CASC9 and miR-424-5p are closely related with hepatocellular carcinoma (HCC) progression. This study aimed to evaluate the effect of CASC9 involved with miR-424-5p on the development of HCC.
Materials and methodsqRT-PCR was performed to determine the mRNA expressions of CASC9 and miR-424-5p in HCC tissues/cells and adjacent normal tissues/human hepatic epithelial cells, and to analyze the relationship of CASC9 with the clinico-pathological characteristics and prognosis of HCC patients. Then, cell proliferation was measured by CCK-8 and11 LncRNAs, long non-coding RNAs; HCC, hepatocellular carcinoma; AFP, alpha-fetoprotein; CASC9, cancer sensitivity 9; ESCC, esophageal squamous cell carcinoma; qRT-PCR, quantitative reverse transcription polymerase chain reaction; ATCC, American Type Culture Collection; DMEM, Dulbecco's Modified Eagle Medium; CCK-8, cell counting kit-8; RIPA, radio-immunoprecipitation assay; SDS-PAGE, sodium dodecylsulphate polyacrylamide gel electrophoresis; PVDF, polyvinylidene fluoride; GAPDH, glyceraldehyde-3-phosphate dehydrogenase; siNC, small interfering RNA of negative control; M: miR-424-5p mimic; IC, inhibitor control; I, miR-424-5p inhibitor.
Higher expression of CASC9 was observed in HCC tissues/ cells than in adjacent normal tissues/ human hepatic epithelial cells, and was closely linked to poor prognosis of HCC, tumor size, TNM stage, differentiation degree, lymph node metastasis and alpha-fetoprotein (AFP). Down-regulation of CASC9 decreased the proliferation, invasion and migration of HCC cells while enhancing apoptosis. Besides, CASC9 was negatively correlated with miR-424-5p. MiR-424-5p inhibitor enhanced cell proliferation, invasion and migration while decreasing apoptosis. Interestingly, siRNA-CASC9 partially offset the effects of miR-424-5p inhibitor on HCC cells.
ConclusionCASC9 promoted proliferation, invasion and migration and inhibited apoptosis in HCC cells by inhibiting miR-424-5p.
Liver cancer is one of the most common cancers that seriously threaten human health and life [1,2]. According to a statistical analysis of cancer in 2018, liver cancer has become a common cancer (∼841,000 new cases each year) and a leading cause of death (∼ 782,000 deaths occur each year) in the world [3]. Hepatocellular carcinoma (HCC) is the main type of liver cancer, which accounts for about 90% of primary liver cancer [4]. However, most HCC patients were already in the advanced stages when diagnosed, and thus lose the opportunity for surgery [5,6]. Therefore, the search for valuable diagnostic markers for HCC may significantly enhance the overall survival of HCC patients.
Long non-coding RNAs (LncRNAs) have aroused wide concern in recent years owing to their regulatory functions of epigenetic, transcriptional and post-transcriptional processes [7]. Interestingly, recent research has shown that non-coding RNA molecules that coordinate many biological functions account for about 98% of the human genome, while coding RNA molecules only account for about 2% [8]. Cancer Sensitivity 9 (CASC9) is an RNA gene which belongs to lncRNA [9]. CASC9 was initially identified to be associated with four transcriptional variants in esophageal squamous cell carcinoma (ESCC), which were subsequently found to be related with a variety of cancers, including liver cancer [9]. Zeng et al. showed that CASC9 plays a vital role in the diagnosis and prognosis of HCC [10]. In addition, studies have shown that CASC9 can be secreted by exosomes, and a high circulating CASC9 level is associated with tumor size and postoperative HCC recurrence, suggesting that CASC9 may be used as a non-invasive prognostic biomarker for recurrence in HCC [11]. Although these studies have demonstrated that CASC9 is involved in the HCC process, its specific mechanism is still unclear.
miR-424-5p has been shown to be implicated in the development of HCC [12,13]. There is substantial evidence that lncRNAs regulate miR-424-5p and affect the HCC process. Li et al. proved that DLX6 AS1 promotes HCC progression by targeting miR-424-5p [14]; Wang et al. suggested that the increase of LINC00511 was associated with the poor prognosis of HCC, and it promoted cell proliferation and metastasis by regulating miR-424 [15]. In this study, bioinformatics analysis revealed the targeting relationship between CASC9 and miR-424-5p. However, the hypothesis that CASC9 may affect the HCC process by regulating miR-424-5p is still inconclusive.
