It is well-known that signaling mediated by the hepatocyte growth factor (HGF) and its receptor c-Met in the liver is involved in the control of cellular redox status and oxidative stress, particularly through its ability to induce hepatoprotective gene expression by activating survival pathways in hepatocytes. It has been reported that HGF can regulate the expression of some members of the NADPH oxidase family in liver cells, particularly the catalytic subunits and p22phox. In the present work we were focused to characterize the mechanism of regulation of p22phox by HGF and its receptor c-Met in primary mouse hepatocytes as a key determinant for cellular redox regulation.
Materials and methodsPrimary mouse hepatocytes were treated with HGF (50 ng/mL) at different times. cyba expression (gene encoding p22phox) or protein content were addressed by real time RT-PCR, Western blot or immunofluorescence. Protein interactions were explored by immunoprecipitation and FRET analysis.
ResultsOur results provided mechanistic information supporting the transcriptional repression of cyba induced by HGF in a mechanism dependent of NF-κB activity. We identified a post-translational regulation mechanism directed by p22phox degradation by proteasome 26S, and a second mechanism mediated by p22phox sequestration by c-Met in plasma membrane.
ConclusionOur data clearly show that HGF/c-Met exerts regulation of the NADPH oxidase by a wide-range of molecular mechanisms. NADPH oxidase-derived reactive oxygen species regulated by HGF/c-Met represents one of the main mechanisms of signal transduction elicited by this growth factor.
The liver is perhaps the major organ capable to efficiently handling profound changes in cellular redox status and oxidative stress [1]. In fact, hepatocytes have gained advantages using reactive oxygen species (ROS) generated as by-products in many metabolic and biotransformation processes [2]. But some ROS-generating systems function exclusively for ROS production, particularly for cell signaling or in the respiratory burst in phagocytic cells. The nicotinamide adenine dinucleotide phosphate (NADPH) oxidase is a remarkable system dedicated to the production of ROS. This system is ubiquitously distributed, not only in phagocytic cells, but in practically all cell types in mammals [3].
In the liver, the NADPH oxidase plays outstanding function in normal physiology and its dysfunction is involved in virtually all liver diseases [4]. A system with these characteristics must be adequately regulated, particularly since NADPH oxidases play a role in signal transduction in non-phagocytic cells as we and others have reported [2,3,5].
The hepatocyte growth factor (HGF) is a multitask receptor tyrosine kinase that in addition of its well-known antiapoptotic and mitogenic properties, has the ability to exerts other functions such as motogenesis, morphogenesis, and metabolic control. In all cases its function is regulated by a fine-tuned ROS production [6–8].
HGF and its receptor c-Met display protective effects in the liver regulating the cellular redox status in apoptosis [2], fibrosis [9], cancer [7,10], drug-induced liver injury [11,12], NASH [6,13], cholestasis [14], and alcoholic liver disease [15], even more this protective response has been observed in the pancreas [16], lungs [12], among other organs.
We have previously reported that HGF/c-Met exerts regulatory properties in the NADPH oxidase complex in hepatocytes. The c-Met KO mouse liver exhibits oxidative stress at basal conditions [7], and even sublethal dose of FAS agonist such as Jo2 antibody induces massive apoptosis in the liver of these animals [1]. Hepatocytes from c-Met KO mice are susceptible to apoptosis due to the oxidative stress produced by the overactivation of the NADPH oxidase, in fact, treatment of wild type hepatocytes with HGF induces the downregulation of the catalytic subunit Nox2 and the regulatory subunit p22phox. The p22phox subunit of the oxidase complex is a key regulatory protein which permanently resides in membranes, and it is a common member of at least four members of the NADPH oxidase family [17,18]. HGF/c-Met as a master regulator of the cellular redox state in the liver, takes control of the pro-oxidative system by regulating p22phox. In the present work we provide mechanistic evidence regarding the regulation of p22phox by HGF/c-Met in primary mouse hepatocytes, particularly related to the transcriptional and post-translational regulation of this relevant subunit of the NADPH oxidases.
2Materials and methods2.1MaterialsAll reagents were purchased from Sigma-Aldrich Inc. (Saint Louis, MO, USA) except where noted. The human recombinant hepatocyte growth factor (HGF) and transforming growth factor beta (TGF-β) were obtained from PeproTech (Rocky Hill, NJ, USA).
2.2Primary mouse hepatocytes isolation, purification and culture8−10-weeks-old C57BL/6 J male mice were obtained from the Universidad Autonoma Metropolitana Iztapalapa (UAM-I) animal facility and were maintained in pathogen-free conditions with controlled temperature and humidity on a 12 h light-dark cycle in the same facility. The experimental protocols used were approved and carried out in accordance with the UAM-I experimental animal guidelines and NIH Guide for the Care and Use of Laboratory Animals. Hepatocytes were isolated by a two-step collagenase perfusion, as we previously described [5]. The viability was >90% as assessed by trypan blue exclusion test. Hepatocytes were seeded at 2.13 × 105 cells/cm2 in 10-cm dishes (Thermo Scientific, Waltham, MA, USA) in the Ham´s F-12/Dulbecco´s modified Eagle´s basal hepatocyte growth medium supplemented with 10% fetal bovine serum. After 4 h of attachment, the medium was replaced to a serum-free basal hepatocyte growth medium. The following day, cells were treated with 50 ng/mL HGF, 10 ng/mL TGF-β, 200 nM epoxomicin (epox, 30 min before HGF treatment), 30 µM of SN50 (Merck and Co., Kenilworth, NJ, USA. 30 min before HGF treatment) or 2.5 mM Sulfasalazine (30 min before HGF treatment). Time of treatment was according to each experiment.
2.3HepG2 cell line cultureHepG2 cell line was obtained from American Type Culture Collection, cells were grown in DMEM media supplemented with 10% heat-inactivated fetal bovine serum, 100 U/mL penicillin and 100 μg/mL streptomycin, at 37 °C in a 5% CO2 atmosphere as previously published [19].
2.4Real-time RT-PCRAfter treatments, cells were subjected to RNA isolation, and DNase digestion with a DNA-free kit (Ambion, Inc, Austin TX, USA), 1 μg of total RNA was reverse transcribed in 20 μL reaction volume with a SuperScript (Invitrogen Corp, Carlsbad CA, USA) first-strand synthesis kit according to the manufacturer’s instructions. The qRT-PCR analysis was performed with a CFX96 Touch (Bio-Rad, Hercules, CA, USA) thermal cycler in a 96-wells reaction plate. The 10 μL PCR reaction mix contained 5 μL 2X SYBR Green PCR Master Mix (Bio-Rad), 200 nM of each primer, and 1 μL cDNA template. Reactions were incubated for 10 min at 95 °C followed by 40 cycles of 30 s at 95 °C and 60 s at specific primer temperature. Melting analysis of the PCR products was also conducted to validate the amplification of the specific product. The expression level of mouse rps18 was used as an internal reference. Relative gene expression level was calculated with the 2−ΔΔCT method. Primer sequences are listed in Supplementary Table 1.
2.5In silico analysis of cyba gene promoterFor the identification of canonical bindings sites for transcription factors (TFs) in the cyba promoter (2000 base pairs upstream of the start of transcription), the JASPAR platform was used (http://jaspar.genereg.net) with a relative profile score threshold of 95%.
