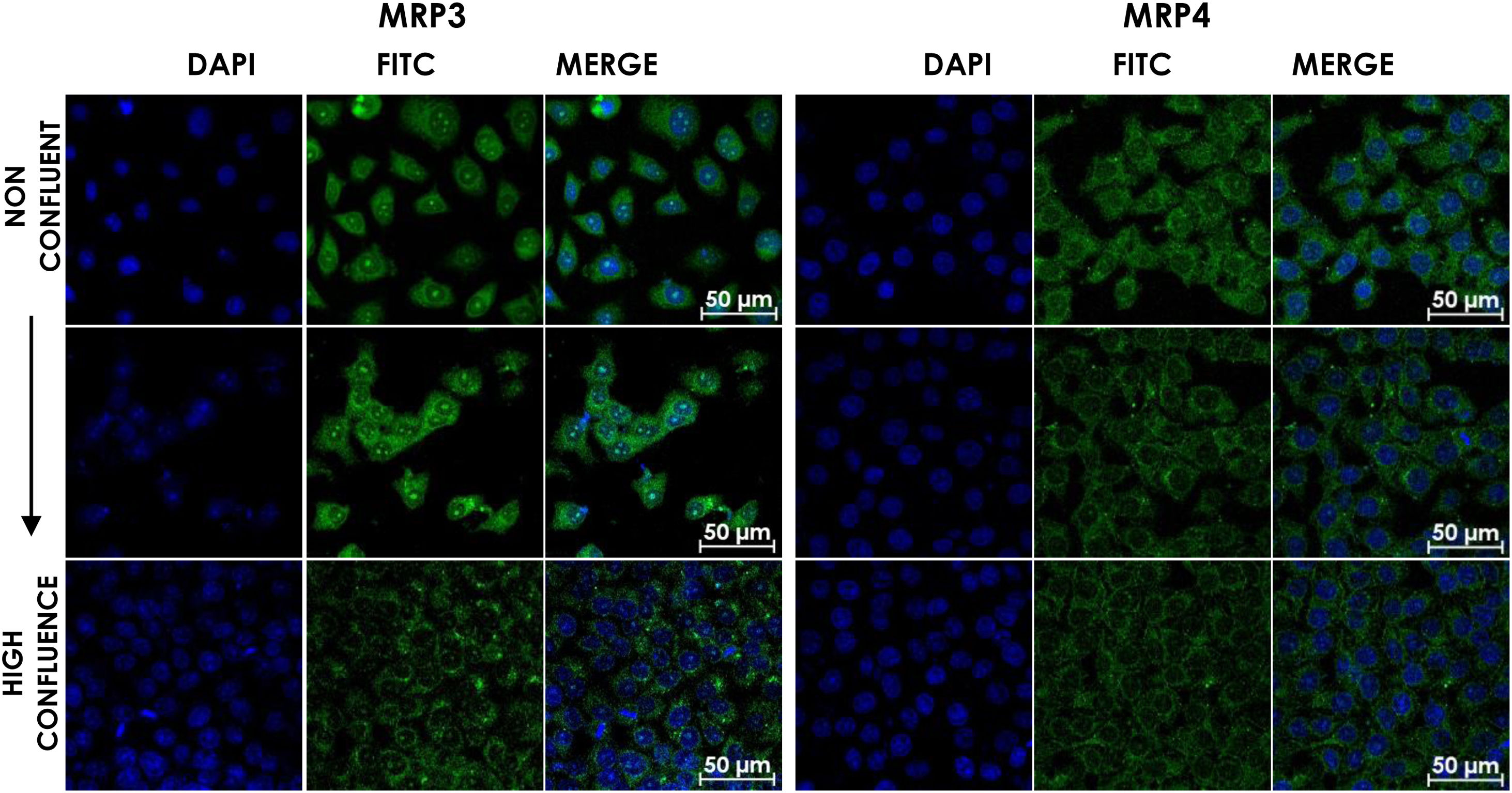
HepG2 cells were seeded at different densities on coverslips and were grown to confluence and fixed and stained for MRP3 y MRP4 as described above. In non-confluent cultures, the antibody label was distributed throughout the cell, especially in the area corresponding to endoplasmic reticulum and nucleolus for MRP4 (Fig. 2A) and MRP3 (Fig. 2B) as the confluence increased. Then, the antibody label moved toward the area belonging to the basolateral membrane and endoplasmic reticulum, as commonly described in the literature in the case of MRP4; for MRP3 at zones of high cell confluences, intense dot labels near the endoplasmic reticulum were observed in some cells, and in others, the label was seen in the basolateral membrane. This transition of the transporters can be observed in the set of images of Fig. 2. As the cell culture's confluence increased, the antibody label migrated toward the basolateral membrane in both cases. The expression of these transporters was correlated with the cell growth or with cell polarization of hepatocytes.