Considering the pathological characteristics of HCC, this study explored the role of CASC9 in HCC from both the clinical and experimental perspectives. We first analyzed the relationship between CASC9 expression and HCC prognosis. Further, the molecular mechanism of CASC9 in promoting HCC cell proliferation, metastasis and invasion was explored, and the relationship between CASC9 and miR-424-5p was verified. Our study enriches the understanding of the development of HCC and may provide a potential therapeutic strategy for HCC treatment.
2Materials and methods2.1Ethics statementSee in the title page.
2.2Quantitative reverse transcription polymerase chain reaction (qRT-PCR)The miRNAs of HCC cells or tissues were quantified by qRT-PCR. Briefly, the total microRNAs in HCC cells or tissues were first isolated using the miRcute miRNA Isolation Kit (DP501, TianGen, China). Then the total microRNAs were reversely transcribed into cDNA. The 10 μL RT reaction system was as follows: 1 μg of RNA Template; 1 μL of Bulge-Loop™ miRNA RT Primer (5 μM); 2 μL of 5×Reverse Transcription Buffer; 2 μL of RTase Mix; with the rest being Rnase-free Water (C10211, RIBOBIO, China). Then, the expressions of microRNA in HCC tissues or cells were analyzed by 7900 Real-Time PCR System (Biosystems, Foster City, USA), and U6 was used as a normalized control for miR-424-5p detection. The 20 μL qPCR reaction system was as follows: 10 μL of 2 × SYBR Green Mix (C10211, RIBOBIO, China); 2 μL of RT Product; 0.8 μL of Bulge-Loop™ miRNA Forward Primer (5 μM) and 0.8 μL Bulge-Loop™ Reverse Primer (5 μM) (MQPS0001272-1-100, RIBOBIO, China); with the rest being ddH2O. The PCR cycle conditions were as follows: 95 °C for 10 min; 40 cycles of 95 °C for 2 s, 60 °C for 20 s and 70 °C for 10 s. The primer sequences were listed in Table 2.
Specific primer sequences for qRT-PCR.
| Gene | Primer sequence | Species |
|---|---|---|
| miR-424-5p | 5’-GTCGTATCCAGTGCGTGTCGTGGAGTCGGC AATTGCACTGGATACGACTTCAAA-3’ (RT) 5’-CAGCAGCAATTCATGTTTTGAA-3’ 5’-AACGCTTCACGAATTTGCGT-3’ | human |
| U6 | 5’-GTTGGCTCTGGTGCAGGGTCCGAGGTATTCG CACCAGAGCCAACAAAATATGG-3’ (RT) 5’-ATTGGAACGATACAGAGAAGATT-3’ 5’-GGAACGCTTCACGAATTTG-3’ | human |
| CASC9 | 5’-TTGGTCAGCCACATTCATGGT-3’ 5’-AGTGCCAATGACTCTCCAGC-3’ | human |
| GAPDH | 5’-CCACTCCTCCACCTTTGAC-3’ 5’-ACCCTGTTGCTGTAGCCA-3’ | human |
The lncRNAs of HCC cells or tissues were determined by qRT-PCR. To put it simply, lncRNAs in HCC tissues or cells were first extracted with the Total RNA Purification Kit (17250, Norgen biotek Corp, CAN, https://norgenbiotek.com/). Then the total lncRNAs were reversely transcribed into cDNA. The 10 μL RT reaction system was as follows: 1 μg of RNA Template; 1 μL of Random Primer (200 μM); 1 μL of Oligo (dT) 18 (25 μM); 2 μL of 5×Reverse Transcription Buffer; 2 μL of RTase Mix; with the rest being Rnase-free Water (C11030-2, RIBOBIO, China). Then, the expressions of CASC9 in HCC tissues and cells were analyzed by 7900 Real-Time PCR System (Biosystems, Foster City, USA), and GAPDH was used as a normalized control for CASC9 detection. The 20 μL qPCR reaction system was as follows: 10 μL of 2 × SYBR Green Mix (C3010, RIBOBIO, China); 2 μL of RT Product; 0.8 μL of Ribo™ mRNA/ lncRNA Forward Primer (5 μM) and 0.8 μL of Ribo™ mRNA/ lncRNA Reverse Primer (5 μM); with the rest being ddH2O. The PCR cycle conditions were as follows: 95 °C for 10 min; 40 cycles of 95 °C for 10 s, 58 °C for 20 s and 72 °C for 10 s. The primer sequences were listed in Table 2.
