This study was undertaken to demonstrate a promising approach for detection and differentiation the serum immunoglobulin G (IgG) against hepatitis E virus (anti-HEV IgG) using a competitive binding assay established with known genotype-specific monoclonal antibodies (mAbs) 2B1 and 4C5.
Materials and methodsThe mAb 2B1 derived from genotype 1 hepatitis E virus (HEV) antigen and specifically reacted with genotype 1, 2 antigens; 4C5 induced by genotype 4 HEV antigen was specific to genotypes 3, 4 antigens. The 2B1 and 4C5 were labeled with Horseradish peroxidase (HRP), respectively. Subsequently, the titers of coated antigens and HRP-conjugated mAbs for establishment of competitive binding assay were determined by enzyme linked immunosorbent assay (ELISA). And then, the competitive binding assay was performed to assess the inhibition percentage of mAbs binding to antigens inhibited by different genotypes anti-HEV IgG.
ResultsThe results of competitive binding assay revealed that genotype 1 anti-HEV IgG could inhibit the binding of mAb 2B1 to genotype 1 antigen more strongly than that of mAb 4C5 to genotype 4 antigen. Whereas, the genotype 3 or 4 anti-HEV IgG could inhibit the binding of mAb 4C5 to genotype 4 antigen more remarkably than that of mAb 2B1 to genotype 1 antigen.
ConclusionsThese findings provided us a valuable approach for detection and differentiation the HEV infection derived from genotypes 1, 2 (human) or genotypes 3, 4 (zoonosis).
HEV is a small, non-enveloped, positive-strand RNA virus known to causes hepatitis E (HE), which can lead to detrimental prognostics, like sever acute hepatitis, chronic infection in the organ transplanted and immunosuppressed patients, high mortality among pregnant women and live failure [1,2]. The genomes of HEV strains consist of three overlapping open reading frames (ORFs), one of which is ORF2 that encodes the capsid protein of virus, C-terminal region of capsid protein has been localized as predominant immunogenic domain [3,4].
HEV strains infecting humans are classified into four genotypes [5], genotypes 1 and 2 are associated with large waterborne epidemic and isolated exclusively from humans, and these two genotypes HEV failed infect pigs in experimental studies, suggesting unlikely potential risk of zoonotic transmission to humans [6]. Whereas, genotype 3, 4 viruses are mainly responsible for sporadic cases, recognized as zoonotic pathogens. Because genotypes 3, 4 strains have been isolated both from humans and many wild and domestic animals, and have been found cross-species transmission from animals to humans [7–9]. Therefore, genotypes 1–4 HEV strains are classified into human(H) (genotypes 1 and 2)and zoonosis (Z) (genotypes 3 and 4) groups according to theirs compatible hosts by some people [10]. So, the confirmation of source of HEV infection from genotypes 1, 2 or zoonotic genotypes 3, 4 is an essential part of hepatitis E (HE) diagnosis and epidemiological investigation. The diagnosis of HE is mainly based on the results of laboratory test, mainly containing the detection of stool virus RNA and serum antibodies. The most widely used method for HE diagnosis is ELISA for its rapid and convenient characteristics [11]. As we all known, Wantai assay (ELISA) for HEV IgG detection is one of the most commonly used commercial kits, and has the highest sensitivity and specificity [12]. But till now, there is no simply serodiagnostic approach can disclose the origin of HEV isolates from human or zoonosis, except for genetic sequencing.
Previous study has reported that the recombinant protein p166 (452–617 aa of ORF2 protein) could model HEV conformational neutralizing epitope(s) [13]. In our previous study, we have found the different sensitivity between p166 proteins of different genotypes in detection of human serum anti-HEV antibodies after different genotypes HEV infection [14,15]. Moreover, the immunogenicity difference has also been found between the HEV p179 vaccine derived from genotype 4 strains and p239 vaccine (Hecolin) from genotype 1 HEV, the immunogenicity difference may attribute to the genotype-specific epitope(s) assessed by mAbs [16]. Therefore, we tested the hypothesis in this study that whether the genotype-specific mAbs could be used to detect and differentiate the source of HEV infection, from human or zoonosis.