Confocal microscopy analysis MRP4 (A) and MRP3 (B) expression in normal cells at different confluence states. HepG2 cells at different confluences were analyzed for the subcellular distribution with either anti-MRP4 or anti-MRP3 antibodies. In the set of images, cell confluence affects the expression of the transporters MPR3 and MRP4. In non-confluent states, both transporters’ presence is low, while at high confluence, MRP4 is expressed as described in the literature: in the basolateral cell membrane; while MRP3 is expressed in an atypical way in patches distributed randomly in the cytoplasm. Control: untreated cells; DMSO: vehicle control (3.3%, v/v); CA750μM; GCA100μM; DCA:50μM; CDCA150μM; GCDCA100μM; LCA50μM.
Based on previous data, we proceeded to seed cells on glass-slides at high densities and incubate for 4h with different concentrations of bile acids (as shown in captions from Figs. 3 and 4), and the MRP3 and MRP4 transporters were analyzed trough immunofluorescence with confocal microscopy, RT-qPCR and Western Blot.
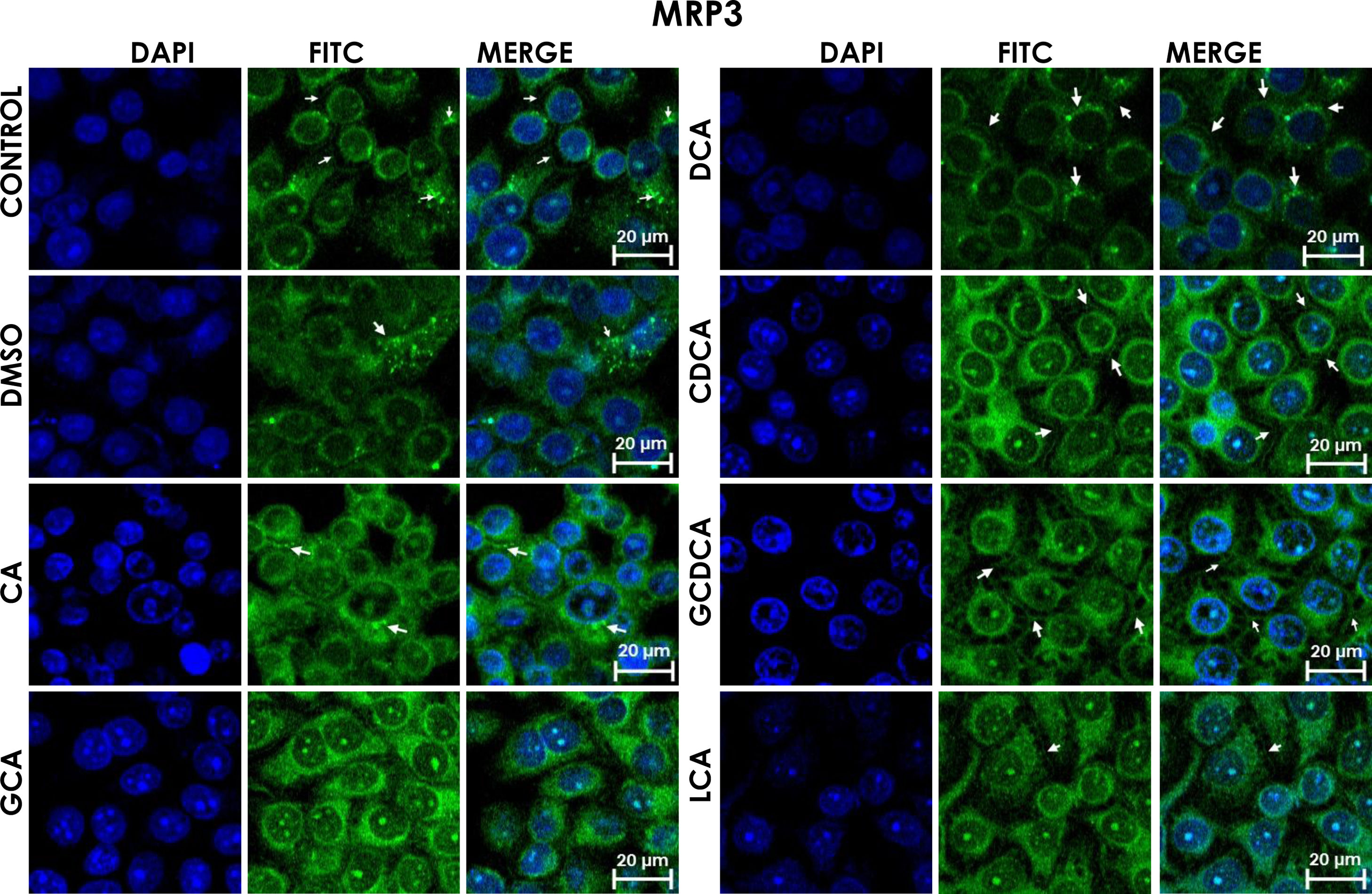
Subcellular distribution of MRP3 in bile acids treated cells. HepG2 cells treated for 4h with bile salts. MRP3 transporter is usually reported in literature in basolateral membrane, however heterologous expression is appreciated in confluent HepG2 cells. In some cells the MRP3 label antibody is located at the basolateral membrane, and in other cells it appears inside of cytoplasm in patches or spots in intracellular compartments and exhibit different fluorescence intensity according to treatment. CA followed by GCA registered more intensity of fluorescence than other treatments. DCA even with low intensity of fluorescence compared with other treatments also showed the atypical expression forming clusters or cell membranes labeled in a heterogeneous distribution (arrows indicate the location of MRP3 protein). Fluorescence quantification is reported in Graph 2A. Control: untreated cells; DMSO: vehicle control (3.3%, v/v); CA750μM; GCA100μM; DCA:50μM; CDCA150μM; GCDCA100μM; LCA50μM.
