Introduction and aim. Occult hepatitis B virus (HBV) infection (OBI) represents a state without detectable hepatitis B surface antigen, but positive for HBV DNA. The correlation between OBI and hepatocellular carcinoma (HCC) carcinogenesis is controversial. We studied the frequency and characteristics of OBI among HCC patients and metastatic liver cancer patients.
Material and methods. DNA was obtained from tumor and non-tumor tissues from 75 HCC patients (15 chronic hepatitis B (CHB), 39 chronic hepatitis C (CHC), 21 cryptogenic) and 15 metastatic liver cancer patients who underwent liver resection. HBV DNA and covalently-closed circular (ccc) DNA were detected using real-time polymerase chain reaction (PCR), and four HBV DNA regions were detected by nested PCR. Clinicopathological factors were compared between patients with and without OBI.
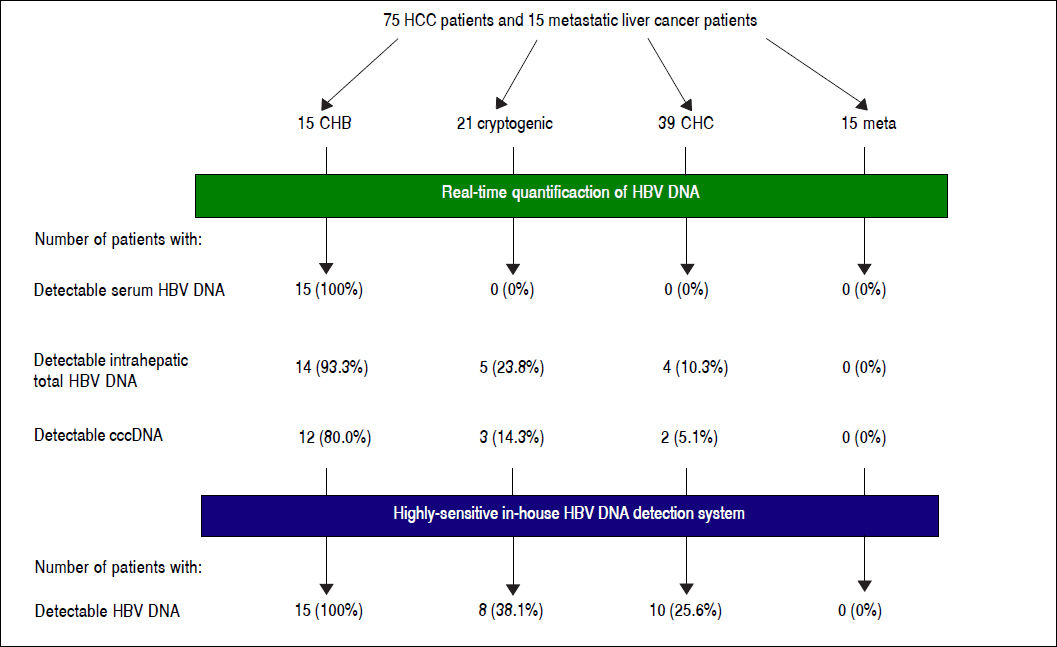
Results. HBV DNA was detected in 14 (93.3%) CHB, five (22.7%) cryptogenic and four (10.3%) CHC patients. cccDNA was detected in 12 (80.0%) CHB, three (14.3%) cryptogenic and two (5.1%) CHC patients. All CHB, eight (38.1%) cryptogenic and ten (25.6%) CHC patients tested positive with nested PCR. No metastatic liver cancer patients were positive for any HBV DNA regions. OBI patients had shorter prothrombin times (P = 0.0055), and lower inflammation activity score in non-tumor liver (P = 0.0274). There were no differences in anti-HBV antibodies.
Conclusions. OBI was detected in 38% of cryptogenic and 25.6% of CHC patients. There was no correlation between OBI and anti-HBV antibodies, but fewer patients with OBI had high inflammatory activity, suggesting that factors other than inflammation may be involved in HCC carcinogenesis in patients with OBI.
Hepatocellular carcinoma (HCC) is the sixth most common cancer and the third most common cause of cancer-related deaths worldwide.1 Chronic hepatitis B (CHB) infection with detectable circulating hepatitis B surface antigen (HBsAg) is a common cause of HCC. Approximately 60% of the annual total of 530,000 HCC cases worldwide are related to CHB.2 In addition to CHB, a past history of hepatitis B virus (HBV) infection among patients with chronic hepatitis C (CHC), and positivity for hepatitis B core antigen (HBcAg) have also been reported as risk factors for HCC carcinogenesis.3,4 HBV infects hepatocytes. For further investigations, specimens from the liver have to be used for the studies.