2.3Clinical parametersVarious clinico-pathological data of HCC patients such as age, sex, tumor size, combined hepatitis, TNM stage, lymph node metastasis, differentiation and AFP were collected from medical records to analyze the relationship between HCC and CASC9 expressions. The known parameters including CASC9 mRNA expression were used to correlate with the prognosis.
2.4Cell cultureHuman liver epithelial cells (THLE-2) and HCC cells (PLC/ PRF/ 5, SNU-387 and SK-Hep1) were obtained from the American Type Culture Collection (ATCC) (Shanghai, China). All cells were cultured in Dulbecco's Modified Eagle Medium (DMEM, 12491-015, Gibco, USA) containing 10% fetal bovine serum (FBS, 10099-141, Gibco, USA) and 1% penicillin-streptomycin (15070063, Gibco, USA) in an incubator (5% CO2, 37 °C).
2.5Cell transfectionSK-Hep1 and PLC/PRF/5 cells were maintained in 6-well plates and divided into siNC transfection group, siRNA-CASC9 (siCASC9) transfection group, miR-NC transfection group and miR-424-5p inhibitor transfection group. siNC and siCASC9 were designed and constructed by GenePharma (Shanghai, China), while miR-NC and miR-425-5p inhibitor (miR20003393-1-5, RIBOBIO, China) were obtained from RIBOBIO (Guangzhou, China). Ten μL of Lipofection 2000 (Lipo-2000) (11668-027, Invitrogen, USA) and 4 μg of si-NC, siCASC9, miR-NC and miR-424-5p inhibitor were separately diluted with 250 μL of serum-free Opti-MEM. After incubation for 5 min, siNC, siCASC9, miR-NC, miR-424-5p inhibitor and Lipo-2000 were mixed at 25 °C for 20 min. After 6 h, the transfection medium was changed.
2.6Cell counting kit-8 (CCK-8) assayCell viability was measured by CCK-8 reagent (C0038, Beyotime Biotechnology, China). The transfected cells were cultured at 2000 cells/ well in 96-well plates. After 0 and 48 h, CCK-8 solution (10 μL) was added to the cells and incubated at 37 °C for 3 h. The 96-well plates after incubation were transferred to a microplate reader (Molecular Devices, Shanghai, China) to measure OD value at 450 nm.
2.7Colony formation assayThe cells were planted in a 6-well plate at 500 cells/ well. Afterwards, cells were maintained in DMEM medium (including 10% FBS) and placed in an incubator (5% CO2, 37 °C) for 7 days. The medium was then removed and the cells were fixed with 4% paraformaldehyde solution (P0099, Beyotime Biotechnology, China) for 30 min. The fixed cells were dyed blue-purple with crystal violet (C0121, Biotechnology, China). Colonies consisting of 50 or more cells were considered positive and counted under an inverted microscope (NIB620, Boshida, China).
2.8Cell apoptosis assayCell apoptosis assay was performed to determine the apoptosis rate with a Annexin V-FITC Apoptosis Detection Kit (556547, Becton, Dickinson and Company, Franklin Lakes, NJ, USA). Briefly, the treated cells were collected in a centrifuge tube and washed with PBS for three times (1000 rpm, 5 min). The cells were suspended in 100 μL of binding buffer. Then, 5 μL of fluorescein isothiocyanate and 5 μL of propidium iodide were added to the cells in the dark for 30 min at room temperature. Finally, cell apoptosis was detected by flow cytometry (Beckman Coulter, CA, USA).
2.9Wound healing assayThe transfected cells were further cultured on a 6-well plate at 2 × 105 cells/ well. When the cells reached 80–90 % fusion, the non-adherent cells were then washed twice with PBS. The monolayer cells were scratched with a pipette tip (200 μL) to form 3 scratch wounds, and the damaged cells were removed by 2 PBS washes. The scratch wound area was imaged with an inverted microscope at 0 and 48 h, and the timing began after the wound was formed. Cell migration was observed with an inverted microscope (100×) (NIB620, Boshida, China).