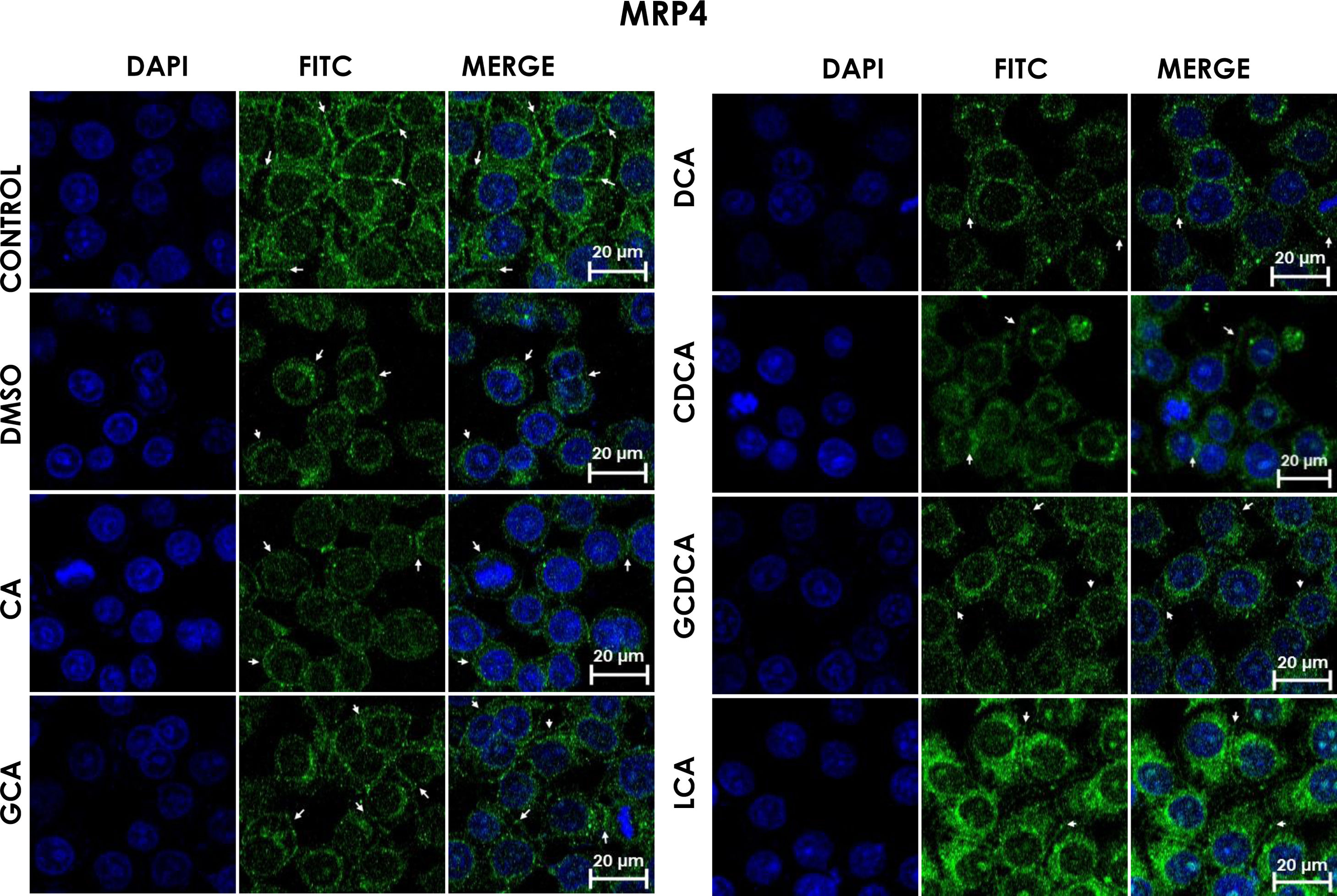
Subcellular distribution of MRP4 in bile acids treated cells. HepG2 cells were treated for 4h with bile salts. As described usually in literature, the label of MRP4 antibody was observed in control cells toward the plasma membrane with an empty cytoplasm as seen in DMSO, CA, DCA treatments; the label of MPR4 antibody was also found in cell membranes with different intensities as compared with LCA treatment, where the MRP4 antibody was predominant in the cytoplasm. It also can be appreciate MRP4 in the basolateral membrane, where GCDCA treatment showed the lowest fluorescence intensity (arrows indicate the location of MRP4 protein). Fluorescence quantification is reported in Graph 2B. Control: untreated cells; DMSO: vehicle control (3.3%, v/v); CA750μM; GCA100μM; DCA:50μM; CDCA150μM; GCDCA100μM; LCA50μM.
