Various studies have identified single-nucleotide polymorphisms (SNPs) associated with nonalcoholic fatty liver disease (NAFLD) and related traits, including ones located in or near the LYPLAL1, GCKR, PPP1R3B, TM6SF2, MBOAT7, and PNPLA3 genes. However, these SNPs were identified primarily in populations of European ancestry. This study examined the associations of these previously identified SNPs with hepatic steatosis in a sample of Mexican-origin adults living in Southern Arizona.
Materials and MethodsA total of 307 Mexican-origin adults between the ages of 18 and 64 with a body mass index (BMI) of 25 kg/m2 or higher were genotyped at the following SNPs: rs12137855 (LYPLAL1), rs1260326 (GCKR), rs4240624 (PPP1R3B), rs58542926 (TM6SF2), rs641738 (MBOAT7), and rs738409 (PNPLA3). Hepatic steatosis was assessed by transient elastography (FibroScan®) with controlled attenuation parameter. Regression models examined the association between each of the six SNPs and hepatic steatosis. BMI was examined as a potential modifier of the genetic associations.
ResultsParticipants were, on average, 45 years old and mostly female (63%) with an overall mean hepatic steatosis of 288.1 dB/m. Models showed no associations between LYPLAL1, GCKR, PPP1R3B, TM6SF2, or MBOAT7 and hepatic steatosis. Only PNPLA3 was statistically significantly associated with hepatic steatosis in both unadjusted and adjusted models (p<0.01). There was no effect modification observed with BMI.
ConclusionsSNPs associated with NAFLD in populations of European descent did not strongly contribute to hepatic steatosis in individuals of Mexican-origin, except for rs738409 (PNPLA3). Further efforts are necessary to explore additional SNPs that may be associated with NAFLD in this high-risk population.
Non-alcoholic fatty liver disease (NAFLD) is the most common chronic liver disease among Hispanics/Latinos in the United States (US) and worldwide [1]. NAFLD encompasses a number of progressive conditions ranging in severity from non-alcoholic fatty liver through non-alcoholic steatohepatitis and eventually, if left untreated, hepatocellular carcinoma [2,3]. NAFLD is commonly defined as the presence of ≥5% hepatic steatosis, without any competing condition (or medication use) that would result in increased hepatic fat accumulation, or significant alcohol use [2,4]. While progression of NAFLD to hepatocellular carcinoma occurs in only a small subset of individuals, progression to liver cirrhosis and/or liver decompensation are more common and can lead to reduced function of the liver and result in the need for a liver transplant [2]. With rates of liver cirrhosis and liver cancer on the rise among Hispanics/Latinos in the US, NAFLD can be an important early target for interventions given the availability of liver imaging tools to assess hepatic steatosis such as ultrasound, computed tomography (CT), magnetic resonance imaging (MRI), and transient elastography [5,6].
While the most important risk factors for NAFLD include obesity and insulin resistance, there also appears to be increased risk for Hispanics/Latinos, specifically those of Mexican-origin [7]. Lazo et al. reported that Mexican-Americans had higher prevalence of NAFLD compared to non-Hispanic Whites (NHWs) (24.1% vs.17.8%) [8]. Genetic variation may help elucidate the reasons behind the varying burden of NAFLD across racial and ethnic subpopulations. For example, an allele in PNPLA3 (patatin-like phospholipase domain-containing protein 3), rs738409 [G], which has been found to be associated with a greater risk of NAFLD, has also been found at much higher frequencies among Hispanics/Latinos than among NHWs (49% vs. 23%) [9]. Identifying genetic variants that confer additional risk in certain populations may not only help in further elucidating mechanisms of NAFLD development and progression but inform personalized clinical recommendations such as decisions related to patient monitoring or the implementation of invasive diagnostic testing for those deemed at greater genetic risk [10].