2Materials and methods2.1Serum samplesFifty five human serum samples with high antibody level were collected from year 2001 to 2006 [14]. The year of collection, HEV genotypes and sample size are shown as below: Chad (2004, genotype 1, n=10), Hungary (2001–2006, genotype 3, n=9), and China (2005–2006, genotype 4, n=36). The Chad samples were first sent to the Centers for Disease Control and Prevention (CDC) in Atlanta in 2004, serum samples from Hungary were initially referred to the Regional Laboratory for Virology in Pécs, and those from China were referred to the Southeast University School of Medicine and the Second Hospital in Nanjing. Personal identifiers of all serum samples in this study were completely stripped of before being tested at Southeast University. Another eight serum samples were obtained from rhesus monkeys which have been inoculated intravenously with HEV strain W01 (genotype 1, JX857689) or NJ703 (genotype 4, AY789225). Viruses inoculation and sera sample collection were in the charge of GuangXi Center for Disease Prevention and Control [16]. All serum samples were stored at −80°C until further use. The procedures involving animals in this study were approved by the local committee of Animal Use and Protection.
2.2Recombinant proteinsRecombinant plasmid containing p166 nucleotide sequence was constructed and expressed in E. coli according to previous study [17]. Four His-tagged p166 were respectively generated from W01 (genotype 1, JX857689), Mexico-14 (genotype 2, M74506), US-1 (genotype 3, AF060668) and China-9829 (genotype 4, AY789225) strains, designated as p166W01, p166Mex, p166Us and p166Chn. And two GST-tagged proteins, p166Sar and p166Chn were respectively generated from the HEV Pakistani Sar-55 strain (genotype 1, M80581) and the China-9829 strain (genotype 4, AY789225).
2.3Generation of mAbs against p166Sar and p166ChnThe mAbs 2B1 and 4C5 were prepared as previously described [16], produced by injecting 106 hybridoma cells into the peritoneal cavity of the BALB/c mice. 7–10 days later, ascites fluid was harvested, filtered, centrifuged, and then purified with protein G affinity column. The purified mAbs were stored at −80°C until further use. All experimental procedures were approved by the Ethics Review Committee of Anhui Medical University, which comply with the Guide for the Care and Use of laboratory Animals published by the US National Institutes of Health (NIH publication no. 85-23, revised 2011).
2.4Labeling mAbs with horseradish peroxidase (HRP)Purified mAbs were dialyzed against 100mM Carbonate/Bicarbonate buffer (pH 9.3) at 4°C over night. And then, antibodies were labeled with HRP according to the manufacturer's instructions (KPL). Methods are briefly described as follows: 1.5mg of HRP and 0.5mg of mAb were added into HRP conjugation buffer at a final volume of 0.5ml. After gently stirring at room temperature for 1h, the mixture was left standing at room temperature for 15min. And then, 10μl of the reducing agent was added into the mixture. At last, the same volume of distilled glycerol was added into the mixture for storage and further use.
2.5Competitive binding assay2.5.1Determination of antigens and mAbs titersThe titers of coated antigens and HRP-conjugated mAbs were determined by ELISA. Initially, HRP-conjugated mAbs of two-fold dilutions in 1% casein PBS, ranging from 1:400 to 1:25,600 were prepared and then added in wells, which pre-coated with serial two-fold dilutions (from 1:200 to 1:6400) of p166W01 or p166Chn antigen. After incubating at 37°C for 45min, the ELISA plate was washed for five times with PBS containing 0.5% Tween-20 (PBST). And then, TMB liquid substrate was added for color development, which was stopped by adding stop solution. Finally, the Optical Density (OD) of each well was read at 450nm, with a 630nm reference wavelength to determine the dilution of coated antigen and HRP-conjugated mAb that give an OD value reading of approximately 1.0.
2.5.2Competitive binding assay [18]The p166W01 and p166Chn were used as coating antigens to establish the competitive binding assay, respectively. Anti-HEV sera of saturating concentration were used to assess the inhibition percentage of the binding of HRP-mAb to antigen. The procedure was similar to our previous study [19], the p166W01 or p166Chn of optional dilution was used as the coating antigen respectively, the mixture of HRP-conjugated mAb at twice the optional dilution and anti-HEV serum of saturated concentration was added to antigen coated well. The OD value was read at 450nm, with a 630nm reference wavelength. The inhibition percentage was calculated by the following formula: 100×[1−(A450,630 of HRP-conjugated mAb and serum anti-HEV IgG)/(A450,630 of HRP-conjugated mAb)].
3Results3.1Isolation and characterization of mAbs against p166Sar or p166ChnThe mAbs were purified using immobilized protein G, and the specificity of mAbs was identified by ELISA with 4 genotypes p166 antigens (p166W01, p166Mex, p166Us, p166Chn), respectively. The results of ELISA revealed that mAb 2B1 reacted only with p166W01 and p166Mex, but not with p166US or p166Chn; whereas, mAb 4C5 reacted only with p166US and p166Chn, but not with p166W01 or p166Mex, suggesting that 2B1 and 4C5 were genotypes-specific mAbs [16].