2.6cyba promoter analysisFor the amplification of the different segments of the cyba promoter, we used the oligonucleotides listed in Supplementary Table 2 which have cut-off sequences for restriction enzymes (HindIII in the forward oligonucleotides; and EcoRI in the reverse oligonucleotides). Products were cloned in the promoterless pmCherry plasmid (Life Technologies, CA, US). Plasmid were transfected as stated below in HepG2 cells. mCherry fluorescence was recorded in a DTX800 Multimode detector microplate reader (Beckman Coulter, Brea, CA, USA) at 587 nm of excitation and 610 nm of emission. Data are reported as absolute fluorescence.
2.7Transfection with decoy oligodeoxynucleotides (ODN)HepG2 cells were seeded in 24-wells plates at a density of 3 × 105 cells/cm2 and transfected with ODN 100 nM. (NF-κB: 5′-AGA GGA ATT TCC ACG ATT-3′. AP-1: 5′-TGT GAT GAC TCA GGT TTG-3′, Mock 5′-CAC AAA GTG TAA CAG TCT-3′) using the cationic liposome vector Lipofectamine 2000 (Invitrogen) according to the manufacturer’s instructions [20]. After 6 h incubation, cells were washed twice with serum-free culture medium and left at 37 °C and 5% CO2 for at least 4 h before HGF treatment (50 ng/mL) for a further 12 h.
2.8ImmunofluorescenceImmunofluorescence was performed as previously published [6,9]. Slides were examined using the Carl Zeiss LSM 780 NLO confocal microscope (Carl Zeiss, Oberkochen, Germany).
2.9Plasmids transfectionPlasmids were transfected by using Lipofectamine 2000 (Invitrogen Inc.) in HepG2 cells at 60% of confluence. We transfected adherent cells with 100 ng of DNA according to the manufacturer's instructions. After transfection, the cells were incubated for further 2–3 days.
2.10Western blotWestern blotting was performed following the protocol previously reported [2,21], using different antibodies as shown in Supplementary Table 3.
2.11Immunoprecipitation1−2 mg of whole cell lysate were subjected to immunoprecipitation (IP), as previously reported [5], SDS-PAGE, using anti-p22phox, anti-c-Met or anti-polyUb antibodies (Supplementary Table 3).
2.12Chemical crosslinkingTo determine spatial protein association between p22phox and c-Met, isolated mice hepatocytes were exposed to crosslinking reagents. Chemical crosslinker stock solutions were freshly prepared before use. The 3,3-dithiobis (sulfosuccinimidyl propionate) (DTSSP) cell permeable crosslinker agent and the dithiobis (succinimidyl propionate) (DSP) cell impermeable crosslinker agent were added to 3 × 106 hepatocytes to a final concentration of 1 mM and 0.5 mM respectively. Crosslinking was performed for 25 min at room temperature. The reaction was terminated by the addition of ice-cold 50 mM Tris/HCl. Next the samples were immunoprecipitated as mention above. Dithiothreitol (DTT) was used to reverse the crosslinking as control.
2.13Chimeric fluorescent proteins productionThe p22phox and c-Met-containing plasmids were generated by PCR based amplification of p22phox and c-Met ORF from mouse cDNA by using primers listed in Supplementary table 4, which have cut-off sequences for restriction enzymes (XhoI in forward oligonucleotide and EcoRI in the reverse oligonucleotide) followed by enhancer Cyan Fluorescence Protein (eCFP) with an excitation wavelength of 405 nm and emission wavelength of 485 nm and enhancer Yellow Fluorescence Protein (eYFP) with an excitation wavelength of 514 nm and emission wavelength of 527 nm respectively sequence to frame into pcDNA3.1 vector (Life Technologies Co.). Cells transfected with plasmids encoding these chimeric proteins were used for FRET analysis.
FRET measurements by sensitized emission. The Förster resonance energy transfer (FRET) measurements between the p22phox and c-Met were obtained using the sensitized emission (SE) protocol. Briefly, FRET was obtained by measuring the acceptor emission resulting from donor excitation. To avoid overestimation of FRET first we evaluated the bleed-trough between both fluorescence channels for donor and acceptor. After subtracting the bleed-through from the donor emission we calculate FRET by using the following equation:
Using an excitation wavelength that excites only the donor, the emission for the acceptor (FexD,mA) and donor (FexD,emD) channels are obtained. Next, fluorescence is measured in the acceptor channel (FexA,mA) at an excitation wavelength that only excites the acceptor. The amount of donor bleed-through into the acceptor channel is determined by a donor-only measurement, which provides the calibration constant β=FexD,mAD∣FexD,emDD. By measuring only the acceptor channel we obtained the constant α=FexD,mAA∣FexA,emAA [22].
2.14Reactive oxygen species determinationReactive oxygen species (ROS) content was determined by dihydroethidium (DHE, 50 μM) fluorescence as previously reported [14]. DHE-derived fluorescence was recorded in a Multimodal reader (DTX880, Beckman Coulter, Inc. Brea, CA, USA) at 520 nm for excitation and 570 nm for emission. Data are reported in arbitrary fluorescence units (AFU).
2.15Statistical analysisEach experiment was performed at least by triplicate. Data are reported as average ± standard error (SEM). Statistical analysis was performed using GraphPad Prism v 8.2.1 software for iOS. Comparisons between groups were made using Student’s t-test or Mann-Whitney-U test. Differences were considered significant at p ≤ 0.05.
3ResultsOur group has proved that HGF and its receptor c-Met are regulators of cellular redox status in the liver [7], particularly regulating the activity of the multiprotein NADPH oxidase complex as we previously reported [2,5]. In the present work, we were aimed to gain mechanistic information about this regulation. Previously, we observed a decrease in the content of the main protein components of the system induced by the signaling of HGF, mainly the p22phox subunit encoded by cyba gene. p22phox which is a conserved subunit in the different complexes of NADPH oxidase, (NOX1–4) [23].