Many studies have examined the relationship between various single nucleotide polymorphisms (SNPs) and NAFLD through both genome-wide association studies (GWAS) and candidate SNP studies [11–14]. Although various research definitions for NAFLD (ranging from elevated transaminase levels with and without histological diagnosis to imaging-assessed hepatic steatosis alone) have been used in genetic association studies, SNPs in or near PNPLA3 have been consistently associated with NAFLD [14–16]. However, other SNPs in or near genes including: LYPLAL1 (lysophospholipase like 1, HGNC ID:20440), GCKR (glucokinase regulatory protein, HGNC ID:4196), PPP1R3B (protein phosphatase 1, regulatory subunit 3, HGNC ID:14942), TM6SF2 (transmembrane 6 superfamily 2, HGNC ID:11861), and MBOAT7 (membrane bound o-acyltransferase domain containing 7, HGNC ID:15505) have had contradicting findings [14,17–20]. Also, despite higher rates of NAFLD in Hispanic/Latinos, most GWAS and candidate SNP studies have been conducted among populations of European descent, and it is therefore not clear whether these genetic associations hold in other racial and ethnic subpopulations.
This study examined the associations of rs12137855 (LYPLAL1), rs1260326 (GCKR), rs4240624 (PPP1R3B), rs58542926 (TM6SF2), rs641738 (MBOAT7), and rs738409 (PNPLA3) with hepatic steatosis, as assessed by transient elastography (Fibroscan®), in a sample of Mexican-origin adults living in Southern Arizona. The association for hepatic steatosis with TM6SF2 and MBOAT7, to our knowledge, has not been previously examined among Mexican-origin populations; although they have been studied in relation to a variety of NAFLD definitions [20]. We further examined a possible interaction effect of body mass index (BMI) with each SNP because of previous evidence suggesting that genetic risk was amplified with higher adiposity [21].
2Materials and Methods2.1Study participantsThe current analyses utilized participant data from a cross-sectional study conducted in southern Arizona [22]. Recruitment and study visits occurred between May 2019 and March 2020. The purpose of the cross-sectional study was to examine the association between clinical, behavioral, and genetic (NAFLD-related SNPs) risk factors with hepatic steatosis among Mexican-origin adults with overweight/obesity residing in Southern Arizona. To be eligible for the study, participants needed to: self-identify as Mexican origin, be between 18 and 64 years old, and have a BMI of > 25 kg/m2. This BMI criterion was included in an effort to include adults at greater risk of NAFLD. Exclusion criteria included uncontrolled (self-reported not currently taking medication) vascular or metabolic diseases such as high blood pressure and type 2 diabetes, self-reported significant alcohol consumption (>21 standard drinks on average per week for men and >14 standard drinks for women), taking medications or supplements known to affect body composition or exposure to hepatotoxic drugs, syndromes that affects body composition or fat distribution, major illness at birth, participation in a structured exercise, nutrition or weight loss program in previous 6 months, history of bariatric surgery, pregnant or breast feeding, previous diagnosis of liver disease or liver cancer, or an active gastrointestinal disorder (e.g., inflammatory bowel disease, ulcerative colitis, Chron's disease, or celiac disease).
2.2Clinical and anthropometric measuresAs a part of the cross-sectional study, participants completed a single study visit which occurred in a clinic that specialized in the treatment of liver disease in Tucson, Arizona. During this visit, participants completed the following study procedures: a range of questionnaires that collected information on demographics and medical conditions, an anthropometric assessment that included assessment of height (using stadiometer), weight (using Tabina WB-100A), and waist circumference (using Gulick measuring tape); and transient elastography (FibroScan®) to assess hepatic steatosis based on controlled attenuation parameters (CAP) scores and liver stiffness (used as a proxy for liver fibrosis) in kilopascals (kPa). CAP values ranged from 100 to 400 dB/m with higher values indicating higher levels of hepatic steatosis. Liver stiffness values ranged from 2 kPa to 75 kPa with higher values indicating higher liver stiffness.
2.3Candidate SNP genotypingGenomic DNA was isolated from buccal swabs using the Whatman™ Omniswab™ as instructed by the manufacturer (Whatman, Buckinghamshire, UK) by the University of Arizona Genetics Core. DNA was then quantitated and used as a template in a TaqMan® SNP Genotyping Assay in codon 148 of PNPLA3 for the rs738409 SNP. Participants were also genotyped at five additional SNPs: LYPLAL1-rs12137855, GCKR-rs1260326, PPP1R3B-rs4240624, TM6SF2-rs58542926, MBOAT7-rs641738, and PNPLA3-rs738409.