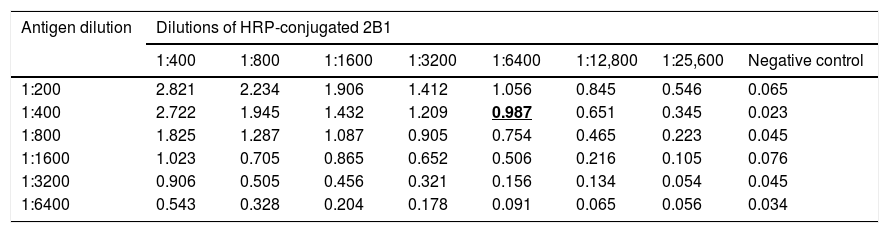
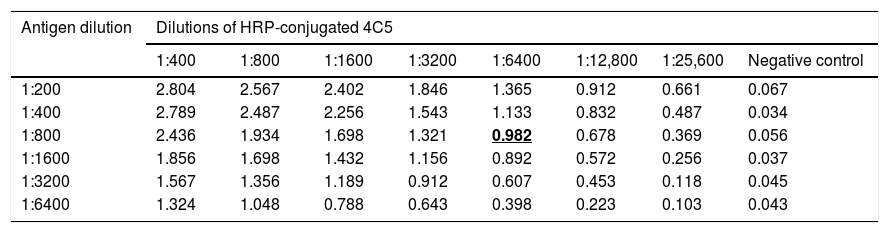
3.2Determination of antigens and mAbs dilutionp166W01 protein was used as coating antigen to establish the competitive binding assay of mAb 2B1 and serum anti-HEV IgG; p166Chn antigen was used to establish the competitive binding assay of mAb 4C5 and serum anti-HEV IgG. As shown in Table 1, the dilution of p166W01 was 1:400, HRP-conjugated 2B1 was 1:6400,the mean OD value was close to 1.0; whereas, the dilution of p166Chn was 1:800, HRP-conjugated 4C5 was 1:6400, the mean OD value was close to 1.0 (Table 2).
Determination of p166W01 antigen and HRP-conjugated 2B1 dilutions (means, n=4).
| Antigen dilution | Dilutions of HRP-conjugated 2B1 | |||||||
|---|---|---|---|---|---|---|---|---|
| 1:400 | 1:800 | 1:1600 | 1:3200 | 1:6400 | 1:12,800 | 1:25,600 | Negative control | |
| 1:200 | 2.821 | 2.234 | 1.906 | 1.412 | 1.056 | 0.845 | 0.546 | 0.065 |
| 1:400 | 2.722 | 1.945 | 1.432 | 1.209 | 0.987 | 0.651 | 0.345 | 0.023 |
| 1:800 | 1.825 | 1.287 | 1.087 | 0.905 | 0.754 | 0.465 | 0.223 | 0.045 |
| 1:1600 | 1.023 | 0.705 | 0.865 | 0.652 | 0.506 | 0.216 | 0.105 | 0.076 |
| 1:3200 | 0.906 | 0.505 | 0.456 | 0.321 | 0.156 | 0.134 | 0.054 | 0.045 |
| 1:6400 | 0.543 | 0.328 | 0.204 | 0.178 | 0.091 | 0.065 | 0.056 | 0.034 |
Bold and underline emphasize that the corresponding dilutions of antigen and HRP-conjugated mAb were used to establish the competitive binding assay.
Determination of p166Chn antigen and HRP-conjugated 4C5 dilutions (means, n=4).
| Antigen dilution | Dilutions of HRP-conjugated 4C5 | |||||||
|---|---|---|---|---|---|---|---|---|
| 1:400 | 1:800 | 1:1600 | 1:3200 | 1:6400 | 1:12,800 | 1:25,600 | Negative control | |
| 1:200 | 2.804 | 2.567 | 2.402 | 1.846 | 1.365 | 0.912 | 0.661 | 0.067 |
| 1:400 | 2.789 | 2.487 | 2.256 | 1.543 | 1.133 | 0.832 | 0.487 | 0.034 |
| 1:800 | 2.436 | 1.934 | 1.698 | 1.321 | 0.982 | 0.678 | 0.369 | 0.056 |
| 1:1600 | 1.856 | 1.698 | 1.432 | 1.156 | 0.892 | 0.572 | 0.256 | 0.037 |
| 1:3200 | 1.567 | 1.356 | 1.189 | 0.912 | 0.607 | 0.453 | 0.118 | 0.045 |
| 1:6400 | 1.324 | 1.048 | 0.788 | 0.643 | 0.398 | 0.223 | 0.103 | 0.043 |
Bold and underline emphasize that the corresponding dilutions of antigen and HRP-conjugated mAb were used to establish the competitive binding assay.