Occult HBV infection (OBI) is defined as a condition with undetectable HBsAg, but detectable HBV DNA in the serum or liver tissues. The incidence of OBI among CHC patients is known to be high, and about a third of CHC patients in the Mediterranean region were reported to be positive for OBI.5,6 OBI is of interest as a cause of de novo hepatitis B and reactivation of hepatitis B during chemotherapy and has also been reported as a cause of hepatitis B after blood transfusion.6 A previous study found that the disease-free survival period and overall survival period of OBI patients were shorter among HCC patients with CHC.7 A study in Hong Kong showed that the OBI rates in patients with cryptogenic HCC and HCC with CHC were 73% and 17%, respectively, and the authors noted a correlation between OBI and HCC.8 According to an 11-year prospective study of CHC patients in Italy, 35% of OBI-positive patients developed HCC, compared with only 9% of OBI-negative patients; the incidence of HCC was significantly higher among OBI-positive patients.9,10 In contrast, the Hepatitis C Antiviral Long-term Treatment against Cirrhosis (HALT-C) trial in the United States failed to find any correlation between HCC carcinogenesis and OBI.11 They detected HBV DNA in liver specimens of patients with and without HCC enrolled in the HALT-C trial and found no significant difference in positivity rates between the groups (10.7% vs. 23.6%, respectively). There is thus no consensus regarding the potential correlation between HCC carcinogenesis and OBI.
OBI is defined as negative HBsAg with positive detection of HBV DNA in serum or liver tissue. Some studies have detected HBV DNA in serum, while others have detected HBV DNA in liver tissue.7,8,10–12 In addition, some studies have detected HBV DNA using real-time polymerase chain reaction (PCR), and others have used nested PCR. The detection sites of HBV DNA also differ among studies. The detection sensitivities of different studies thus differ widely.
Whole-genome sequencing of HCC patients has recently revealed the integration sites of HBV DNA into the human genome.13,14 Based on these reports, we developed a sensitive in-house detection system for HBV DNA, using primers designed based on the reported integration sites of HBV DNA. In this study, we investigated the incidence of OBI among HCC patients and metastatic liver cancer patients using this highly-sensitive, in-house detection system, and revealed the characteristics of OBI-positive patients.
Material and MethodsPatientsTissue specimens were obtained from 90 patients who had undergone liver resection at the Department of Surgery and Science, Kyushu University Hospital, between March 2005 and January 2012. Fifteen HCC patients with CHB, 39 HCC patients with CHC, 21 patients with cryptogenic HCC, and 15 patients with metastatic liver cancer (13 colon cancer, 1 gastric cancer and 1 renal cell carcinoma) were enrolled in this study. No patients were diagnosed with alcoholic liver disease. DNA was extracted from exenterated HCC and peripheral liver tissues after surgery using a QIAamp DNA Mini Kit (Qiagen KK, Tokyo, Japan). No patients from the cryptogenic, CHC or metastatic liver tumor groups were positive for HBsAg or HBV DNA according to real-time PCR analysis of serum. Samples were collected according to an established protocol approved by the Ethics Committee of Kyushu University.
Pathological examination of the liverAll liver tissue specimens were evaluated by pathologists, who was blinded to the clinical condition of the patient. Inflammatory activity and fibrosis was graded and staged according to the New Inuyama classification system as follows: A0 (no necro-inflammatory reaction), A1 (mild), A2 (moderate), and A3 (severe) for inflammatory activity and F1 (periportal expansion), F2 (portoportal septa), F3 (portocentral linkage or bridging fibrosis), and F4 (cirrhosis) for fibrosis.
Quantification of intrahepatic HBV DNAIntrahepatic total HBV DNA, covalently-closed circular DNA (cccDNA), and human RNaseP contents in liver tissues were measured by real-time PCR.15 Briefly, intrahepatic total HBV DNA and cccDNA were measured using the primer-probe sets shown in table 1. Plasmid safe DNase I (Epicentre Inc. Chicago, IL) was used to digest the single-stranded region of the HBV genome, thus allowing enrichment of cccDNA for subsequent real-time PCR detection. Real-time PCR experiments were performed in a Light-Cycler (Roche Diagnostics, Basel, Switzerland) in 100-µL reaction volume. Amplification was performed as follows: 95 °C for 10 min, followed by 50 cycles of 95 °C for 10 s, 62 °C for 10 s, and 72 °C for 20 s. RNaseP amplification was performed using a Control Kit (Applied Biosystems inc., Caelsbad, CA). Serial dilutions of a plasmid containing a monomeric genotype C HBV insert were used as quantification standards. Taking into account the real-time PCR system used, the lower limit of detection of intrahepatic total HBV DNA and cccDNA quantification was 1×104 copies/cell.