2.4Statistical analysisFor each SNP, Chi-square tests were used to assess departures from Hardy-Weinberg Equilibrium (HWE). Unadjusted and adjusted linear regression models were used to examine the association between each of the six SNPs and hepatic steatosis (CAP score). Models were adjusted for age (years), sex (female or male), and BMI. Furthermore, we examined whether an interaction effect with BMI was present in the relationship between each of the six SNPs and hepatic steatosis. While our primary models coded SNP genotypes as additive, we also constructed dominant and recessive inheritance models.
Finally, an unweighted polygenic risk score was developed consisting of the count of risk alleles according to larger previous across all six SNPs [11,13,23–26]. The risk alleles were T for LYPLAL1, T for GCKR, G for PPP1R3B, T for TM6SF2, T for MBOAT7, and G for PNPLA3. Unadjusted and adjusted (age, sex, and BMI) linear regression models examined the association between the risk score and hepatic steatosis. A P<0.05 was considered statistically significant. We estimated that with our sample size and a type 1 error rate of 5%, we have 80% statistical power to detect an effect with an r2 as small as 0.038 (3.8%). All analyses were carried out with SAS 9.4 (Cary, NC).
2.5Ethical statementWritten informed consent was obtained from each participant included in the study and the study protocol conforms to the ethical guidelines of the 1975 Declaration of Helsinki as reflected in a priori approval by the University of Arizona Institutional Review Borad (IRB #1902380787). In addition, a Certificate of Confidentiality (CC-OD-19-293) was obtained due to the ethical considerations of our population (e.g., citizenship status) and the sensitive research information (genetics) collected.