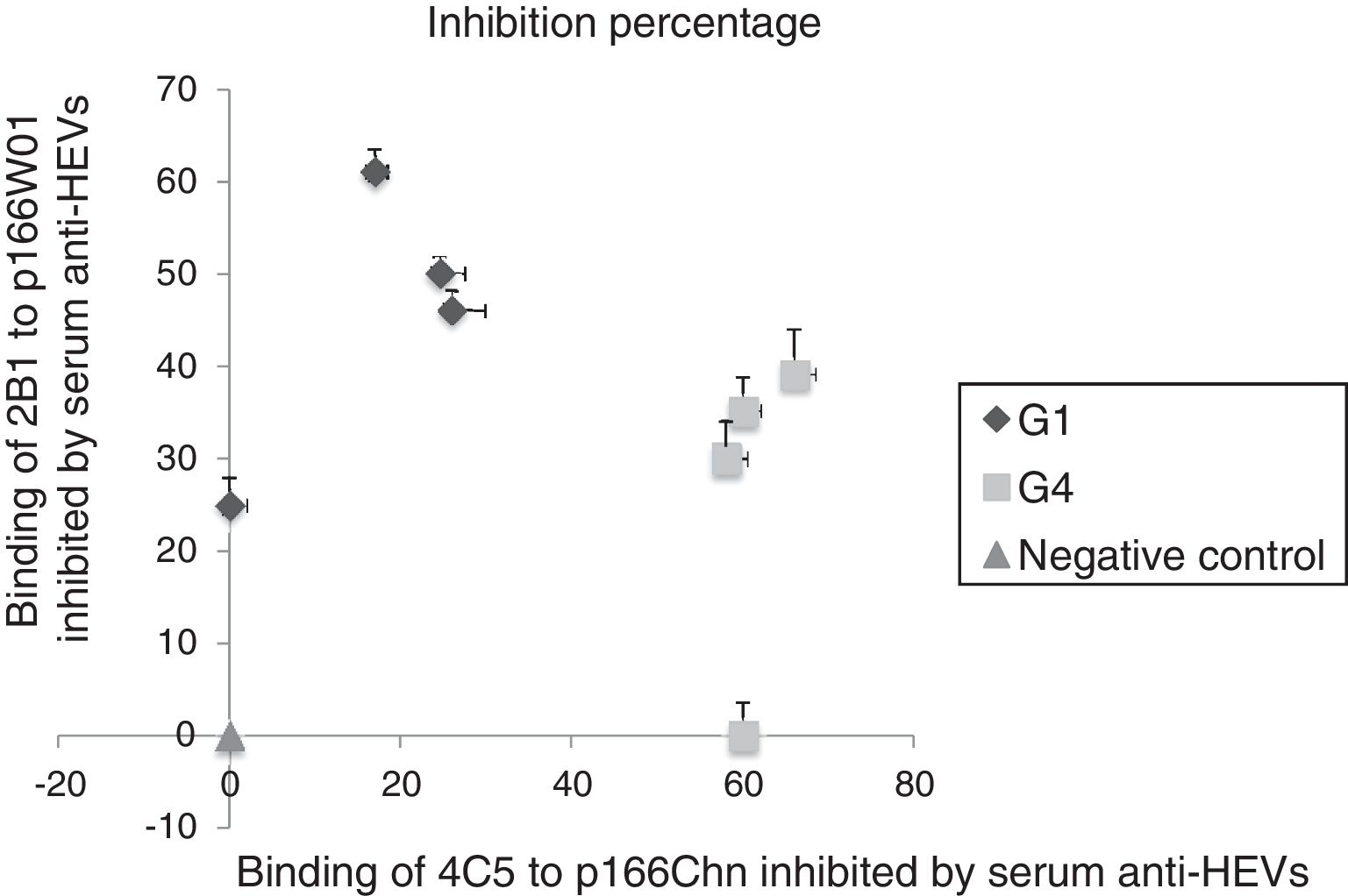
Sera sample containing anti-HEV IgG were collected from rhesus monkeys challenged with genotype 1HEV strains (W01) or genotype 4 HEV strains (NJ703) [16], used to evaluate the binding inhibition of mAb 2B1 to p166W01, mAb 4C5 to p166Chn. As shown in Fig. 1, both the binding of mAb 2B1 to p166W01 and mAb 4C5 to p166Chn could be inhibited by the serum anti-HEVs against genotype 1 or 4 HEV. But more high inhibition percentages were obtained from the binding of 2B1 to p166W01 inhibited by genotype 1 anti-HEV IgGs, from 25% to 61%. Whereas, the inhibition percentage of mAb 4C5 to p166Chn inhibited by genotype 1 anti-HEV IgG strain was not more than 26%. On the contrary, the binding of mAb 4C5 to p166Chn was inhibited more remarkably by the gentoype 4 anti-HEV IgG (from 60% to 70%) than that by the genotype 1 anti-HEV IgG (from 0% to 38%).