Oligonucleotides used for real-time PCR and nested PCR identification of occult HBV infection.
| Region | Round | Forward primer | Reverse primer | Probe 1 | Probe 2 | |
|---|---|---|---|---|---|---|
| S | real-time PCR | ATCCTGCTGCTATGCCTCAT | GGGAAAGCCCTRCGAACCACTGAACAA | FAM-TGCACTTGTATTCCCATCCC-MGB | FAM-TGAGGCCCACTCCCATAGGA-MGB | |
| cccDNA | real-time PCR | TCCCCGTCTGTGCCTTCTC | GCACAGCTTGGAGGCTTGA | FAM-CCGTGTGCACTTC-MGB | FAM-CCGTGAACGCCCA-MGB | |
| S | nested PCR | 1st 2nd | AGGTATGTTGCCCGTTTGTC GTATGTTGCCCGTTTGTCCT | AAAGCCCTACGAACCACTGA AAGCCCTACGAACCACTGAA | ||
| Core | nested PCR | 1st 2nd | TTCAAGCCTCCAAGCTGTGC CAAGCCTCCAAGCTGTGCCT | TCTTCCAAATTACTTCCCACCCA TCTTCCAAATTACTTCCCACCCA | ||
| Pol | nested PCR | 1st 2nd | GGAGTGTGGATTCGCACTCC GGAGTGTGGATTCGCACTCC | CGTCTGCGAGGCGAGGGA CGAGGCGAGGGAGTTCTTCTTC | ||
| X | nested PCR | 1st 2nd | GGGCGCACCTCTCTTTACGC TCTCCCCGTCTGTGCCTTCTC | CATGCGACGTGCAGAGGTGAAG CATGCGACGTGCAGAGGTGAAG |
The targets of the S gene (255 bp, nucleotide positions 458-712), X gene (91 bp, nucleotide positions 1521-1611), C gene (272 bp, nucleotide positions 1863-2134) and Pol gene (137 bp, nucleotide positions 2316-2452) were amplified by nested PCR and sequenced to detect sensitivity. The primers used are shown in table 1. Nested PCR reactions were performed with these primers for 35 cycles (95 °C, 15 s; 58 °C, 30 s; 72 °C, 30 s) in first and second PCR using Veriti (Applied Biosystems, Foster City, CA, USA). The PCR products were sequenced on both strands using a BigDye Terminator V3.1 cycle sequencing kit (Applied Biosystems), with the same primers used for the second PCR. The sequencing products were analyzed using an ABI 3130xl DNA analyzer (Applied Biosystems). The obtained sequences were aligned with GenBank sequences corresponding to HBV genotypes. The lower limit of detection was 1-10 copies/mL.
Statistical analysisNon-parametric tests (χ2 and Fisher’s exact probability tests) were used to compare the characteristics of the groups. Univariate logistic regression analysis was used to identify the factors that contributed significantly to early viral dynamics. The odds ratios and 95% confidence intervals were also calculated. All p values < 0.05 using two-tailed tests were considered significant. Analyses were performed using JMP (version 9.0.2, SAS, Inc., Cary, NC, USA).
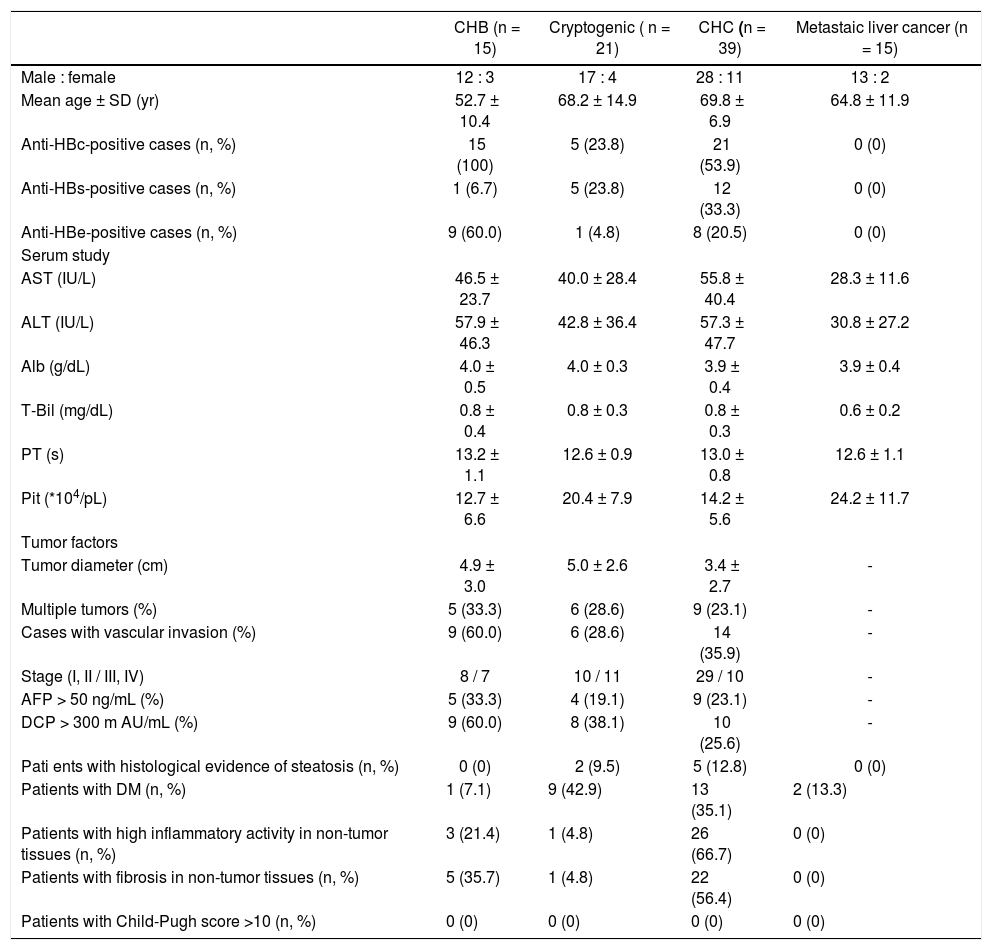
ResultsPatient backgroundsThe demographic data for the 90 patients are shown in table 2. Five patients with cryptogenic HCC (23.8%) and 21 with CHC (53.9%) were anti-HBc antibody (HBcAb)-positive. Five patients with cryptogenic HCC (23.8%) and 12 with CHC (33.3%) were anti-HBs antibody (HBsAb)-positive. No serological HBV infection markers were detected in patients with metastatic liver cancer. Two patients with cryptogenic HCC (9.5%) showed histological evidence of steatosis. One patient with cryptogenic HCC (4.8%), 26 with CHC (66.7%), and three with CHB (21.4%) showed high inflammatory activity in non-tumor tissue. Four patients with cryptogenic HCC (18.2%), 22 with CHC (56.4%), and five with CHB (35.7%) demonstrated fibrosis of non-tumor tissue. No patients with metastatic liver cancer showed high inflammation or fibrosis in non-tumor liver tissue.