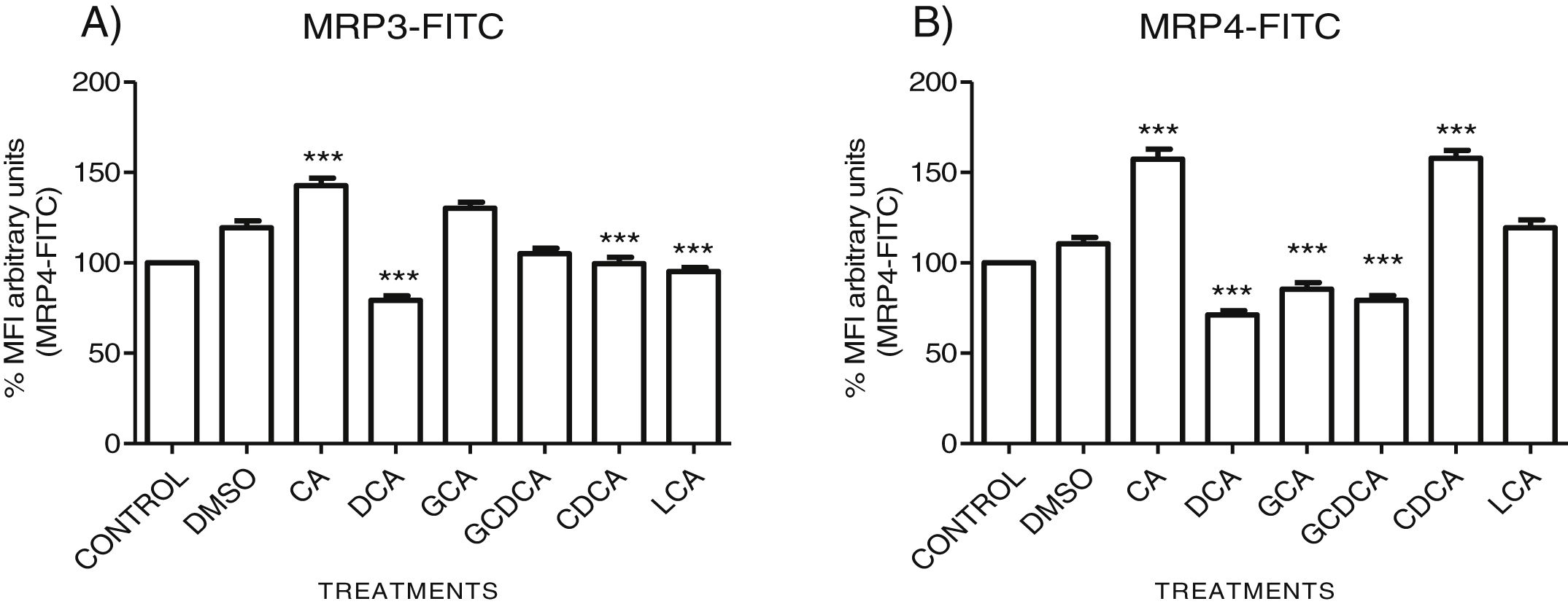
The localization of MRP3 in untreated cells was mislocalized and displayed a heterogeneous behavior. Some cells showed intracellular localization in compartments surrounding the nucleus, forming a halo and clusters in the cytoplasm with atypical distribution and in other cells, as expected, in the basolateral membrane. DMSO treatment and untreated cells showed similar behavior. CA, GCA, CDCA, LCA treatments also caused similar effects in the localization. However, these treatments strongly increased fluorescence intensity, especially in the cell cytoplasm, endoplasmic reticulum, and nucleolus in some cells (Fig. 3). DCA treatment also showed the atypical expression of MRP3, forming clusters or cell membranes labeled in a heterogeneous manner; although in this case, the fluorescence intensity was too low compared with other treatment groups. GCDCA treatment produced the typical expression of MRP3 in the basolateral membrane in most of the cells, including intense labeling in the endoplasmic reticulum. Quantifying the fluorescence intensity of CA and GCA treatments showed that these were the main inducers of the transporter expression while DCA decreased the expression of MRP3 to more than other treatments (Graph 2A).