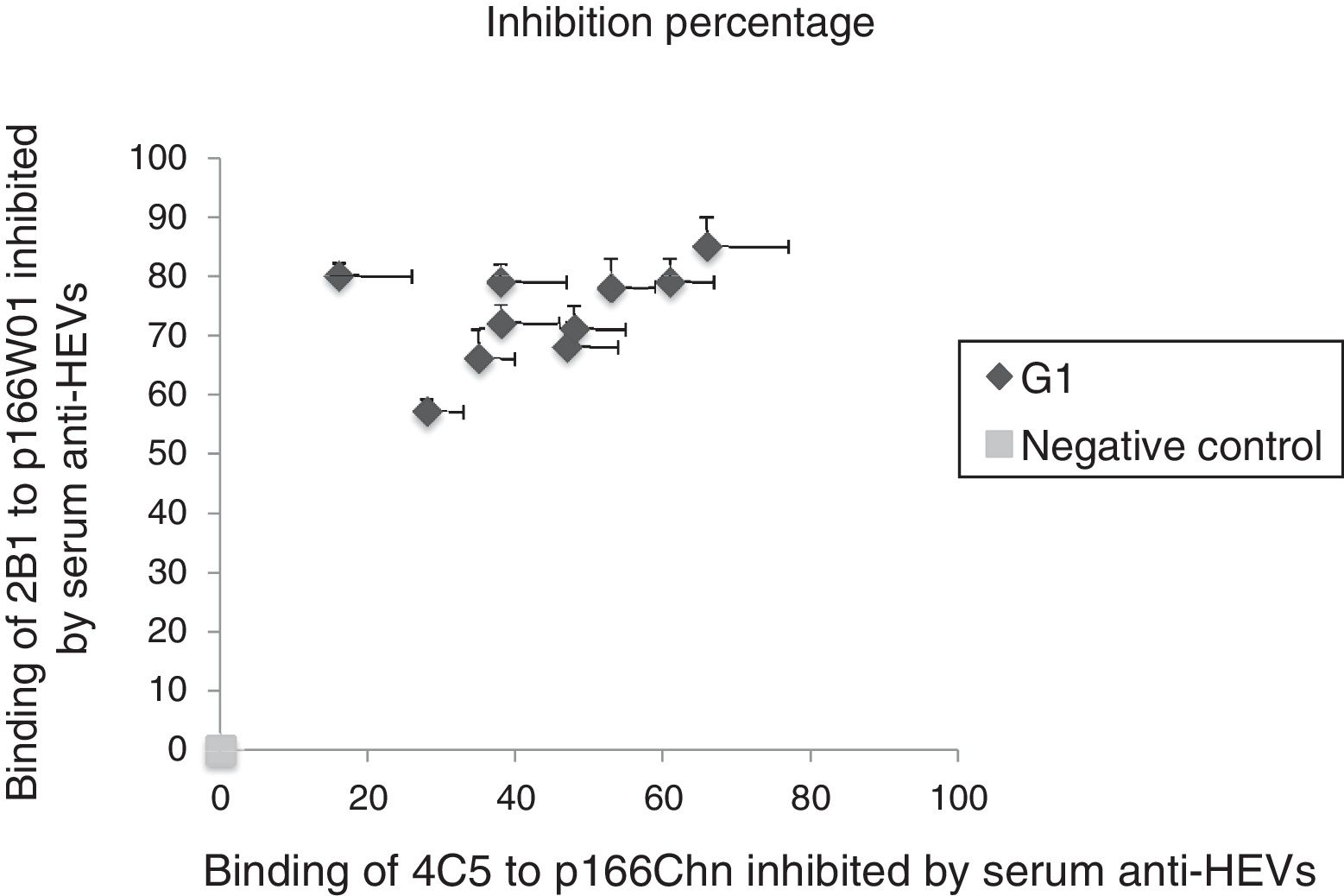
3.4Binding of mAbs to antigens inhibited by anti-HEV IgG in human serum against genotype 1 HEVAnti-HEV IgG in human sera infected by genotype 1 strains were also used to further demonstrate the binding inhibition of mAbs to antigens. As shown in Fig. 2, the binding of 2B1 to p166W01 was inhibited remarkably by the genotype 1 anti-HEV IgG, the inhibition percentages were from 57% to 85%. Whereas, genotype 1 anti-HEV IgG could only partially inhibit the binding of mAb 4C5 to p166Chn, the inhibition percentages were not more than 66%.