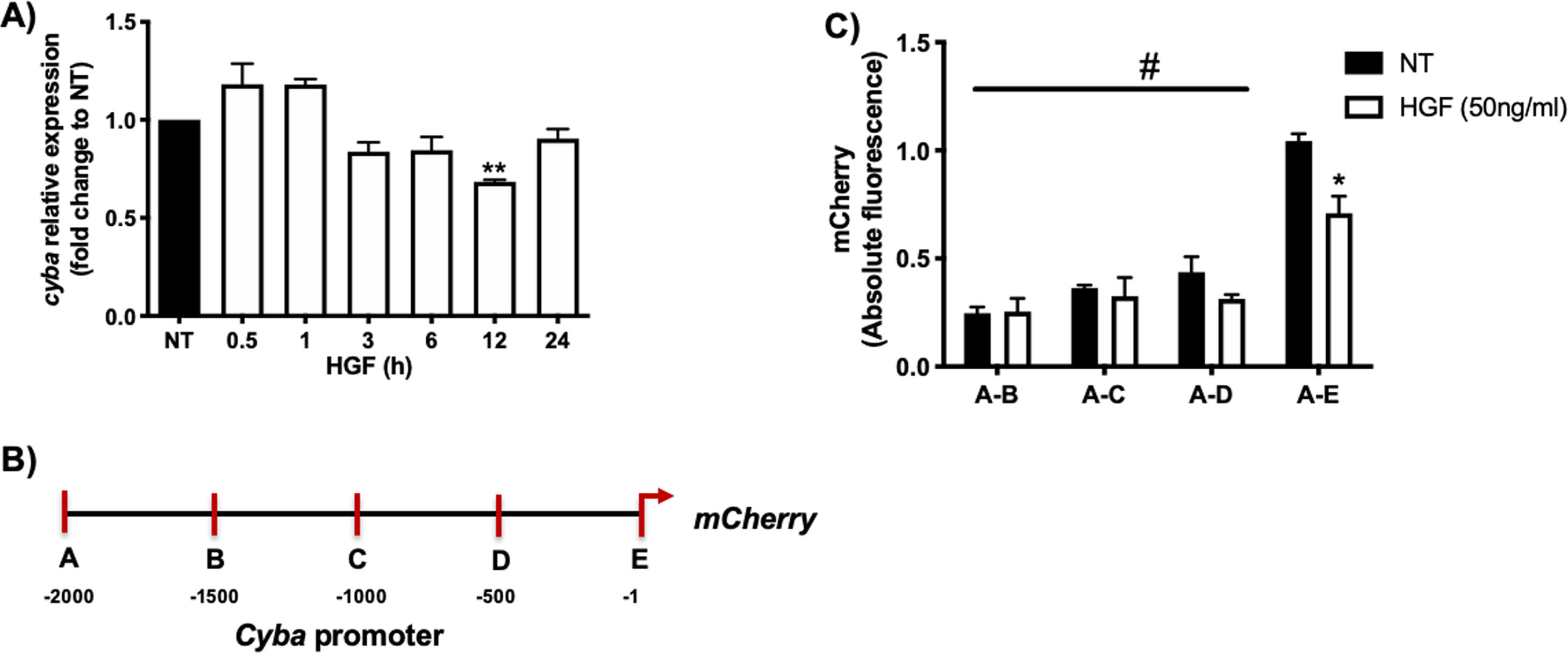
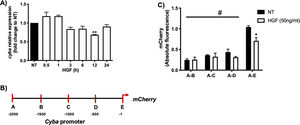
Fig. 1A shows that HGF treatment in primary mouse hepatocytes significantly decreases the expression of cyba gene at 12 h of treatment, data consistent with our previous finding at the protein level. In order to gain mechanistic evidence of the cyba repression we constructed and cloned different cyba promoter sections (Fig. 1B) by using the promotorless pmCherry vector. Fig. 1C shows the different cyba promotor activity (judged by mCherry reporter fluorescence) induced by HGF treatment, indicating that the segment A–E (−1 to −500 bp) owns the transcriptional repression of cyba gene, remaining segments of the promotor exhibited no changes.
cyba (p22phox) expression is regulated by HGF. A) Primary mouse hepatocytes were treated with HGF (50 ng/mL) for different times. cyba expression was addressed by qRT-PCR. B) Graphical representation of cyba promoter and different sections produced for, C) Cloning studies in the promoterless mCherry vector and its activation induced by HGF treatment. Each bar represents the average of at least three independent experiments ± SEM, * p < 0.05 vs NT; ** p < 0.01 vs NT; # p < 0.05 vs A-E promoter section.
Interestingly, this cyba promoter section contains a NF-κB (p65 subunit) consensus sequence determined by JASPAR software (Supplementary Figs. 1 and 2), it has been reported that NF-κB could exert gene repression in some genes [24,25].
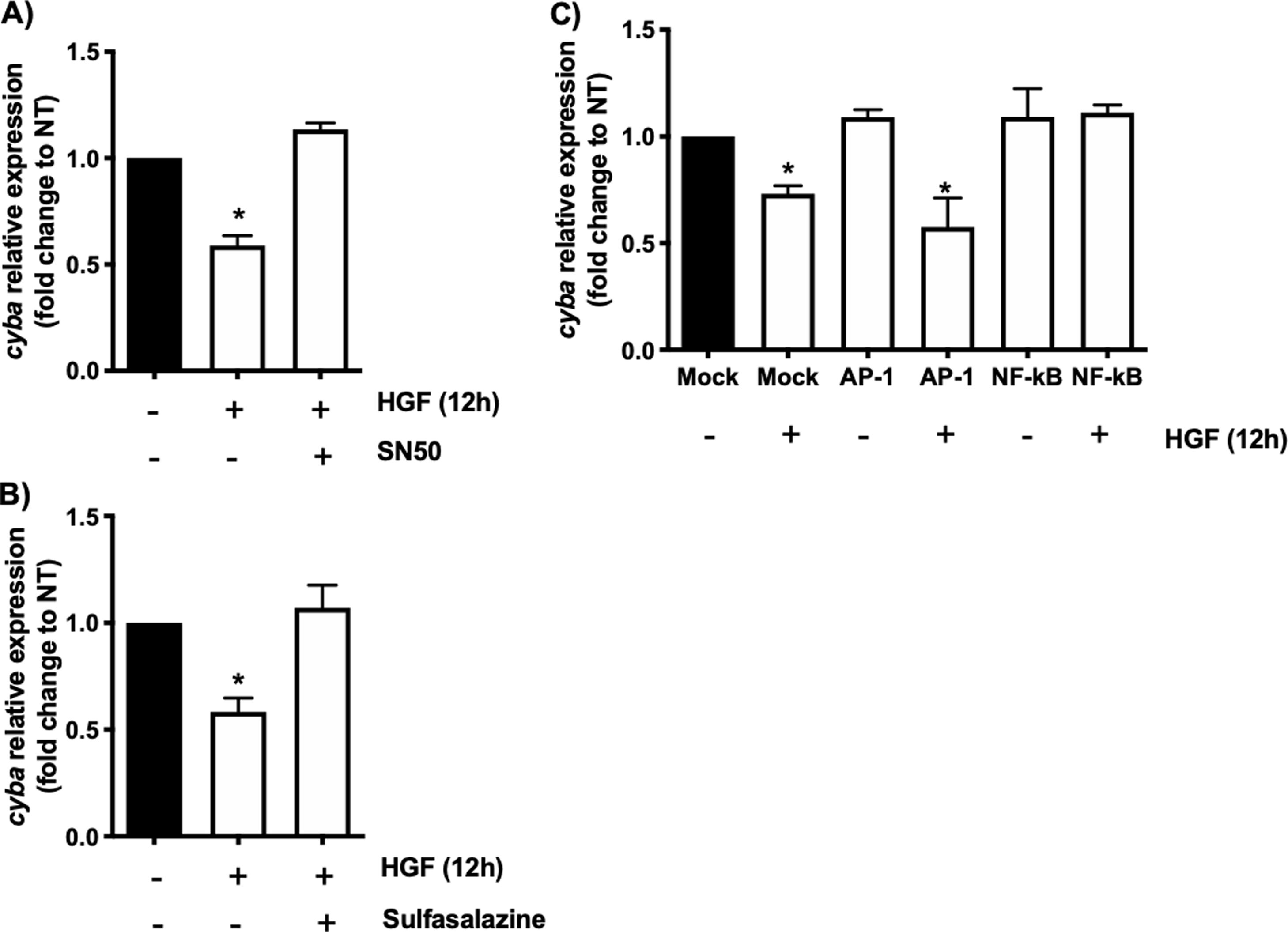
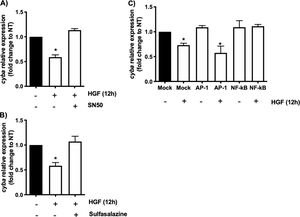
In order to address whether the cyba transcriptional repression could be instigated by NF-κB we used the well-known specific NF-κB peptide inhibitor SN50 [2], Fig. 2A shows that the decrement in the expression of the gene induced by HGF treatment is abrogated by the SN50, similar data were observed by the NF-κB chemical inhibitor sulfasalazine (Fig. 2B). The decrement of the cyba gene is evaded by the inhibition of the transcription factor, clarifying that NF-κB drives the repressive activity exerted by HGF over the p22phox gene in hepatocytes.