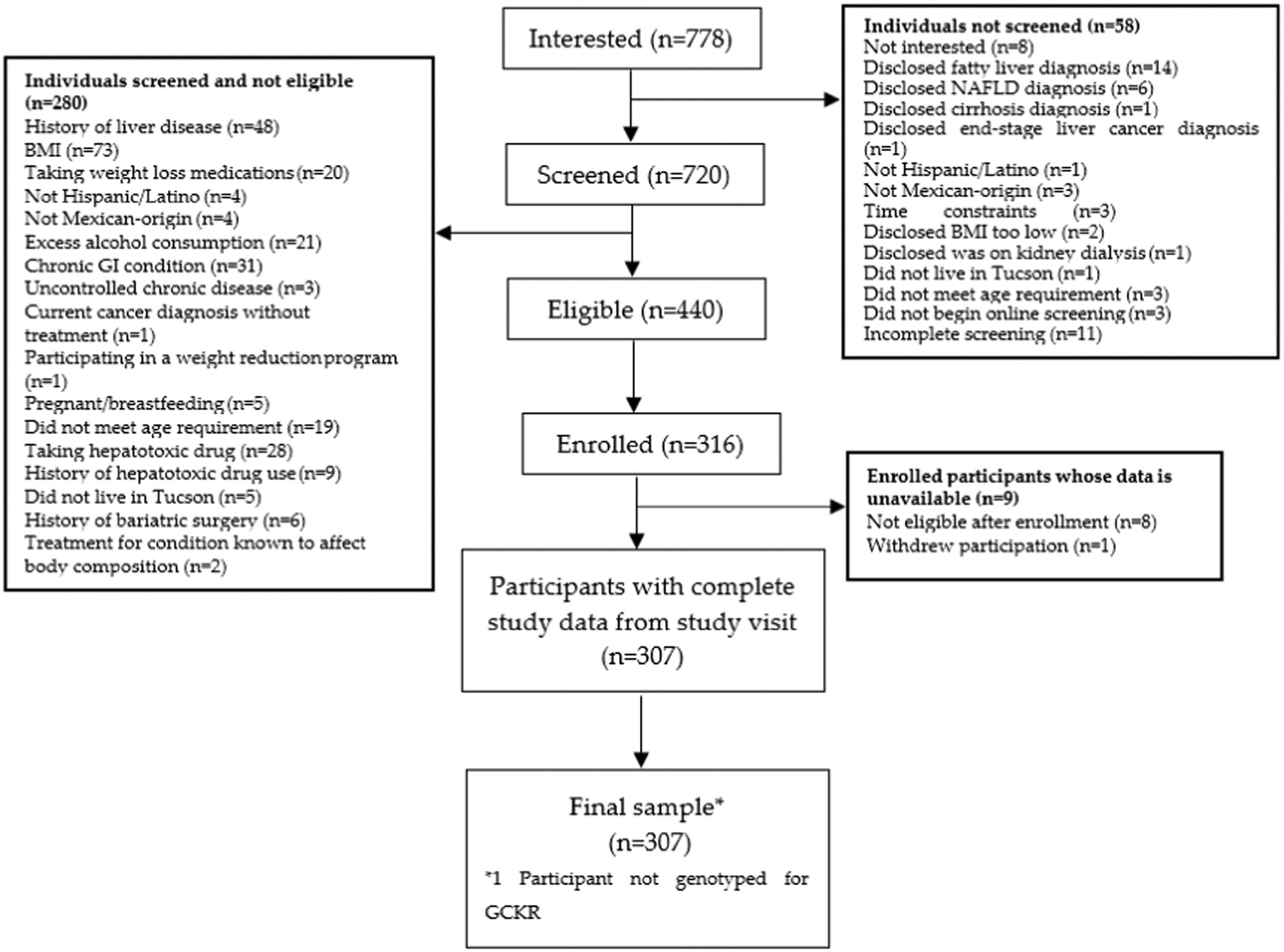
3Results3.1Participant characteristicsA total of 307 participants were included in this study (Fig. 1). Study participants were, on average, 45 years old and mostly female (Table 1). The median BMI was approximately 32 kg/m2 (32 kg/m2 for females and 31 kg/m2 for males), about 10% of all participants had self-reported type 2 diabetes, and most (51.1%) participants had an income under $29K. Mean hepatic steatosis was 288.1 dB/m, representing moderate levels of steatosis, and median liver fibrosis was 5.0 kPa, representing no or mild liver scarring.
Characteristics of Mexican-origin study participants (n=307).
| Trait | Female n=194 | Male n=113 | All Participants n=307 |
|---|---|---|---|
| Age in years, mean (SD) | 45.5 (10.6) | 43.0 (11.8) | 44.6 (11.1) |
| Waist Circumference (cm), median (IQR) | 103.1 (17.0) | 106.8 (14.6) | 105 (16.1) |
| BMI (kg/m2), median (IQR) | 32.3 (6.4) | 30.7 (5.6) | 31.7 (6.4) |
| Diabetes, n (%) | 21 (10.8) | 10 (8.9) | 31 (10.1) |
| Income Level, n (%) | |||
| <$29K | 100 (51.6) | 57 (50.4) | 157 (51.1) |
| $30K-59K | 68 (35.1) | 37 (32.7) | 105 (34.2) |
| >$60K | 26 (13.4) | 19 (16.8) | 45 (14.7) |
| Hepatic Steatosis1, (db/m), mean (SD) | 288.4 (48.6) | 287.8 (54.2) | 288.1 (50.6) |
| Liver Fibrosis2 (kPa), median (IQR) | 5.0 (1.8) | 5.0 (2.2) | 5.0 (2.0) |
All SNPs were in HWE, and all participants were genotyped for the LYPLAL1 rs12137855, PPP1R3B rs4240624, TM6SF2 rs58542926, MBOAT7 rs641738, and PNPLA3 rs738409 variants; however, GCKR rs1260326 allele carrier status was not available for one participant (Table 2). Participants who were homozygous for the G allele of PNPLA3 had the highest mean hepatic steatosis of 301.7 dB/m (Table 2).