Patient backgrounds
| CHB (n = 15) | Cryptogenic ( n = 21) | CHC (n = 39) | Metastaic liver cancer (n = 15) | |
|---|---|---|---|---|
| Male : female | 12 : 3 | 17 : 4 | 28 : 11 | 13 : 2 |
| Mean age ± SD (yr) | 52.7 ± 10.4 | 68.2 ± 14.9 | 69.8 ± 6.9 | 64.8 ± 11.9 |
| Anti-HBc-positive cases (n, %) | 15 (100) | 5 (23.8) | 21 (53.9) | 0 (0) |
| Anti-HBs-positive cases (n, %) | 1 (6.7) | 5 (23.8) | 12 (33.3) | 0 (0) |
| Anti-HBe-positive cases (n, %) | 9 (60.0) | 1 (4.8) | 8 (20.5) | 0 (0) |
| Serum study | ||||
| AST (IU/L) | 46.5 ± 23.7 | 40.0 ± 28.4 | 55.8 ± 40.4 | 28.3 ± 11.6 |
| ALT (IU/L) | 57.9 ± 46.3 | 42.8 ± 36.4 | 57.3 ± 47.7 | 30.8 ± 27.2 |
| Alb (g/dL) | 4.0 ± 0.5 | 4.0 ± 0.3 | 3.9 ± 0.4 | 3.9 ± 0.4 |
| T-Bil (mg/dL) | 0.8 ± 0.4 | 0.8 ± 0.3 | 0.8 ± 0.3 | 0.6 ± 0.2 |
| PT (s) | 13.2 ± 1.1 | 12.6 ± 0.9 | 13.0 ± 0.8 | 12.6 ± 1.1 |
| Pit (*104/pL) | 12.7 ± 6.6 | 20.4 ± 7.9 | 14.2 ± 5.6 | 24.2 ± 11.7 |
| Tumor factors | ||||
| Tumor diameter (cm) | 4.9 ± 3.0 | 5.0 ± 2.6 | 3.4 ± 2.7 | - |
| Multiple tumors (%) | 5 (33.3) | 6 (28.6) | 9 (23.1) | - |
| Cases with vascular invasion (%) | 9 (60.0) | 6 (28.6) | 14 (35.9) | - |
| Stage (I, II / III, IV) | 8 / 7 | 10 / 11 | 29 / 10 | - |
| AFP > 50 ng/mL (%) | 5 (33.3) | 4 (19.1) | 9 (23.1) | - |
| DCP > 300 m AU/mL (%) | 9 (60.0) | 8 (38.1) | 10 (25.6) | - |
| Pati ents with histological evidence of steatosis (n, %) | 0 (0) | 2 (9.5) | 5 (12.8) | 0 (0) |
| Patients with DM (n, %) | 1 (7.1) | 9 (42.9) | 13 (35.1) | 2 (13.3) |
| Patients with high inflammatory activity in non-tumor tissues (n, %) | 3 (21.4) | 1 (4.8) | 26 (66.7) | 0 (0) |
| Patients with fibrosis in non-tumor tissues (n, %) | 5 (35.7) | 1 (4.8) | 22 (56.4) | 0 (0) |