Fluorescence quantification from IFI images of (A) MRP3 and (B) MRP4. Fluorescence quantification from Figs. 3 and 4 as expressed in percentage in relation to control. Treatment with CA increases the expression of both transporters, while DCA decreases in both cases. Asterisks indicate statistically significant differences between control vehicle and treatments (*** P<0.001, T3-Dunnette).
The features were similar to those described previously in the literature about the expression of MRP4 in confluence control cells (untreated). Cell rounding with perimeter limited by a membrane with an empty cytoplasm as seen in DMSO, CA, DCA treatments; the label of anti-MPR4 was also found in cell membranes with different intensities in comparison with LCA treatment, where the anti-MRP4 label was predominant in the cytoplasm (Fig. 4). The fluorescence intensity quantification showed that the CA and CDCA treatments were the main inducers of the transporter's expression, while DCA decreased notably its expression (Graph. 2B).
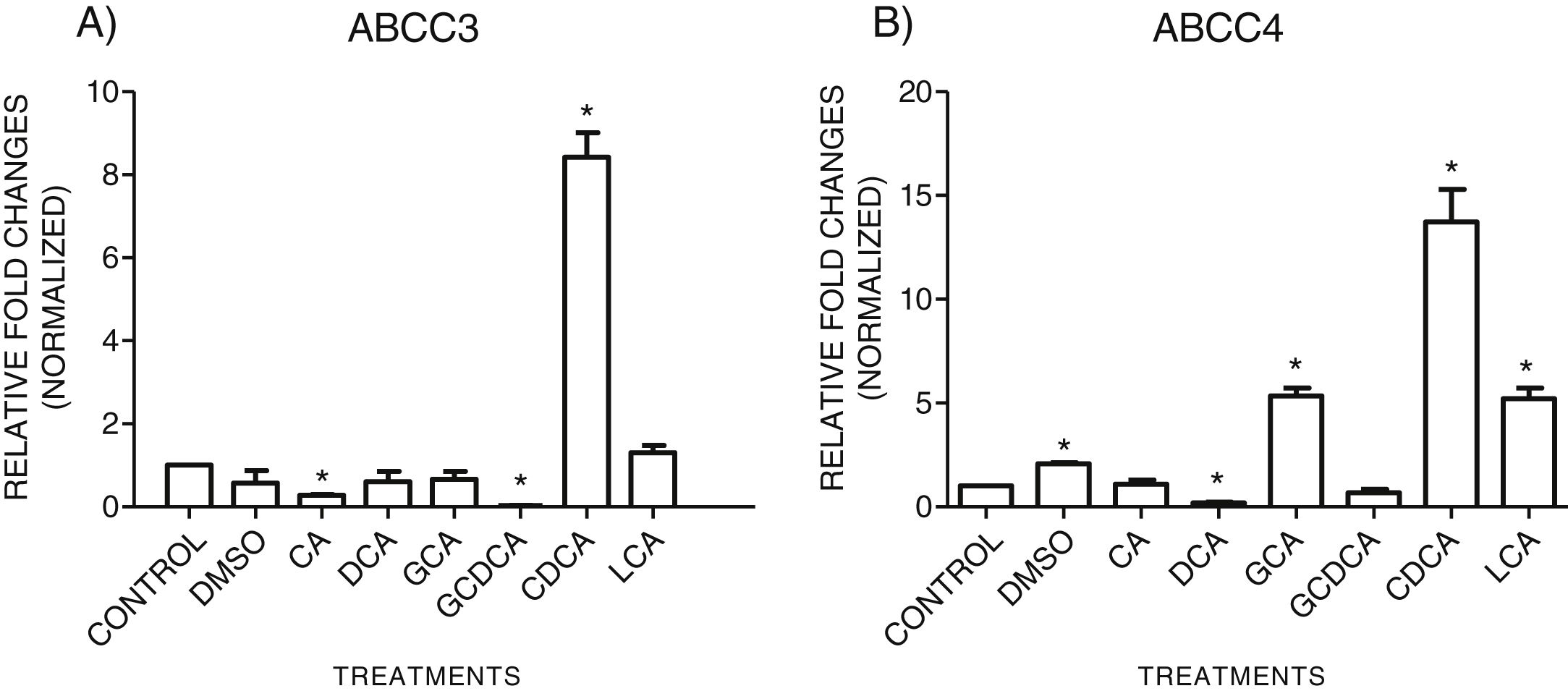
3.3Expressions of MRP3 and MRP4 after treatment with bile acidsTo evaluate the expression of mRNAs from MRP3 and MRP4 after treatment with different concentrations of bile acids, we performed RT-qPCR assays in confluent cell cultures. After exposition to bile salts, we observed that the expression of the MRP4 mRNA increased mainly due to CDCA, GCA, and LCA, while DCA drastically decreased its expression. For MRP3 mRNA expression, CDCA also induced a strong overexpression, and the induction was slight with LCA, while the other treatments decreased the expression, as shown in Graph 3.