cyba (p22phox) transcriptional repression is driven by NF-κB. Primary mouse hepatocytes were pretreated with A) SN50, NF-κB peptide inhibitor or, B) sulfasalazine, NF-κB chemical inhibitor, for 30 min previous to HGF (50 ng/mL) treatment for 12 h. C) Binding competence of NF-κB by using decoy oligodeoxynucleotides (ODN). Each bar represents the average of at least three independent experiments ± SEM, * p < 0.05 vs NT.
To gain confidence we performed a competitive assay transfecting NF-κB consensus sequence (ODS) at high concentration (100 nM) in order to compete with natural sequence in the genome. Fig. 2C shows that NF-κB sequence abrogated the effect of HGF, confirming previous results obtained in Fig. 2A and B. As a control we used AP-1 transcription factor consensus sequence, which had no effect, corroborating the irrelevance of AP-1 in the repression of cyba in hepatocytes.
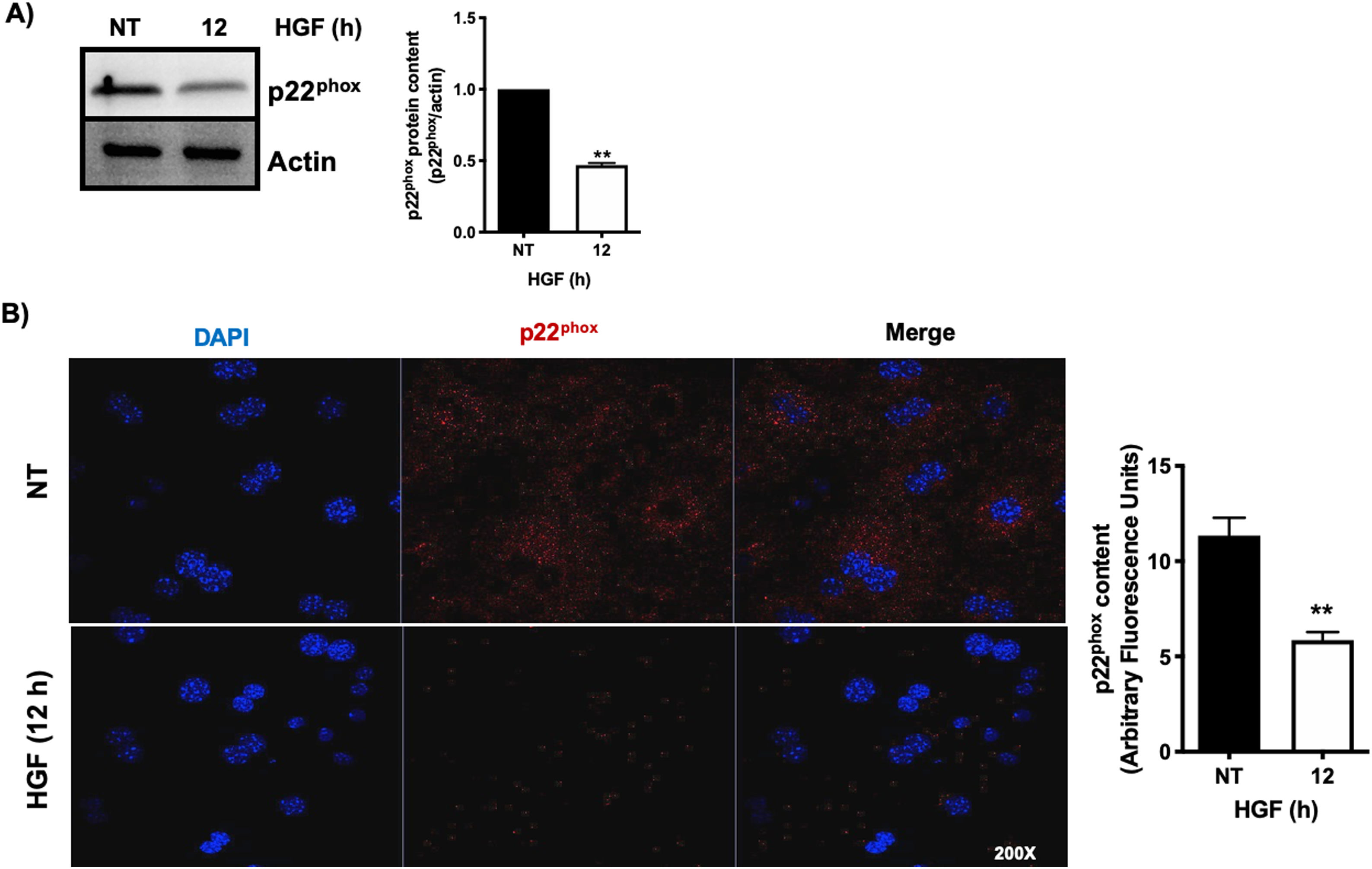
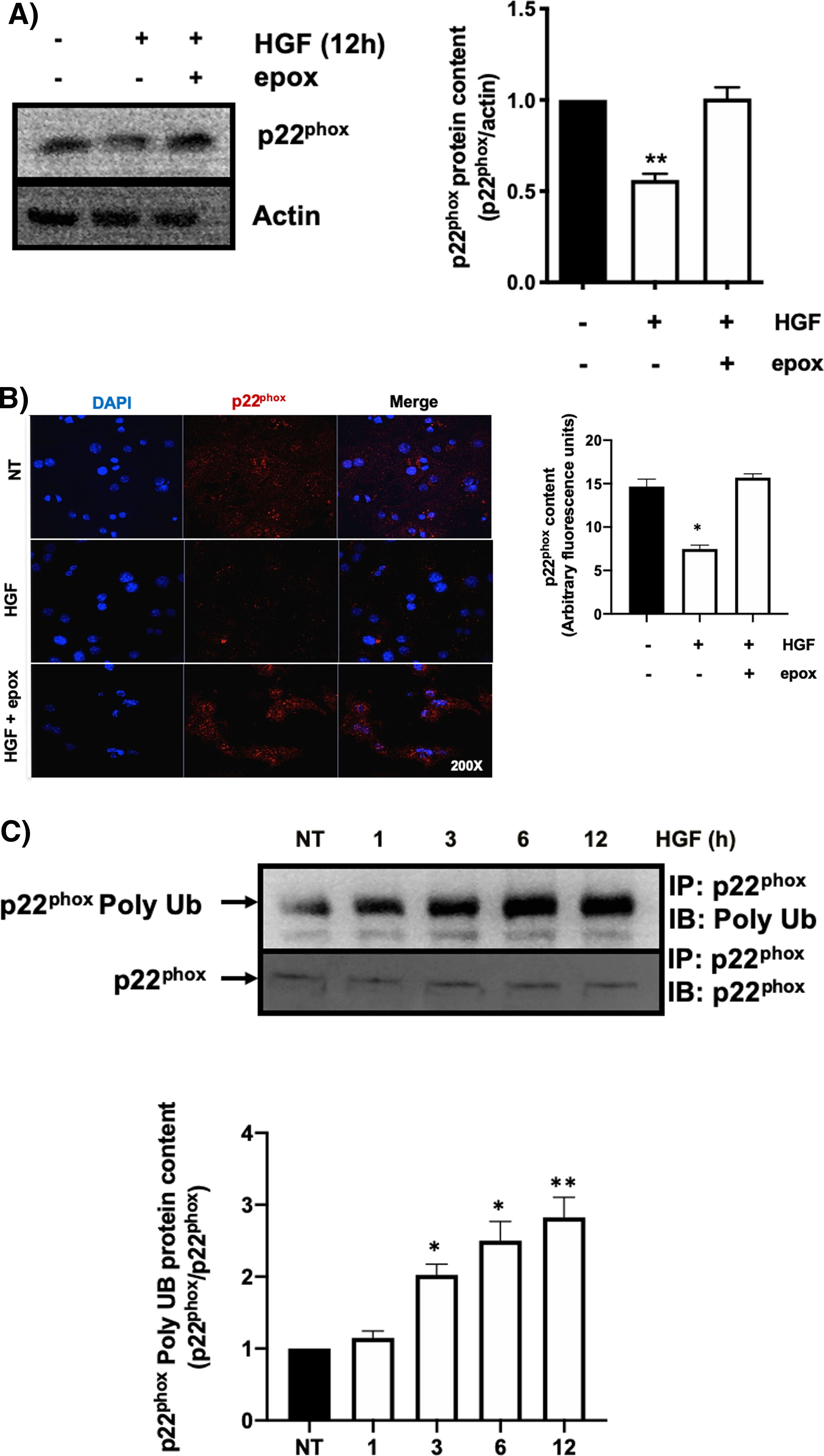
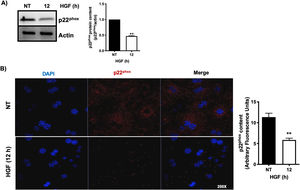
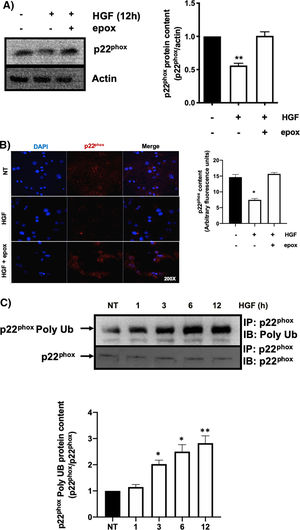
3.1HGF induces proteasome 26S-dependent degradation of p22phoxAs we previously reported, HGF effect on cyba gene is also observed at the protein level at 12 h of treatment, addressed by both Western blot and immunofluorescence of p22phox (Fig. 3A and B, respectively). The overlapping of cyba mRNA and p22phox protein decrement at 12 h suggests different mechanism of regulation. Data suggest that the decrement in protein content is driven by a post-translational mechanism. Then, we decided to address the