| Patients with Child-Pugh score >10 (n, %) | 0 (0) | 0 (0) | 0 (0) | 0 (0) |
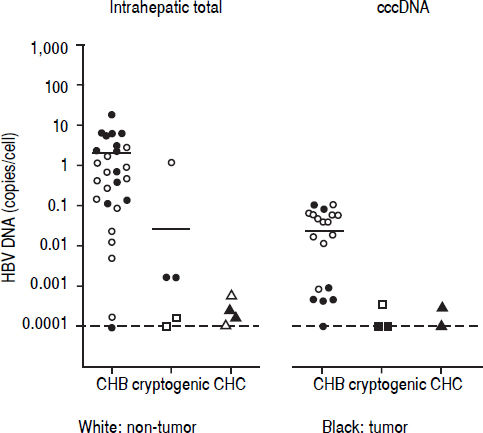
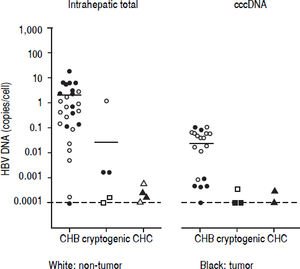
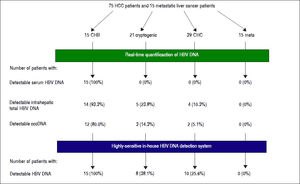
Detection of HBV DNA and cccDNA in tumor and peripheral non-tumor tissues were performed using realtime PCR. HBV DNA was detected in 14 (93.3%) patients with CHB (median, 0.6867 copies/cell; range, < 0.000118.12 copies/cell) and cccDNA was detected in 12 (80.0%) patients with CHB (median, 0.0398 copies/cell; range, < 0.0001-0.1035 copies/cell), respectively (Figure 1). HBV DNA was detected in five (22.7%) patients with cryptogenic HCC (median, 0.0002 copies/cell; range, < 0.0001-1.1735 copies/cell) and four (10.3%) patients with CHC (median, 0.0002 copies/cell; range, < 0.0001-0.0006 copies/cell), and cccDNA was detected in three (14.3%) patients with cryptogenic HCC (median, 0.0001 copies/cell; range, < 0.0001-0.0004 copies/cell) and two (5.1%) patients with CHC (median, 0.0002 copies/cell; range, < 0.0001-0.0003 copies/cell).
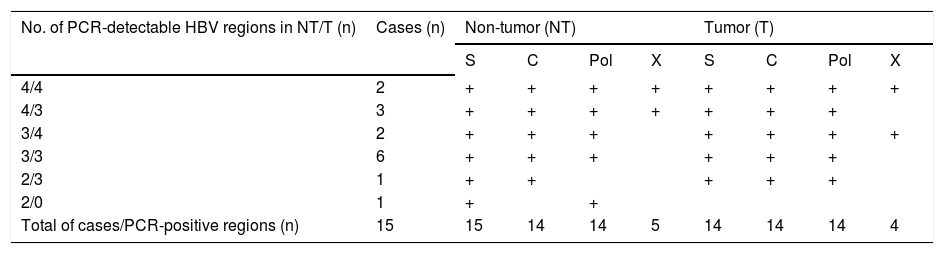
Detection of intrahepatic HBV DNA using a highly-sensitive in-house detection systemDetection of HBV DNA in tumor and non-tumor tissues was performed using a highly-sensitive in-house HBV DNA detection system, using nested PCR on four regions (S, C, Pol and X) of the HBV DNA. Five patients with CHB were positive for all four regions in non-tumor tissue (Table 3). All CHB patients were positive for at least two regions in non-tumor tissue, and 14 patients were positive for at least three regions in tumor tissue.
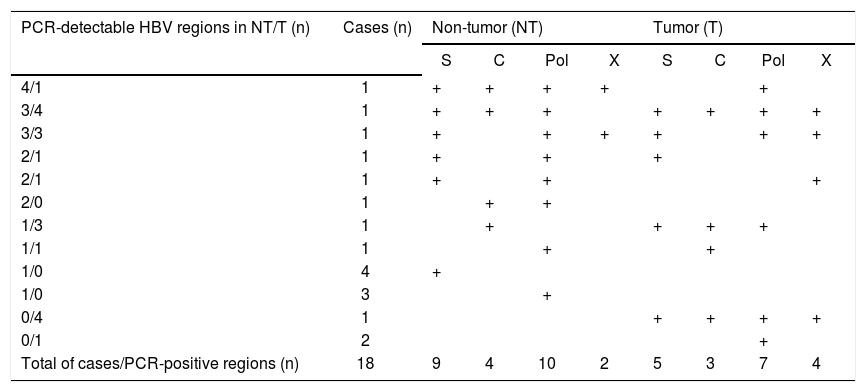
Distribution of HBV regions detectable by highly-sensitive in-house detection system in non-tumor and tumor tissues in 18 HCC patients with OBI.