qPCR assay. Relative expression of MRP3 (A) and MRP4 (B) in HepG2 cells that were treated for 4h with the different types of bile salts, using as a constitutive control. In the expression of GAPDH by qPCR, CDCA strongly increased in both transporters mRNA (* P<0.05, U-Mann Whitney) (Software Applied Biosystem, Pfaffl modification of the ΔΔCt method).
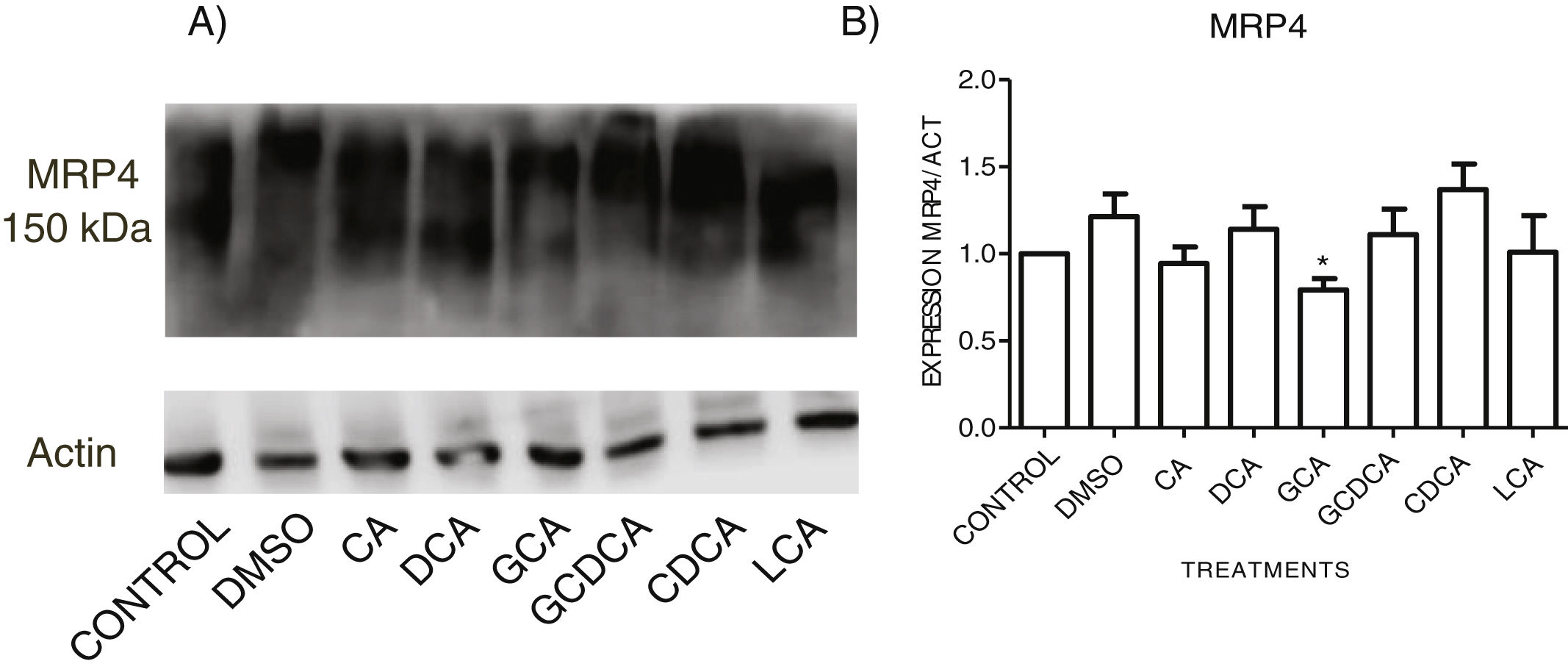
When evaluating the expression of MRP4 in the cytoplasmic fraction through Western Blot assay after exposure to different bile acids in confluent culture cells, it was found that CDCA treatment slightly increased the expression. However, no significant differences were found in the expression than the other treatments; only GCA decreased its expression significantly (Fig. 5 and Graph 4). Western Blot assay for MRP3 detection was negative.
Western Blot analysis of MRP4 expression in bile salts treated cells. Cells were treated with bile salt as described, GRAPH. 4. Relative expression of MRP4. Densitometric quantification from Fig. 5. CDCA increased the expression of MRP4 transporter (no significative difference was found in most cases) and the GCA treatment significantly decreased the expression of MRP4 (* P<0.05, U-Mann Whitney).