involvement of the proteasome 26S in degradation of p22phox. Hepatocytes were pretreated with epox for 30 min, after that, cells were treated with HGF for 12 h, Fig. 4 shows the decrement of p22phox, determined by Western blot (Fig. 4A) and IF (Fig. 4B). To gain more confidence we studied the ubiquitination of p22phox induced by HGF treatment. Fig. 4C shows that ubiquitination of the protein was time-dependent manner, peaking at 12 h of treatment.
HGF induces the decrease of the protein content of p22phox. Primary mouse hepatocytes were treated for 12 h with HGF (50 ng/mL). A) Western blot of p22phox and densitometric analysis. B) Immunofluorescence of p22phox (in red), nuclei judged by DAPI fluorescence (blue). Images are representative of at least three independent experiments. Each bar represents the average ± SEM, ** p < 0.01 vs NT.
HGF induces the decrease of p22phox by a mechanism of proteasome 26S. Primary mouse hepatocytes were pretreated with epoxomicin (epox, 200 mM) for 30 min, followed of HGF treatment (50 ng/mL) for 12 h and p22phox was evaluated by A) Western blot and densitometric analysis; B) Immunofluorescence and fluorescence quantification of p22phox (in red), nuclei judged by DAPI fluorescence (blue). Images are representative of at least three independent experiments. Original magnification 200 × . C) Immunoprecipitation of p22phox and immunoblot of poly ubiquitination (poly Ub) and densitometric analysis. Each bar represents the average of at least three independent experiments ± SEM. * p < 0.05 vs NT. ** p < 0.01 vs NT.