| PCR-detectable HBV regions in NT/T (n) | Cases (n) | Non-tumor (NT) | Tumor (T) | ||||||
|---|---|---|---|---|---|---|---|---|---|
| S | C | Pol | X | S | C | Pol | X | ||
| 4/1 | 1 | + | + | + | + | + | |||
| 3/4 | 1 | + | + | + | + | + | + | + | |
| 3/3 | 1 | + | + | + | + | + | + | ||
| 2/1 | 1 | + | + | + | |||||
| 2/1 | 1 | + | + | + | |||||
| 2/0 | 1 | + | + | ||||||
| 1/3 | 1 | + | + | + | + | ||||
| 1/1 | 1 | + | + | ||||||
| 1/0 | 4 | + | |||||||
| 1/0 | 3 | + | |||||||
| 0/4 | 1 | + | + | + | + | ||||
| 0/1 | 2 | + | |||||||
| Total of cases/PCR-positive regions (n) | 18 | 9 | 4 | 10 | 2 | 5 | 3 | 7 | 4 |
Eight patients with cryptogenic HCC (38.1%) and 10 with CHC (25.6%) were positive for at least one region of HBV DNA (Table 4). HBV DNA was detected in 15 nontumor and 10 tumor tissues from non-CHB patients. The Pol region of HBV DNA was detected in 10 non-tumor and seven tumor tissues, while the HBx region was only detected in two non-tumor and three tumor tissues. The sequences of the nested PCR products were checked to rule out any contamination. We, therefore, defined OBI as positivity for at least one region of HBV DNA. Eighteen patients were diagnosed with OBI-positive (Figure 2).
Distribution of HBV regions detectable by highly-sensitive, in-house detection system in non-tumor and tumor tissues from 15 HCC patients with CHB
| No. of PCR-detectable HBV regions in NT/T (n) | Cases (n) | Non-tumor (NT) | Tumor (T) | ||||||
|---|---|---|---|---|---|---|---|---|---|
| S | C | Pol | X | S | C | Pol | X | ||
| 4/4 | 2 | + | + | + | + | + | + | + | + |
| 4/3 | 3 | + | + | + | + | + | + | + | |
| 3/4 | 2 | + | + | + | + | + | + | + | |
| 3/3 | 6 | + | + | + | + | + | + | ||
| 2/3 | 1 | + | + | + | + | + | |||
| 2/0 | 1 | + | + | ||||||
| Total of cases/PCR-positive regions (n) | 15 | 15 | 14 | 14 | 5 | 14 | 14 | 14 | 4 |
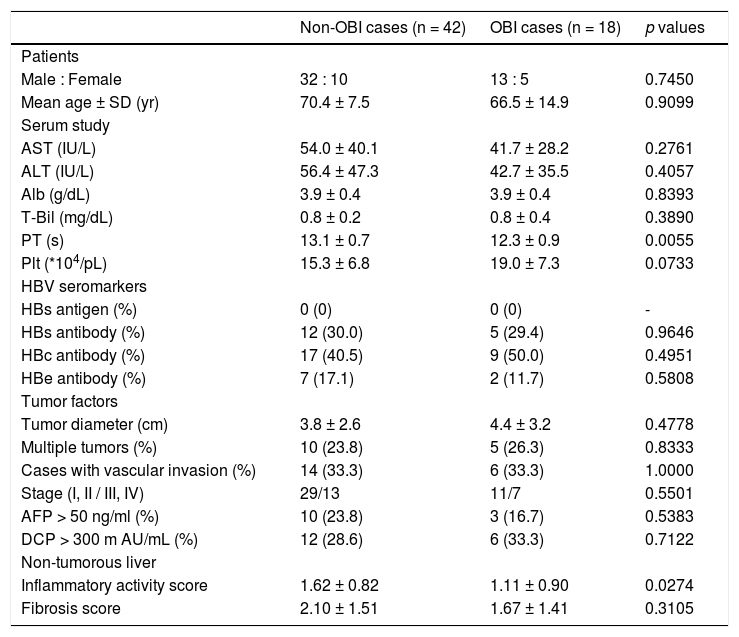
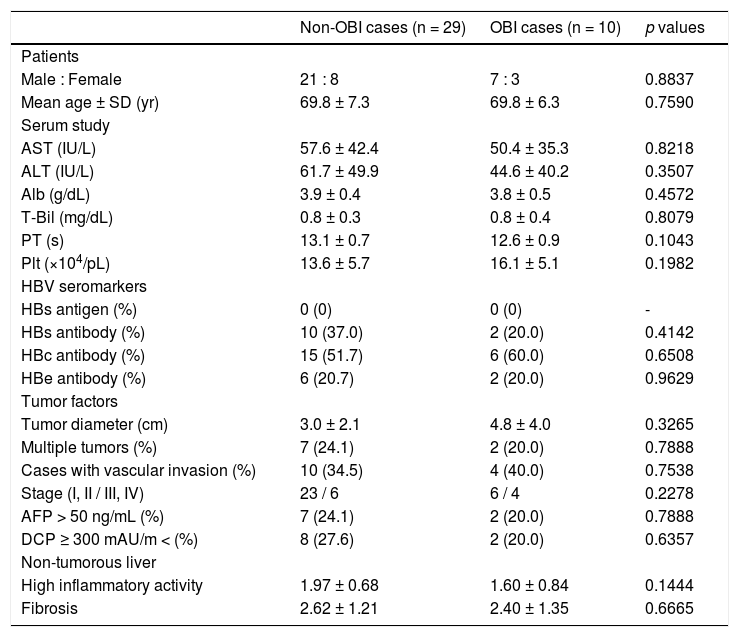
We compared the clinicopathological findings of OBI-positive and OBI-negative patients (Table 5). There was no significant difference in terms of sex or age of the patients between the two groups. Prothrombin time (PT) was significantly shorter (p = 0.0055), and transaminases and platelets tended to be lower in OBI cases. Inflammatory activity score in non-tumor liver tissue was significantly lowere in OBI cases (p = 0.0274). There were no differences in HBV seromarkers, including HBsAb and HBcAb, or in tumor factors, including tumor diameter and tumor staging, between OBI-positive and -negative cases. We also compared clinicopathological findings between OBI-positive and OBI-negative CHC patients (Table 6), which showed the same tendencies in terms of serological liver function markers. Similarly, OBI-positive cases had lower inflammatory activity in non-tumorous liver (p = 0.1444).