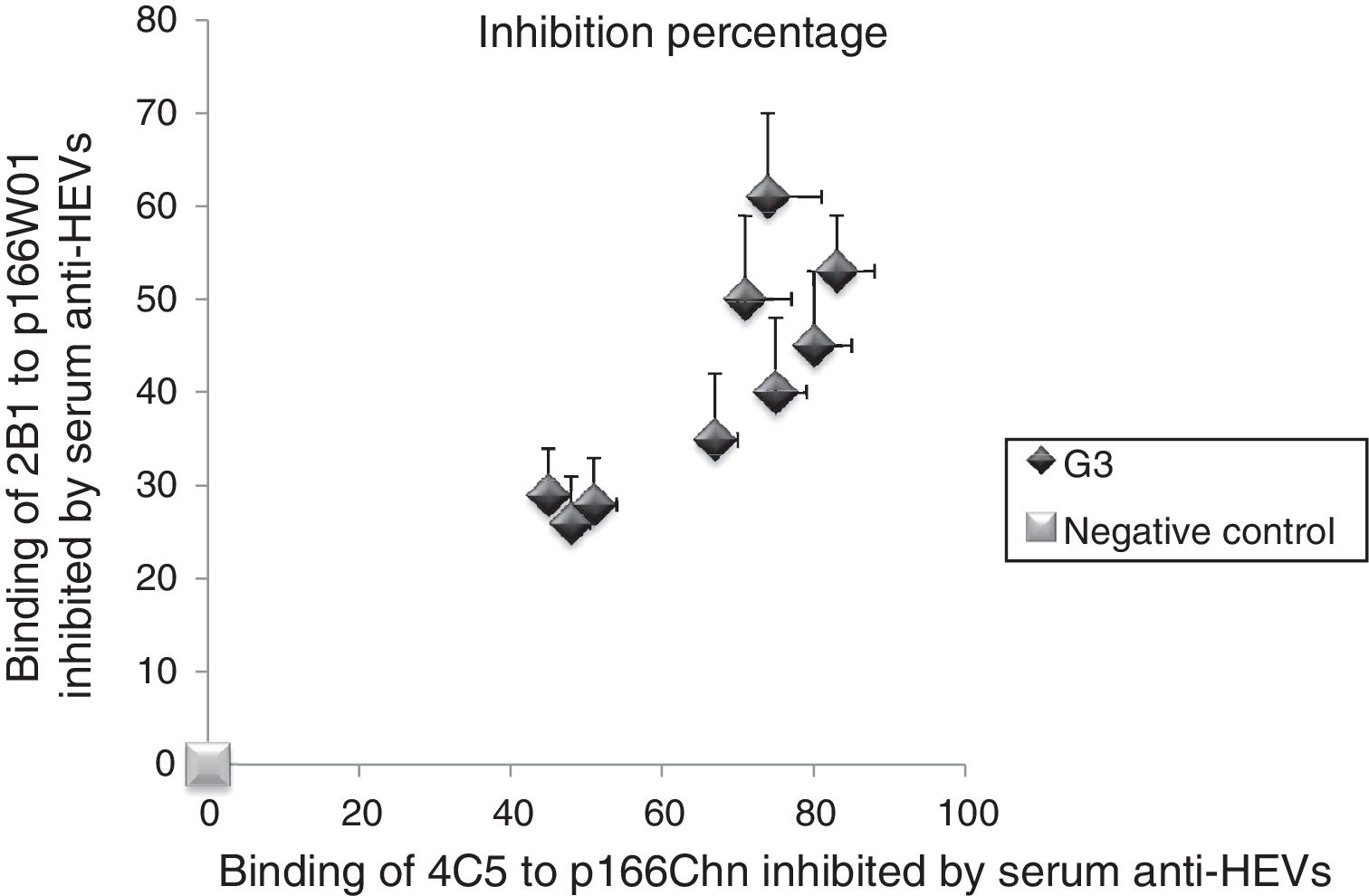
3.5Binding of mAbs to antigens inhibited by human serum anti-HEV IgG against genotype 3 HEVWe followed the same approach to investigate the inhibition percentage of mAbs to antigens by serum anti-HEV IgG against the genotype 3 HEV. As shown in Fig. 3, the binding of 4C5 to p166Chn antigen was inhibited more remarkably by genotype 3 anti-HEV IgG, the inhibition percentages were from 45% to 83%. Whereas, each of the same serum could only partially inhibit the binding of 2B1 to p166W01, the inhibition percentages were not more than 61%.