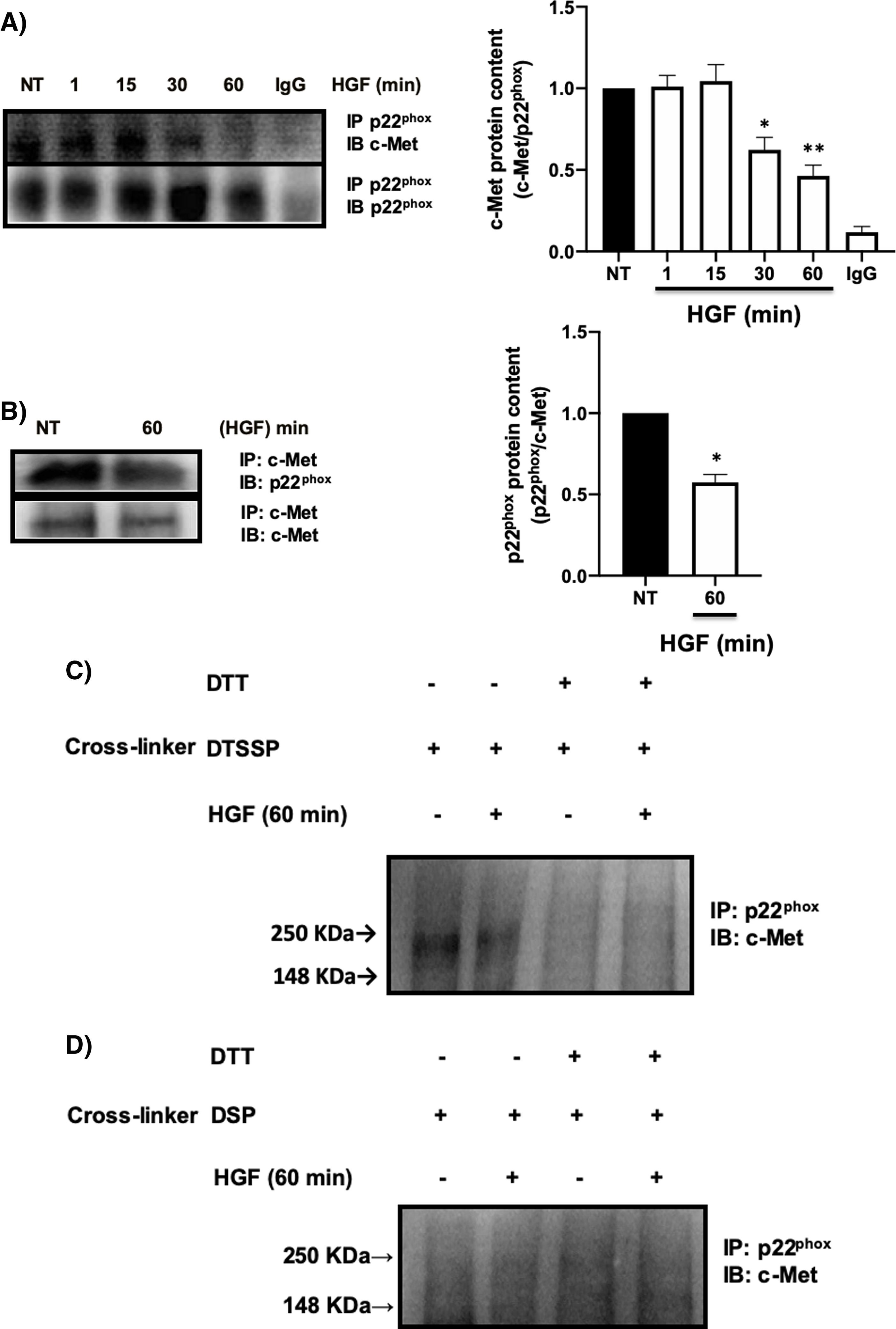
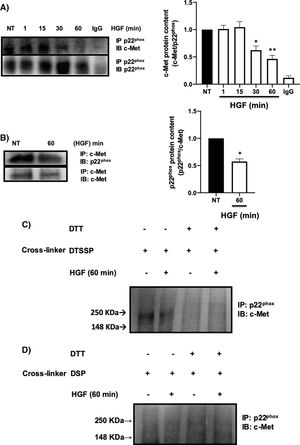
It has been reported the peculiar mechanism of regulation exerted by c-Met over some key membrane proteins such as FAS [26], plexin B1, CD44, among others [27], by physical interaction each other. We explored the possible interaction between c-Met and p22phox by IP, identifying an interaction between both proteins at basal levels (NT) being stable up to 30 min after treatment with HGF, presenting a decrease in the interaction at later times, reaching its maximum point at 60 min (Fig. 5A). To validate these results, we performed the reverse IP (Fig. 5B) corroborating the interaction of both proteins and its decrement at 60 min. To gain further insight into the mechanism of c-Met-p22phox interaction or association, chemical crosslinking was performed. Hepatocytes culture was subjected to the cell-impermeable thiol-cleavable crosslinking agent DSSTP, which only crosslinks proteins on the extracellular space and previously proved in other c-Met interactions [26]. Fig. 5C shows the detection of a strong band consisting of c-Met plus p22phox, in the non-treated cells (basal interaction) which was decreased with HGF treatment for 60 min. The use of DTT as agent of crosslinking reversion, induced the disappearance of such band indicating that the band was product of the crosslinking. No detectable bands were observed by using the cell-permeable DSP crosslinker agent (Fig. 5D) clearly indicating that c-Met and p22phox interaction is on the cell membrane.
c-Met interacts with p22phox in primary mouse hepatocytes. Primary mouse hepatocytes were treated with HGF (50 ng/mL) for different times. A) Immunoprecipitation of p22phox and immunoblot of c-Met, and B) Immunoprecipitation of c-Met and immunoblot of p22phox. C) Crosslinking using 3,3-dithiobis (sulfosuccinimidyl propionate) (DTSSP, 1 mM) cell permeable crosslinker agent and, D) Crosslinking using the dithiobis (succinimidyl propionate) (DSP, 0.5 mM) cell impermeable crosslinker agent. Crosslinking experiments were subjected to immunoprecipitation of p22phox and immunoblot of c-Met. The product band at 250 KDa represents the molecular weight of c-Met plus p22phox. Images representative of at least three independent experiments.
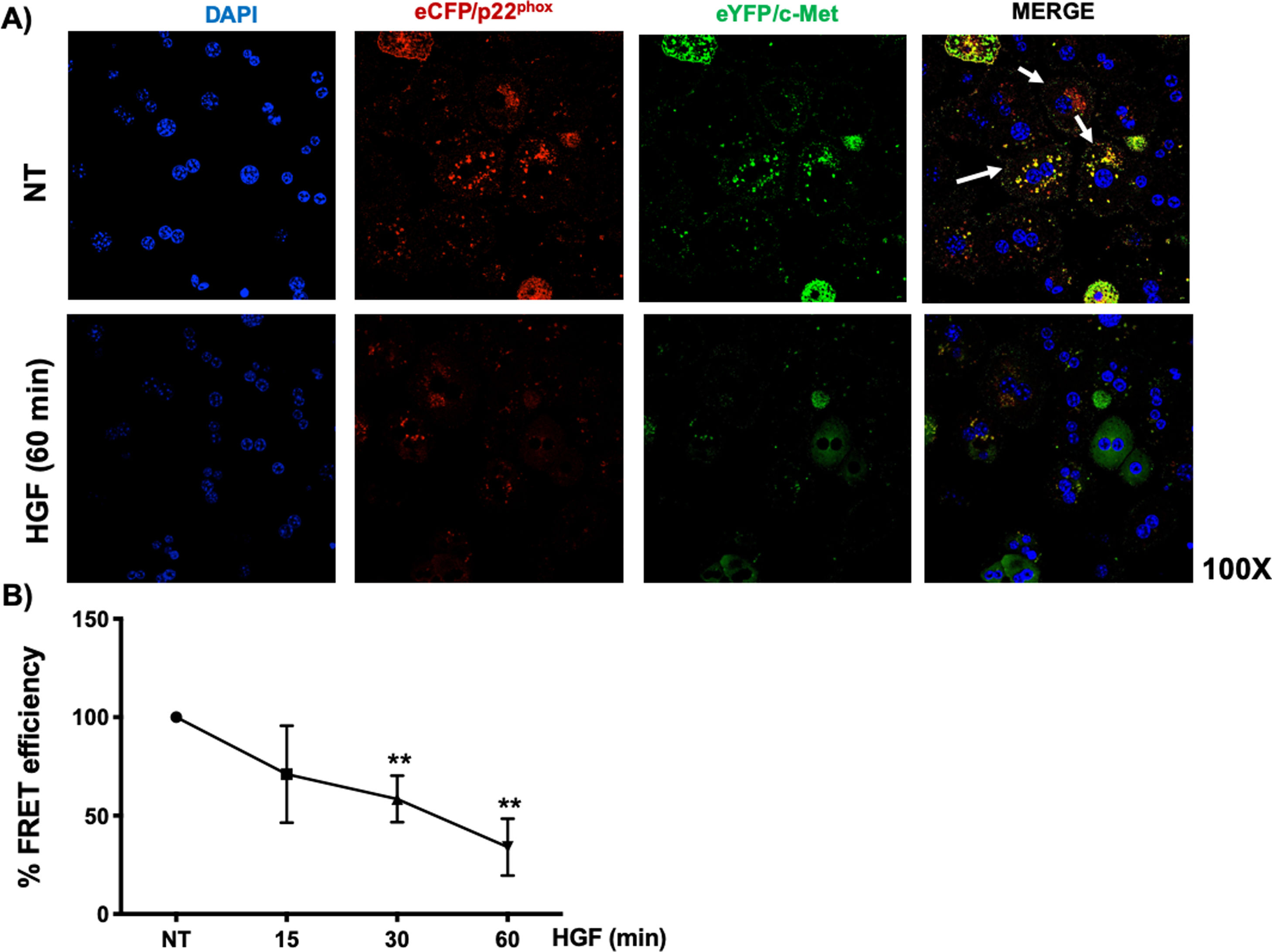
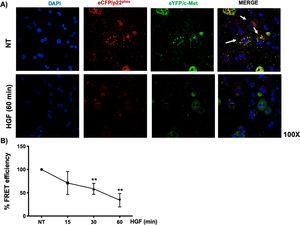
Interestingly, we could not identify the interaction of c-Met and p22phox by regular immuno-colocalization using fluorescence antibodies. The first antibody used impeded the attachment of the second one (data not shown), we decided to construct plasmids encoding sequences for the production of chimeric proteins coupled to fluorescence markers eCFP/p22phox and eYFP/c-Met, as stated under Material and Methods section. HepG2 cells were co-transfected with both plasmids and after verification of proper production of fluorescent proteins, were treated with HGF. Fig. 6A shows the basal co-localization (green fluorescence, arrows) in no treated cells, which disappears after 60 min of HGF treatment, confirming the c-Met and p22phox interaction.