Characteristics of OBI-positive HCC patients.
| Non-OBI cases (n = 42) | OBI cases (n = 18) | p values | |
|---|---|---|---|
| Patients | |||
| Male : Female | 32 : 10 | 13 : 5 | 0.7450 |
| Mean age ± SD (yr) | 70.4 ± 7.5 | 66.5 ± 14.9 | 0.9099 |
| Serum study | |||
| AST (IU/L) | 54.0 ± 40.1 | 41.7 ± 28.2 | 0.2761 |
| ALT (IU/L) | 56.4 ± 47.3 | 42.7 ± 35.5 | 0.4057 |
| Alb (g/dL) | 3.9 ± 0.4 | 3.9 ± 0.4 | 0.8393 |
| T-Bil (mg/dL) | 0.8 ± 0.2 | 0.8 ± 0.4 | 0.3890 |
| PT (s) | 13.1 ± 0.7 | 12.3 ± 0.9 | 0.0055 |
| Plt (*104/pL) | 15.3 ± 6.8 | 19.0 ± 7.3 | 0.0733 |
| HBV seromarkers | |||
| HBs antigen (%) | 0 (0) | 0 (0) | - |
| HBs antibody (%) | 12 (30.0) | 5 (29.4) | 0.9646 |
| HBc antibody (%) | 17 (40.5) | 9 (50.0) | 0.4951 |
| HBe antibody (%) | 7 (17.1) | 2 (11.7) | 0.5808 |
| Tumor factors | |||
| Tumor diameter (cm) | 3.8 ± 2.6 | 4.4 ± 3.2 | 0.4778 |
| Multiple tumors (%) | 10 (23.8) | 5 (26.3) | 0.8333 |
| Cases with vascular invasion (%) | 14 (33.3) | 6 (33.3) | 1.0000 |
| Stage (I, II / III, IV) | 29/13 | 11/7 | 0.5501 |
| AFP > 50 ng/ml (%) | 10 (23.8) | 3 (16.7) | 0.5383 |
| DCP > 300 m AU/mL (%) | 12 (28.6) | 6 (33.3) | 0.7122 |
| Non-tumorous liver | |||
| Inflammatory activity score | 1.62 ± 0.82 | 1.11 ± 0.90 | 0.0274 |
| Fibrosis score | 2.10 ± 1.51 | 1.67 ± 1.41 | 0.3105 |
Characteristics of OBI-positive HCC patients with CHC.
| Non-OBI cases (n = 29) | OBI cases (n = 10) | p values | |
|---|---|---|---|
| Patients | |||
| Male : Female | 21 : 8 | 7 : 3 | 0.8837 |
| Mean age ± SD (yr) | 69.8 ± 7.3 | 69.8 ± 6.3 | 0.7590 |
| Serum study | |||
| AST (IU/L) | 57.6 ± 42.4 | 50.4 ± 35.3 | 0.8218 |
| ALT (IU/L) | 61.7 ± 49.9 | 44.6 ± 40.2 | 0.3507 |
| Alb (g/dL) | 3.9 ± 0.4 | 3.8 ± 0.5 | 0.4572 |
| T-Bil (mg/dL) | 0.8 ± 0.3 | 0.8 ± 0.4 | 0.8079 |
| PT (s) | 13.1 ± 0.7 | 12.6 ± 0.9 | 0.1043 |
| Plt (×104/pL) | 13.6 ± 5.7 | 16.1 ± 5.1 | 0.1982 |
| HBV seromarkers | |||
| HBs antigen (%) | 0 (0) | 0 (0) | - |
| HBs antibody (%) | 10 (37.0) | 2 (20.0) | 0.4142 |
| HBc antibody (%) | 15 (51.7) | 6 (60.0) | 0.6508 |
| HBe antibody (%) | 6 (20.7) | 2 (20.0) | 0.9629 |
| Tumor factors | |||
| Tumor diameter (cm) | 3.0 ± 2.1 | 4.8 ± 4.0 | 0.3265 |
| Multiple tumors (%) | 7 (24.1) | 2 (20.0) | 0.7888 |
| Cases with vascular invasion (%) | 10 (34.5) | 4 (40.0) | 0.7538 |
| Stage (I, II / III, IV) | 23 / 6 | 6 / 4 | 0.2278 |
| AFP > 50 ng/mL (%) | 7 (24.1) | 2 (20.0) | 0.7888 |
| DCP ≥ 300 mAU/m < (%) | 8 (27.6) | 2 (20.0) | 0.6357 |
| Non-tumorous liver | |||
| High inflammatory activity | 1.97 ± 0.68 | 1.60 ± 0.84 | 0.1444 |
| Fibrosis | 2.62 ± 1.21 | 2.40 ± 1.35 | 0.6665 |
We performed real-time PCR on two regions and nested PCR on four regions of HBV DNA, and checked the sequences of the nested PCR products. To the best of our knowledge, the sensitivity of this in-house HBV DNA detection system is higher than those used in previous studies. Among 60 HCC cases, excluding patients with CHB, 18 (30.0%) were diagnosed as OBI-positive. Ten of 38 CHC cases (25.6%) and eight out of 21 cryptogenic cases (38.1%) were also diagnosed as OBI-positive. No patients with metastatic liver cancer were OBI-positive. In a previous study from Hong Kong, one of six patients with CHC and HCC (16.7%) and 24 of 33 with cryptogenic HCC (72.7%) were reported to be OBI-positive.[8] In CHC patients with HCC, 15 out of 80 cases in Taiwan (20.0%), three out of 91 in the United States (3.3%), and 45 out of 73 in Italy (61.6%) were OBI-positive.7–9,11 These differences are thought to reflect differences in the causes of infection and the infection rates of HBV and HCV, and differences in HBV genotypes among different areas. Because of the geographical distance and major HBV genotypes B and C, it was reasonable that the OBI rates in Hong Kong and Taiwan were consistent with those in the current study. The fact that the HBV DNA detection rate in non-tumor tissue was higher than in tumor tissue was compatible with the results of previous reports.