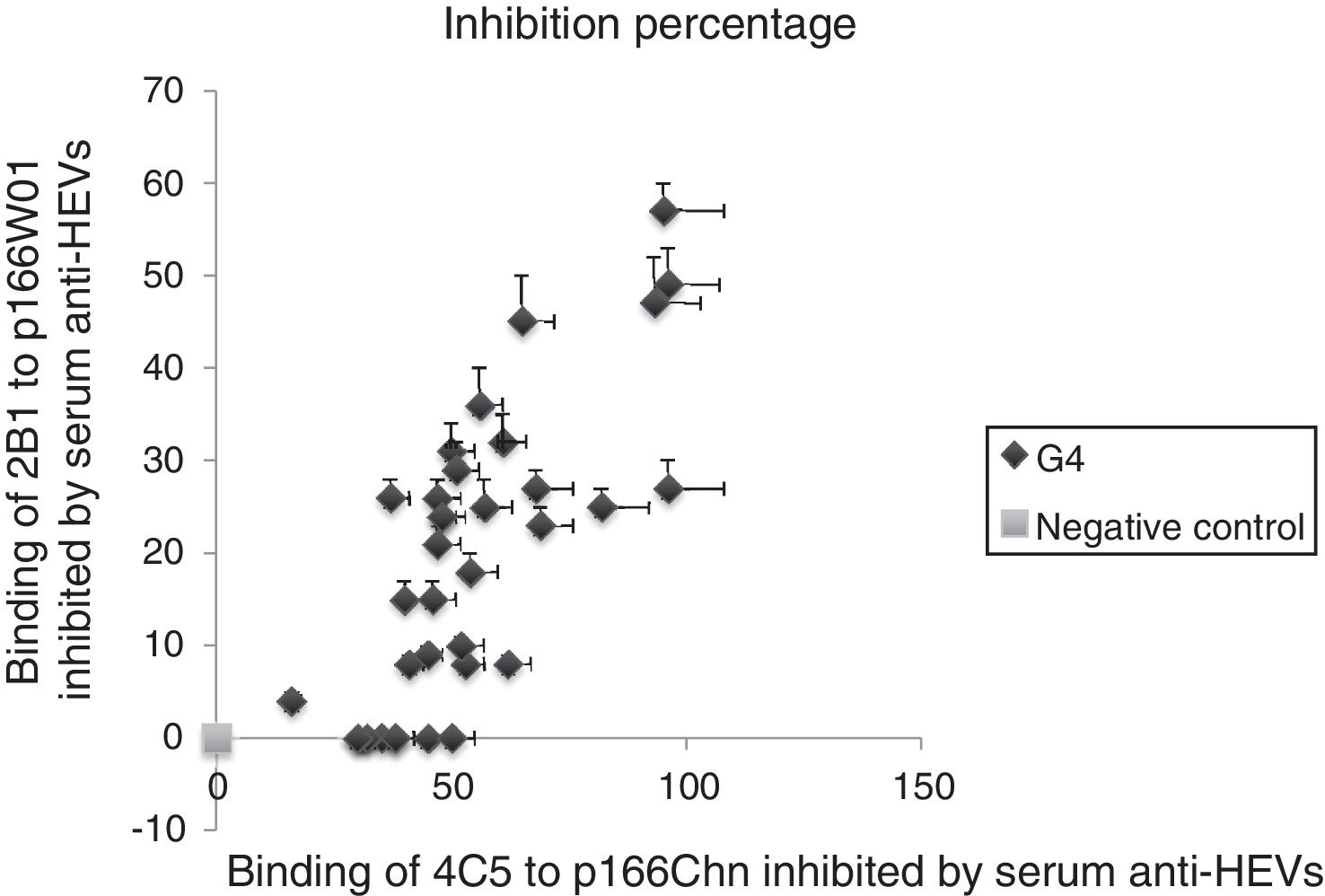
3.6Binding of mAbs to antigens inhibited by human serum anti-HEV IgG against genotype 4 HEVThe serum samples were collected from human infected by genotype 4 HEV strains, the binding inhibition of mAbs to antigens inhibited by serum anti-HEV IgG was evaluated with same approach. The results were similar to previous descriptions, the binding of 4C5 to p166Chn antigen was inhibited more remarkably by genotype 4 anti-HEV IgG, the inhibition percentages were from 30% to 96%. Whereas, each of the same serum could only partially inhibit the binding of 2B1 to p166W01, the inhibition percentages were not more than 57% (Fig. 4).