Bile acids are not capable of crossing cell membranes passively; in hepatocytes, there is an active transport system in the membranes that carry in and out these biliary molecules. For this purpose, the members of the ABC transporter family are required. The hepatocyte membrane contains several active carrier proteins to transport specific substrates to the bile canaliculus and play a vital role in the production of bile in the mammalian liver, and MRP3 and MRP4 provide the pathways for the proper efflux of free or conjugated BA's from hepatocytes into the blood [13].
In the present work, we performed an in vitro cholestasis study using the HepG2 cells, a human hepatocellular carcinoma cell line with epithelial-like morphology and are widely used for toxicology studies. We analyzed the expression of MRP3 and MRP4 transporters at the mRNA and protein level after incubation with the most abundant human bile acids CA and CDCA and their conjugates: GCA, GCDCA, and their secondary BA's deoxycholic acid and lithocholic acid respectably.
To evaluate the effects of BA's viability and toxicity on this cell culture, we implemented MTT tests with the vehicle's maximum concentration used in the treatments (3.3%, v/v) as a control. The highest concentrations assayed are an approximation of the concentrations of BA's that accumulate during cholestasis, consistent with other reports in which this range of concentrations is used for in vitro studies [22,23]. This system has certain limitations because in cholestasis in vivo, they is a pool of many bile acids causing the pathology, and due to the low expression of transporter proteins, this cell line is most sensitive than normal hepatocytes. We observed that the highest concentration assayed tested (750μM) of the DCA, CDCA, and GCDCA bile acids produced the cells’ lysis. In contrast, CA and GCA maintained cell viability above 74% at the same concentrations. In other cases, such as GCA, CDCA, LT, at medium concentrations (150–50μM), an increase of metabolic activity exceeded 100% compared to the untreated control. Considering that there is not enough cell proliferation in such a short time, the data suggest that the BA's at these concentrations may affect the metabolic activity that affects the mitochondria or oxidative stress generation. It has been documented that MTT assay reduction can be significantly affected, including different metabolic perturbations such as oxidative stress, reactive oxygen species (ROS) production, and mitochondrial affectations due to drug treatments. It has been reported that some of these effects are caused by BA's [24,25], which may explain the overvaluation in MTT assays after some treatments (e.g. GCA, CDCA, LT, at medium concentrations). However, no significant morphologic changes were observed in most cell culture treatments, except in CA where the rounding of cells was similar to that observed only in the vehicle (DMSO) group.
In the MRP4 and MRP3 mRNA expression analyses, we found that both transporters were over-expressed after treatment with CDCA (150μM). In decreasing order, the treatments produced overexpression: CDCA>LCA>GCA>CA>GCDCA>DCA for MRP4 and CDCA>LCA>GCA>DCA>CA>GQ for MRP3. This induced over-expression by CDCA could be due to its chemical structure; its first radical, an αOH group, influences bile acids’ hydrophobicity. It has also been reported that CDCA is a potent activator of the nuclear receptor FXR, which plays a significant role in BA's homeostasis. So, CDCA can generate biological effects, such as apoptosis [26,27].
When analyzing the expression of the MRP4 protein by Western Blot in the membrane fractions, only a few differences were found between the control and the treatments (non-statistically significant), only with a significant decrease with GCA. The Western Blot for MRP3 was negative, consistent with previous findings reported by other authors where MRP3 was not found, even in the membrane fraction's mass spectrometry analysis [28]. In the localization of MRP3 and MRP4 by confocal microscopy, we observed that both receptors are not detected in the expected location at low cell densities but were distributed throughout the entire cell. In MRP4, the fluorescence increased as cells reached confluence; the protein level was detected in its classical location in the basolateral membrane. On the other hand, when the culture reaches high confluences, MRP3 was observed in the basolateral membrane of hepatocytes forming clusters or peaks, distributed in the cytoplasm, possibly in vesicles or other cell compartments. A similar phenomenon has been reported in Huh7 culture cells, where at long culture times, MRP4 is localized in the basolateral membrane, and MRP3 showed a mislocalized ubication in intracellular compartments [28,29]. These observations strengthen the importance of the cell culture polarization or maturity acquired in a long-term cell culture upon reaching confluence, enabling the correct expression and function of these transporters and other functions described for this phenomenon [30].
At cell confluence, MRP4 was located on the basolateral membrane in all treatments, except with LCA, where it was in the cytoplasm toward the endoplasmic reticulum (redirection). There were different intensities of fluorescence (protein expression) depending on the treatment, being the degree of overexpression of MRP4 as follows: CA>CDCA>LCA.
On the other hand, MRP3 was observed, both on the plasma membrane and in clusters in intracellular compartments, with different intensity in fluorescence for each treatment (protein expression) where CA>GCA again overexpressed MRP3.