The sensitivities and specificities of HBV detection appear to differ greatly among studies, but this is not surprising given that the HBV DNA detection methods used (real-time PCR or nested PCR), and the regions of HBV DNA detected differ among studies. In most studies, OBI cases were defined by positive detection of at least two regions of HBV DNA, to prevent the occurrence of false positive results due to contamination. However, we checked the sequences of the nested PCR products to rule out the possibility of contamination, and we, therefore, defined OBI cases as those positive for at least one region of HBV DNA.
HBV is a double-stranded DNA virus, the DNA sequence of which is highly variable. We designed the primers based on full-length sequences of HBV DNA in the NCBI database, using lesions with few reported variations. We also referred to studies that performed whole-genome sequencing of HCC patients and identified integration sites of HBV DNA into the host genome, and designed the primers based on these reported integration sites.13,14 However, as shown in table 5, the positivity rates varied between HBV DNA lesions, with the positivity rates of HBc and HBx lesions being lower than others. This may suggest that the HBV responsible for OBI contains variations of HBc and HBx DNA sequences that differ from those in the NCBI database. Further studies are needed to clarify this.
In the current study, the positivity rates for HBsAb, HBcAb and anti-HBe antibody (HBeAb) were 29.4%, 50.0% and 11.7% in the OBI group and 30.0%, 40.5% and 17.1% in the control group, respectively, with no significant differences between the groups. Squadrito et al. reported that HBcAb-positivity was higher in OBI-positive than in OBI-negative CHC patients.10 Wong, et al. reported that HBcAb- and HBsAb-positivity were 70.0% and 60.0%, respectively, in OBI-positive operated cases of HCC. Furthermore, 10.0% of OBI-positive HCC cases were negative for all serum HBV markers.8 Further studies are needed to clarify the relationship between HBV-related antibodies and OBI.
In this study, inflammation in non-tumor tissue was significantly lower, and serum liver function markers tended to be better among OBI-positive patients. This suggests that factors not involving inflammation of the background liver may be involved in carcinogenesis. Further studies are needed to clarify the relationship between OBI and liver carcinogenesis.
Abbreviations- •
ALT: alanine aminotransferase.
- •
AST: aspartate aminotransferase.
- •
CHB: chronic hepatitis B.
- •
CHC: chronic hepatitis C.
- •
HALT-C: Hepatitis C Antiviral Long-term Treatment against Cirrhosis.
- •
HBcAb: anti-hepatitis B core antibody.
- •
HBcAg: hepatitis B core antigen.
- •
HBeAb: anti-hepatitis B envelope protein.
- •
HBsAb: anti-hepatitis B surface antibody.
- •
HBsAg: hepatitis B surface antigen.
- •
HBV: hepatitis B virus.
- •
HCC: hepatocellular carcinoma.
- •
OBI: occult hepatitis B virus infection.
- •
PCR: polymerase chain reaction.
- •
PT: prothrombin time.
- •
TERT: telomerase reverse transcriptase.
The Ministry of Health Labour and Welfare, Health Labour Sciences Research. Grant (H23-Kanen-Ippan-003).
The Ministry of Health Labour and Welfare, Health Labour Sciences Research. Grant (H24-Kanen-Ippan-004).
Conflicts of InterestThe authors have no conflicts of interest.