4DiscussionHEV infection is an important public health concern not only in developing countries but also in developed ones. The ratio of infection-to-mortality of HEV is generally considered to be small, but this significantly increases in pregnant women, in infants under the age of 2 years, and in adults over the age of 56 years. Moreover, the outbreaks of HEV have been repeatedly reported in India, Nepal, Pakistan, Burma, Mexico, and China [20], due to inadequate sanitary conditions. Although only one serotype has been recognized, HEV of genotypes 1, 2 still differed from genotypes 3, 4 in their antigenic and immunogenic characteristics [16]. Especially, genotypes 3, 4 HEV strains may be capable of infecting cross-species, from chicken to wild birds, and even from animals to human beings [21,22]. High prevalence of HEV frequently displayed in Asia and Africa. The anti-HEV-positive rates have been found more than 30% in most Asia and Africa countries, suggesting a high rate of subclinical infection [23–26]. The appearance of anti-HEV antibodies in serum is considered as an epidemiological marker of HEV exposure assessment in a population [6]. Anti-HEV IgG can be detected within two weeks and lasting for more than 10 years after the virus exposure, and can be used as an index of epidemiological investigation [27].
As aforementioned, genotypes 1, 2 HEV only infect humans while genotypes 3,4 HEV are zoonotic agents, the main reservoir of HEV3 and HEV4 is domestic pigs. While, genotypes 1, 2 HEV are believed to transmitted via contaminated water. Furthermore, both genotypes 3 and 4 HEV have been reported in chronic infection, however, neither genotype 1 nor genotype 2 HEV has been shown to associate with chronic HE so far [28]. In addition, previous studies have reported that HEV could transmit by blood transfusion in a number of countries [29,30]. Therefore, HE diagnosis and epidemiological investigation, especially, confirmation the source of HEV from genotypes 1, 2 or 3, 4 are essential parts of prevention of HEV transmission.
In present study, we sought to clarify whether the genotype-specific mAbs 2B1 and 4C5 could be used for HE diagnosis and disclosure the source of HEV from human or zoonosis by establishment of the competitive binding assay. The results of competitive binding assay indicated that both the binding of 2B1 to p166W01, 4C5 to p166Chn could be inhibited by the anti-HEV IgG in rhesus monkey sera against genotypes 1 or 4 strains. But the genotype 1 anti-HEV IgG could inhibit the binding of 2B1 to p166W01 more significantly than that of 4C5 to p166Chn. Whereas, genotype 4 anti-HEV IgG in rhesus monkey sera could block the binding of 4C5 to p166Chn more strongly than that of 2B1 to p166W01.
We next tried to further confirm the differences of competitive inhibition of the binding of mAbs to antigens. Thirty seven human sera samples infected by genotypes 1, 3 or 4 HEV were collected and used to confirm the differently binding inhibition between 2B1 to p166W01 with 4C5 to p166Chn. The results revealed that genotype 1 anti-HEV IgG could inhibit the binding of 2B1 to p166W01 more remarkably than that of 4C5 to p166Chn. Whereas, serum anti-HEV IgG against the genotypes 3 or 4 HEV could inhibit the binding of 4C5 to p166Chn more significantly than that of 2B1 to p166W01. These findings indicated that the binding of genotype-specific mAbs to p166W01 or p166Chn antigen can be blocked at different degree by anti-HEV IgG against genotype 1, 3 or 4. This is the first study that demonstrates the genotype-specific mAbs 2B1 and 4C5 may be used for HE diagnosis and epidemiological investigation of HEV exposure from human or zoonosis by establishment of competitive binding assay. Despite major steps forward in demonstrating a promising approach for detection and differentiation the serum anti-HEV IgG using a competitive binding assay, following questions need to be solved, the clinical phenotype of HE continues to emerges; locally acquired zoonotic hepatitis E is recognized slowly in developed countries. Therefore, further studies are urgently needed to optimize the experimental conditions of competitive binding assay for future development.AbbreviationsHE
hepatitis E
HEVhepatitis E virus
IgGimmunoglobulin G
mAbsmonoclonal antibodies
HRPHorseradish peroxidase
ODOptical Density
ORFsopen reading frames
PBSTPBS containing 0.5% Tween-20
Ethical statementAll experimental procedures were approved by the Ethics Review Committee of Anhui Medical University, which comply with the Guide for the Care and Use of laboratory Animals published by the US National Institutes of Health (NIH publication no. 85-23, revised 2011).
FundingThis study was supported by Grants for Scientific Research of BSKY (No. XJ201612) from Anhui Medical University and from Natural Science Foundation of Hefei Technology College (No. 201914KJA020)
ContributionsParticipated in research design and experiments: Jiyue Wen, Weizhuo Lu.
Contributed new reagents or analytical tools: Weizhuo Lu.
Performed data analysis: Jiyue Wen.
Contributed to writing of the manuscript: Jiyue Wen, Jihong Meng.
Conflict of interestThe authors have no conflict of interest to declare.