Primary bile acids (CDCA) in lower concentrations could induce overexpression of mRNA, probably due to its chemical structure and overload of bile acids, as could be the case of higher concentrations of CA overexpressed protein levels as observed by quantification of immunofluorescence.
This in vitro system has certain limitations as mentioned before [20]. However, it provides better understanding of the individual effects of bile acids over the expression of MRP3 and MRP4 transporters during an overload of BA's, which may lead to the development of new therapeutic uses for theses BA's in cholestasis.
5Conclusions- 1.
The hepatocytes polarization is an essential event for the correct expression and function of MRP3 and MRP4 transporters.
- 2.
MRP3 has an atypical subcellular localization in the HepG2 cells
- 3.
High concentrations of CA induce an increase in protein levels.
- 4.
The chemical structure of CDCA is responsible for MRP3 and MRP4 mRNA overexpression.
The authors have no conflicts of interest to declare.
We would like to thank Mr. Enrique Martinez de Luna and Ms. Silvia Galindo Gómez for their technical assistance, and Dr. Rosa María del Ángel for donating the HepG2 cell line. SIPP was a recipient of a Ph.D. fellowship from Consejo Nacional de Ciencia y Tecnología (CONACyT), México (CVU 636713). This work was supported by grants from Secretaría de Educación Pública-CONACyT (SEP-CONACyT), México (Grants CB 2014-240882-M to JLRE and Grant A1-S-27705 to VT).





 MTT assay. The highest concentrations tested of DC,
MTT assay. The highest concentrations tested of DC, 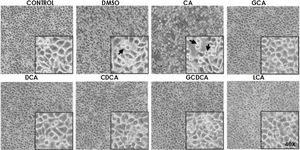 GCA100μM;
GCA100μM; 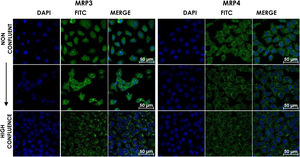 GCA100μM;
GCA100μM; 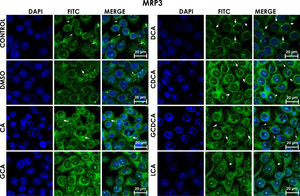 GCA registered more intensity of fluorescence than other treatments.
GCA registered more intensity of fluorescence than other treatments. 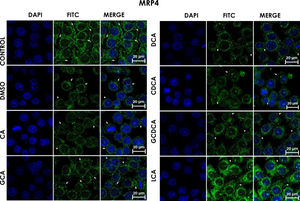 DCA treatments; the label of MPR4 antibody was also found in cell membranes with different intensities as compared with
DCA treatments; the label of MPR4 antibody was also found in cell membranes with different intensities as compared with 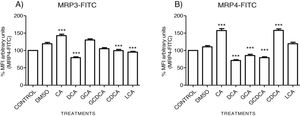 DCA decreases in both cases. Asterisks indicate statistically significant differences between control vehicle and treatments (*** P<0.001, T3-Dunnette).' title='Fluorescence quantification from IFI images of (A) MRP3 and (B) MRP4. Fluorescence quantification from Figs. 3 and 4 as expressed in percentage in relation to control. Treatment with CA increases the expression of both transporters, while
DCA decreases in both cases. Asterisks indicate statistically significant differences between control vehicle and treatments (*** P<0.001, T3-Dunnette).' title='Fluorescence quantification from IFI images of (A) MRP3 and (B) MRP4. Fluorescence quantification from Figs. 3 and 4 as expressed in percentage in relation to control. Treatment with CA increases the expression of both transporters, while 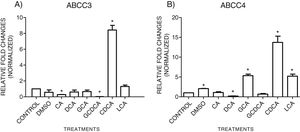 CDCA strongly increased in both transporters mRNA (* P<0.05, U-Mann Whitney) (Software Applied Biosystem, Pfaffl modification of the ΔΔCt method).' title='qPCR assay. Relative expression of MRP3 (A) and MRP4 (B) in HepG2 cells that were treated for 4h with the different types of bile salts, using as a constitutive control. In the expression of GAPDH by qPCR,
CDCA strongly increased in both transporters mRNA (* P<0.05, U-Mann Whitney) (Software Applied Biosystem, Pfaffl modification of the ΔΔCt method).' title='qPCR assay. Relative expression of MRP3 (A) and MRP4 (B) in HepG2 cells that were treated for 4h with the different types of bile salts, using as a constitutive control. In the expression of GAPDH by qPCR, 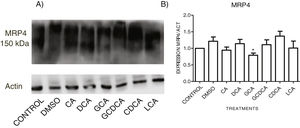 CDCA increased the expression of MRP4 transporter (no significative difference was found in most cases) and the
CDCA increased the expression of MRP4 transporter (no significative difference was found in most cases) and the 