SNP genotype relationships with hepatic steatosis among Mexican-origin study participants.
| Gene (n total) | RA (RAF)1 | SNP (n) | Mean Hepatic steatosis [SD] |
|---|---|---|---|
| LYPLAL1 (n=307) | T (0.31) | rs12137855 | |
| CC (n=211) | 289.3 (51.1) | ||
| CT (n=85) | 286.1 (50.8) | ||
| TT (n=11) | 281.1 (43.5) | ||
| GCKR2 (n=306) | T (0.54) | rs1260326 | |
| CC (n=142) | 290.5 (51.2) | ||
| CT (n=130) | 285.3 (52.0) | ||
| TT (n=34) | 289.5 (44.2) | ||
| PPP1R3B (n=307) | G (0.20) | rs4240624 | |
| AA (n=192) | 285.4 (47.9) | ||
| AG (n=105) | 293.4 (54.7) | ||
| GG (n=10) | 285.8 (58.9) | ||
| TM6SF2 (n=307) | T (0.05) | rs58542926 | |
| CC (n=276) | 287.4 (50.8) | ||
| CT (n=30) | 295.1 (50.4) | ||
| TT (n=1) | 281.0 (N/A) | ||
| MBOAT7 (n=307) | T (0.60) | rs641738 | |
| CC (n=122) | 292.5 (52.9) | ||
| CT (n=140) | 284.5 (48.1) | ||
| TT (n=45) | 287.9 (52.4) | ||
| PNPLA3 (n=307) | G (0.50) | rs738409 | |
| CC (n=80) | 280.9 (58.1) | ||
| CG (n=148) | 284.9 (45.1) | ||
| GG (n=79) | 301.7 (50.5) |
Only rs738409 in PNPLA3 was found to be statistically significantly associated with hepatic steatosis in our unadjusted and adjusted additive models (P=0.01) (Table 3). No SNPs were statistically significantly associated with hepatic steatosis in the dominant models. Only rs738409 was associated with hepatic steatosis in the unadjusted and adjusted recessive models (P=0.01 for both). Only the recessive model for rs4240624 (PPP1R3B) had a statistically significant interaction with BMI (P=0.0154). The polygenic risk score was not statistically significantly associated with hepatic steatosis in adjusted and unadjusted models (Table 4).
Associations between six SNPs and hepatic steatosis and interaction with BMI in a Mexican-origin population.
| SNP (Gene) | Model | RA | SNP Beta (SE) | SNP P-value1 | SNP-Adj Beta (SE) | SNP P-value1 | SNP*BMI Beta (SE) | SNP*BMI P-value2 |
|---|---|---|---|---|---|---|---|---|
| rs12137855 (LYPLAL1) | Additive | T | -3.52 (5.29) | 0.51 | -6.70 (4.79) | 0.16 | 0.26 (0.91) | 0.78 |
| DominantCT+TT vs CC | T | -3.79 (6.24) | 0.54 | -6.43 (5.63) | 0.25 | 0.40 (1.10) | 0.72 | |
| RecessiveTT vs CC+CT | T | -6.84 (15.57) | 0.66 | -17.78 (14.13) | 0.21 | -0.01 (2.64) | 0.99 | |
| rs1260326 (GCKR) | Additive | T | -2.08 (4.33) | 0.63 | -0.26 (3.91) | 0.95 | 0.17 (0.83) | 0.84 |
| DominantCT+TT vs CC | T | -4.37 (5.82) | 0.45 | -1.77 (5.28) | 0.74 | 0.79 (1.02) | 0.44 | |
| RecessiveTT vs CC+CT | T | 1.52 (9.24) | 0.87 | 3.25 (8.34) | 0.70 | -2.29 (2.16) | 0.28 | |
| rs4240624 (PPP1R3B) | Additive | G | 5.34 (5.22) | 0.31 | 0.52 (4.75) | 0.91 | 0.32 (0.93) | 0.73 |
| DominantAG+GG vs AA | G | 7.32 (5.97) | 0.22 | 2.04 (5.43) | 0.71 | 1.19 (1.02) | 0.25 | |
| RecessiveGG vs AG+AA | G | -2.42 (16.31) | 0.88 | -10.00 (14.72) | 0.50 | -8.13 (3.34) | 0.02 | |
| rs58542926 (TM6SF2) | Additive | T | 6.35 (9.15) | 0.49 | -0.86 (8.31) | 0.92 | 0.11 (1.59) | 0.95 |
| DominantCT+TT vs CC | T | 7.24 (9.60) | 0.45 | 0.29 (8.70) | 0.97 | 0.42 (1.68) | 0.80 | |
| RecessiveTT vs CC+CT | T | -7.16 (50.81) | 0.89 | -34.45 (45.91) | 0.45 | n/a | n/a | |
| rs641738 (MBOAT7) | Additive | T | -3.65 (4.17) | 0.38 | -4.10 (3.76) | 0.28 | 0.23 (0.76) | 0.76 |
| DominantCT+TT vs CC | T | -7.17 (5.90) | 0.23 | -7.88 (5.31) | 0.14 | -0.54 (1.06) | 0.61 | |
| RecessiveTT vs CC+CT | T | -0.29 (8.19) | 0.97 | -0.68 (7.39) | 0.93 | 1.85 (1.46) | 0.21 | |
| rs738409 (PNPLA3) | Additive | G | 10.37 (3.98) | 0.01 | 9.32 (3.59) | 0.01 | 0.83 (0.65) | 0.20 |