c-Met interaction with p22phox is disrupted at early times of treatment with HGF in HepG2 cells. HepG2 cells were co-transfected with eCFP/p22phox and eYFP/c-Met chimeric proteins after two days cells were observed by confocal microscopy at different times of incubation with HGF (50 ng/mL). A) representative images of cells treated or not with HGF for 60 min. co-localization of p22phox (red), c-Met (green) and nuclei judged by DAPI fluorescence (blue). Images are representative of at least three independent experiments. Original magnification 200 × . B) Independent experiment for FRET analysis following protocol stated under material and methods section. Cells were treated with HGF for 15, 30 and 60 min. Each point represents the average of at least three independent experiments ± SEM. ** p < 0.01 vs NT.
We selected eCFP/p22phox and eYFP/c-Met because both fluorescent proteins allow FRET studies. Fig. 6B shows the percentage of FRET efficiency in a time course study, where significant change was observed after 30 min and peaked at 60 min, confirming the IP and crosslinking results in Fig. 5.
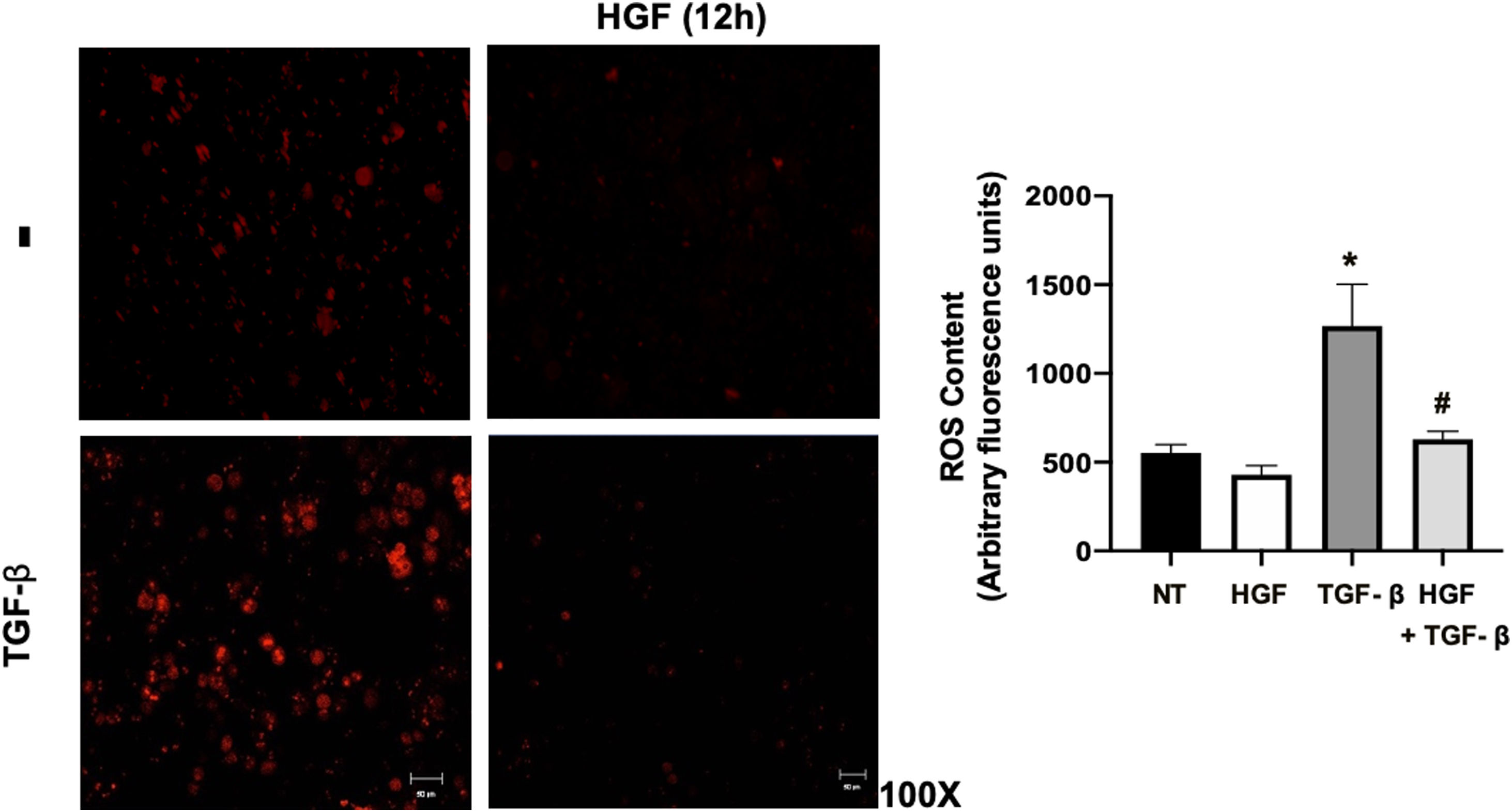
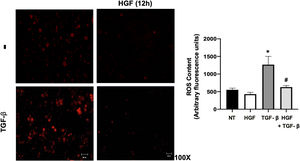
Finally, the ROS content in hepatocytes treated with TGF-β, pretreated or not with HGF was addressed. Fig. 7 shows that TGF-β increases ROS production, judged by DHE fluorescence, and the treatment with HGF significantly decreased the content of ROS, indicating that p22phox repression certainly decreases ROS derived from NADPH oxidase, as previously we published [5].