| DominantCG+GG vs CC | G | 9.82 (6.57) | 0.14 | 10.19 (5.92) | 0.09 | 1.03 (1.06) | 0.33 | |
| RecessiveGG vs GG+CG | G | 18.19 (6.54) | 0.01 | 15.04 (5.92) | 0.01 | 1.42 (1.13) | 0.21 |
RA: Risk Allele. SE: Standard Error. Adj: Adjusted for age, sex, and BMI. 1SNP P-value is the SNP main-effect P-value with BMI or interaction term in the model. SNP*BMI P-value are the interaction between BMI with SNP.
Polygenic risk score association with hepatic steatosis and interaction with BMI in a Mexican-origin population.
| Unadjusted Beta Coefficient PRS | ||||||
|---|---|---|---|---|---|---|
| Model | PRS Beta (SE) | PRS P-value | PRS-Adj Beta (SE) | PRS-Adj P-value | PRS*BMI Beta (SE) | PRS*BMI1 P-value |
| Six SNP PRS | 1.87 (2.01) | 0.35 | 0.41 (1.82) | 0.82 | 0.40 (0.35) | 0.25 |
Adj (Adjusted for age, sex, and BMI); SE (Standard Error); PRS (Polygenic Risk Score); 1PRS*BMI P-value is the interaction between BMI with SNP.
Among a sample of Mexican-origin adults, this study found no association between the following loci previously found to be associated with NAFLD and hepatic steatosis: rs12137855 (LYPLAL1), rs1260326 (GCKR), rs4240624 (PPP1R3B), rs58542926 (TM6SF2) or rs641738 (MBOAT7). Only rs738409 (PNPLA3) was significantly associated with hepatic steatosis. A polygenic risk score was not significantly associated with hepatic steatosis levels. There was also no interaction effect between any SNP and BMI in our population, except for the recessive model for rs4240624 (PPP1R3B).
Among Hispanics/Latinos, while PNPLA3 has been consistently found to be significantly associated with hepatic steatosis, inconsistent findings have been reported regarding PPP1R3B. A 2013 study conducted by Hernaez et al. found that while PPP1R3B and PNPLA3 variants were associated with hepatic steatosis among Mexican American adults, LYPLAL1 and GCKR were not [27]. Another 2013 study by Palmer et al. found that PNPLA3 and PPP1R3B were associated with hepatic steatosis among Hispanic/Latino Americans, but not LYPLAL1 or GCKR[28]. Finally, a 2015 study by León-Mimila et al. showed an association between PNPLA3, LYPLAL1, GCKR, and PPP1R3B polymorphisms and hepatic triglyceride content (HTG) in a population of Mexican individuals classified as morbidly obese [13]. However, when examining hepatic steatosis stage, only PNPLA3 was significantly associated with increased steatosis stage [13]. One potential explanation for the inconsistencies in the association of PPP1R3B and hepatic steatosis is differences in each study's method for measuring hepatic steatosis. Specifically, Hernaez et al. (2013), Palmer et al. (2013), and León-Mimila et al. (2015) measured hepatic steatosis using ultrasound, CT, and histology, respectively. It is possible that PPP1R3B is an indicator for glycogen and not hepatic steatosis in ultrasound and CT [14]. PPP1R3B encodes a glycogen-targeting subunit of PP1 protein phosphatase and is strongly expressed in the liver [29]. PPP1R3B overexpression in hepatocytes increases both basal and insulin-stimulated glycogen synthesis which amplifies hepatic glycogen deposition [29]. This may result in the misclassification of glycogen as steatosis. This is supported by a previous study in a population of European descent which found that PPP1R3B was associated with CT-diagnosed hepatic steatosis, but not histology-diagnosed hepatic steatosis [14], and a small study which did not find PPP1R3B to be associated with biopsy-proven diagnosed NAFLD [30].
A limitation of our study was the small sample size, particularly for TM6SF2 which had a risk allele frequency of 5%. However, the decreased power was partially mitigated by deriving a polygenic risk score. Another limitation was the exclusion of lean participants from the original study. However, it has been shown previously that risk factors for NAFLD in lean participants varies from those with higher BMI [31]. Lean NAFLD is more closely linked to genetic risk factors such as PNPLA3, indicating reduced metabolic adaptability at lower body weights. Moreover, an increase in visceral adipose tissue adds to the reduced metabolic adaptability and the likelihood of developing NAFLD in individuals with lower body weights [32]. Future research in this field should include Mexican-origin adults with lean NAFLD due to high incidence of PNPLA3, visceral adipose tissue, and insulin resistance [33]. Some strengths of our study include the well-defined phenotype of hepatic steatosis using transient elastography and the examination of these SNPs in an understudied population. Although CAP scores calculated from transient elastography are not the gold standard for measuring hepatic steatosis the noninvasive nature and high specificity and sensitivity (ranging from 78% to 100%) make it a good alternative to biopsy [6]. By focusing on hepatic steatosis specifically, rather than a diagnosis of NAFLD, we can elucidate the mechanism by which these SNPs contribute to the NAFLD phenotype (via hepatic steatosis). Furthermore, this study adds to the limited literature examining the associations of SNPs previously found to be associated with NAFLD in a population of Mexican origin adults, a population underrepresented in the liver disease literature despite a high burden of disease. This is also the first study to our knowledge to examine the associations of TM6SF2 and MBOAT7 variants with hepatic steatosis among Mexican-origin adults.
Our findings further support the role of PNPLA3 in hepatic steatosis in Mexican origin populations and expand on the limited literature evaluating the association of SNPs previously identified to be related to NAFLD with hepatic steatosis. We also found support for the hypothesis that previous associations between PPP1R3B and hepatic steatosis may be due to factors other than hepatic steatosis (i.e. glycogen) and may be attributable to imaging type [14]. The same may be said for the other variants for which we found no association either individually or in combination through a polygenic risk score. Future studies should more closely examine the relationship between the various SNPs related to NAFLD and each of the components that comprise this phenotype (hepatic steatosis, ALT, AST, etc). This would help elucidate the specific mechanisms by which each SNP is related to NAFLD. Further investigation of these SNPs in Hispanic/Latino populations could also help inform personalized treatment and prevention approaches to NAFLD.
5ConclusionsIn conclusion, this study found that among a sample of Mexican-origin adults, only PNPLA3 was significantly associated with hepatic steatosis. There was no significant association between any of the other loci previously found to be related to NAFLD in populations of European descent (LYPLAL1, GCKR, PPP1R3B, TM6SF2, MBOAT7). The inconsistent findings regarding PPP1R3B in previous studies among Hispanics/Latinos may be due to differences in measurement of hepatic steatosis using different imaging techniques. This study provides further support for the role of PNPLA3 in hepatic steatosis in Mexican-origin populations and highlights the need for further research to understand the specific mechanisms by which each SNP is related to NAFLD.
CRediT authorship contribution statementMario Jesus Trejo: Formal analysis, Writing – original draft, Writing – review & editing. Kristin E. Morrill: Methodology, Writing – original draft, Writing – review & editing. Yann C. Klimentidis: Formal analysis, Writing – original draft, Writing – review & editing. David O. Garcia: Methodology, Writing – original draft, Writing – review & editing.