HGF treatment decreases reactive oxygen species (ROS) induced by transforming growth factor beta (TGF-β). Primary mouse hepatocytes were treated with HGF (50 ng/mL) for 12 h after that cells were treated with TGF-β (10 ng/mL) and ROS were determined using dihydroethidium (DHE, 50 μM) fluorescence (red), for 15 min, in the dark. Images are representative of at least three independent experiments. Each bar represents the average ± SEM, * p < 0.05 vs NT; # p < 0.05 vs TGF-β.
We have previously published that HGF/c-Met is key regulator of cellular redox status and oxidative stress in many systems such as liver [7,13], pancreas [16], lungs [12], biliary tree [14], among others. Particularly, in the liver, HGF/c-Met has demonstrated to influence the initiation, progression or treatment of alcoholic liver disease [15], cholestasis [14], hepatocellular carcinoma [7,10], fibrosis [9], drug-induced liver injury [11,12], and NASH [6,13]. Data published have provided mechanistic insights indicating that HGF/c-Met could exert these antioxidant/protective responses by multiples mechanisms, such as the activation of antioxidant-related transcription factors such as Nrf2 or NF-κB which, in addition express antioxidant enzymes like catalase, superoxide dismutase 1 and 2, and glutathione-related enzymes [2,15]. Another mechanism of action is directed to eliminate xenobiotics, which are potentially inducers of oxidative stress as we recently reported in cholestasis [14].
In the present work we are characterizing another mechanism of antioxidant action elicited by HGF/c-Met, which is directed to control the ROS-generating systems driven by the NADPH oxidases. The conserved membrane-resident component of the NADPH oxidases p22phox, which is used by at least four complexes (NADPH oxidase 1–4), is particularly affected by HGF signaling, as we previously reported in wild-type primary mouse hepatocytes [5] and in c-Met KO hepatocytes, where we observed a profound oxidative stress in untreated hepatocytes. In fact, this ROS control is key component of the HGF/c-Met signature defining an new aggressive phenotype tumor subtype with poor prognosis as we previously reported [7,10].
Previously we showed the downregulation of some components of the NADPH oxidase by HGF signaling, including the catalytic subunit Nox2; contrary, in c-Met KO hepatocytes we observed the over activation of the enzyme and a prominent oxidative stress [7]. Although, we reported that HGF exerts gene expression repression in NADPH oxidase components, in the present work we found that NF-κB is part of the gene repression machinery since the inhibition of its activity abolished the downregulation of p22phox maintaining basal protein content (Fig. 2), suggesting that this transcription factor is key regulator of the NADPH oxidase, since abrogation was not complete. NF-κB consensus sequence was identified downstream near to the transcription start site. Our current data support previous works reporting a profound effects induced by HGF in downregulation of NADPH oxidase at the transcriptional level, we have now advanced in the knowledge pointing out the relevance of NF-κB in the repressive stimulus of the pro-oxidant systems and in the expression of antioxidant and protective machinery [2,15].
The second mechanism of HGF/c-Met-mediated regulation of NADPH oxidase, particularly in p22phox, is post-translational. In this case, data show a fast decrease in the p22phox content since 3 h of HGF treatment in primary mouse hepatocytes. This effect continued in a time-dependent manner peaking at 12 h [5], so we decided to explore whether this effect could be related to p22phox degradation by proteosome 26S. Our results clearly indicate the polyubiquitination of p22phox induced by HGF, epox, inhibitor of the proteasome activity, avoided the degradation of the target protein. Data suggest that ubiquitin/proteasome 26S system is key determinant in NADPH oxidase activity, recently it has been published that perturbation of ubiquitin homeostasis led to a rapid and transient ROS generation in macrophages cell line [28], supporting this mechanism of regulation of the system, at least partially characterized in hepatocytes in terms of p22phox regulatory subunit.
The relevance of p22phox goes beyond of being a simple scaffolding protein, it displays regulatory functions, particularly at the level of p47phox [17], in fact some polymorphism has been identified in cyba resulting in a significant risk factor for coronary artery disease [29], but the relationship of this polymorphisms remains uncharacterized in liver diseases.
The physical interaction of c-Met with multiple membrane proteins has been positioned as a key regulatory mechanism [27]. It is particularly relevant the interaction or sequestration with FAS, which is a remarkable mechanism of HGF-induced anti-apoptosis response [26], in addition, c-Met interacts with CD44 [30] acting each other as coactivators of signaling; plexin B1 [31] is another membrane protein that interacts with c-Met, in this case antagonizing each other [31]. Our research provides evidence that c-Met is capable to interact with p22phox and repress the NADPH oxidase activity, at least partially, as we previously published [5] and supported in this work (Figs. 5 and 6). In addition, we are reporting that this interaction is intracellular, and after HGF treatment this protein-protein binding is lost, judged by FRET analysis, in order to increase ROS production at early times, for Nrf2 signaling [5].
The relationship of HGF/c-Met and NADPH oxidase has been corroborated by another mechanism, the control of Rac1, a prominent regulator of the oxidase. Ozaki and collaborators proved that HGF and its canonical signaling mediated by PI3K/Akt is necessary and sufficient to suppress intracellular ROS generation and apoptosis by inhibiting Rac1 in a model of hypoxia/reoxygenation in the liver [32], supporting the broad range of action induced by HGF on the modulation of cellular redox state driven by NADPH oxidase in the liver.
This regulation has been observed in other cell types. It has been reported that HGF increases the NADPH oxidase-mediated ROS formation in endothelial progenitor cells and in human umbilical vein cells, thereby inducing cell migration and angiogenesis, a phenomenon not observed in NOX2-deficient cells [33]. Similar findings have been found in the lung endothelial cells, where c-Met signaling induced the NADPH oxidase activation particularly in motile cells; this effect was mediated by p47phox, cortactin, and Rac1 translocation to lamellipodia. The NADPH oxidase inhibition by apocynin, or the siRNA-mediated inactivation of p47phox, or cortactin, significantly decreased the lamellipodia formation and cell migration promoted by HGF [34].
5ConclusionIn conclusion, in the present work we gained mechanistic evidence regarding the regulation of the NADPH oxidase by HGF/c-Met, particularly interfering with the regulatory subunit p22phox that supports the previous relevant physiological findings in many liver diseases (Supplementary Fig. 3), and this positions the axis HGF/c-Met/NADPH oxidase/ROS as a key mechanism of HGF singling in the liver.AbbreviationsHGF
Hepatocyte growth factor
c-MetCellular-mesenchymal epithelial transition factor (HGF receptor)
NADPHNicotinamide adenine dinucleotide phosphate
FRETFörster resonance energy transfer
NF-κBNuclear factor-kappa B
ROSReactive oxygen species
KOKnock out
TGF-βTransforming growth factor beta
EpoxEpoxomicin
DTSSP3,3-dithiobis sulfosuccinimidyl propionate
DSPDithiobis succinimidyl propionate
Conflicts of interestThe authors have no conflicts of interest to declare.
Data availability statementData available on request from the authors.