2.10Transwell assayTranswell inserts (pore size: 8 μm) (BD Biosciences, MA, USA) were used to determine cell invasion. Briefly, matrigel (diluted with DMEM medium at 1:4) was spread on the bottom of the transwell plate upper chamber and placed in the incubator (5% CO2, 37 °C) for 3 h. Then, FBS-free DMEM medium (100 μL) was used to culture the cell (2 × 104) suspension in the upper chamber. DMEM medium (750 μL) containing 20% FBS was added in the lower chamber. After 48 h of culture, cells in the upper chamber were removed and the invasive cells (upper compartment and lower surface) were fixed with 4% paraformaldehyde solution for 30 min. Then the fixed cells were dyed blue-purple with crystal violet. Lastly, cells from 5 randomly selected fields were counted under a microscope (200×) (NIB620, Boshida, China).
2.11Database predictionDiana-LncBase V2 (http://carolina.imis.athena-innovation.gr/diana_tools/web/index.php?r=site%2Ftools) was used to assess the binding sites of CASC9 and miR-424-5p [16].
2.12Dual-luciferase reporter gene assayThe 3'-UTR sequence of human CASC9 was acquired from NCBI (https://www.ncbi.nlm.nih.gov/), and was inserted into the pmirGLO-luciferase vector (E1751, promega, USA) to obtain the pmirGLO-CASC9-3'-UTR recombinant construct. The MUT-CASC9-3'-UTR recombinant construct was produced and used as a negative control, which contained a mutated 3'-UTR sequence of CASC9 in a specific miR-424-5p seeded region (UUGCUGCU to GCGACAGC). PLC/PRF/5 and SK-Hep1 cells were seeded in 96-well plates, and when achieving 60–80 % fusion, the cells were transfected with the reporter gene constructs (pRL-TK-CASC9-3'-UTR or MUT-CASC9-3'-UTR) and/or miR-424-5p mimics. After incubation for 24 h, a fluorescence detection microplate reader (Molecular Devices) was used to measure the fluorescence intensity in the cells.
2.13Western blotThe total protein of cells was extracted using the radio-immunoprecipitation assay (RIPA) reagent (P0013B, Beyotime Biotechnology, China) (containing 1% protease inhibitor (P1030, Beyotime Biotechnology, China) and 2% phosphatase inhibitor (P1081, Beyotime Biotechnology, China)). Then, 10% sodium dodecylsulphate polyacrylamide gel electrophoresis (SDS-PAGE) was performed to isolate the 30 μg protein, and the protein size was marked with marker (PR1910, Solarbio, China). The protein was then transferred to a polyvinylidene fluoride (PVDF) membrane. After being blocked with 5% skimmed milk at 25 °C for 1 h, the membrane was incubated with the following primary antibodies overnight at 4 °C: Bax (#5023, 1:1000, Cell Signaling Technology), Bcl-2 (#4223, 1:1000, Cell Signaling Technology), cleaved caspase-3 (ab2302, 1:500, abcam) and glyceraldehyde-3-phosphate dehydrogenase (GAPDH, ab181602, 1:1000, abcam). The next day, the primary antibodies were recycled and the membrane was cleaned with TBST for 4 times, 5 min for each time. The membrane was transferred to the secondary antibody (ab6721, 1:10000, abcam) and incubated for 1 h (room temperature). Then, the secondary antibody (ab6721, 1:10000, abcam) was recycled and the membrane was cleaned with TBST for 6 times, 5 min for each time. Finally, the enhanced chemiluminescence solution (WBKLS0500, MILLIPORE, USA) was added to the band and the strip was visualized by the detection system. Image J (1.8.0, National Institutes of Health, Germany) was used to analyze the gray value of the strip.
2.14Statistical analysisData were evaluated using SPSS 19.0 software (IBM, NY, USA). Measurement data were represented as means ± standard deviation. Differences between paired HCC samples were analyzed using paired t-test. One-way ANOVA and Tukey’s test were performed for multi-group comparison. Comparison between two groups were performed by Student t-test. Kaplan-Meier was performed to analyze the correlation between CASC9 and the overall survival of HCC patients. The gray value of the protein bands and the migration distance of HCC cells were analyzed by Image J. The correlation between miR-424-5p and CASC9 was analyzed by Pearson correlation. p < 0.05 was accepted as indicative of significant differences. All experiments were performed in at least three independent repeats.
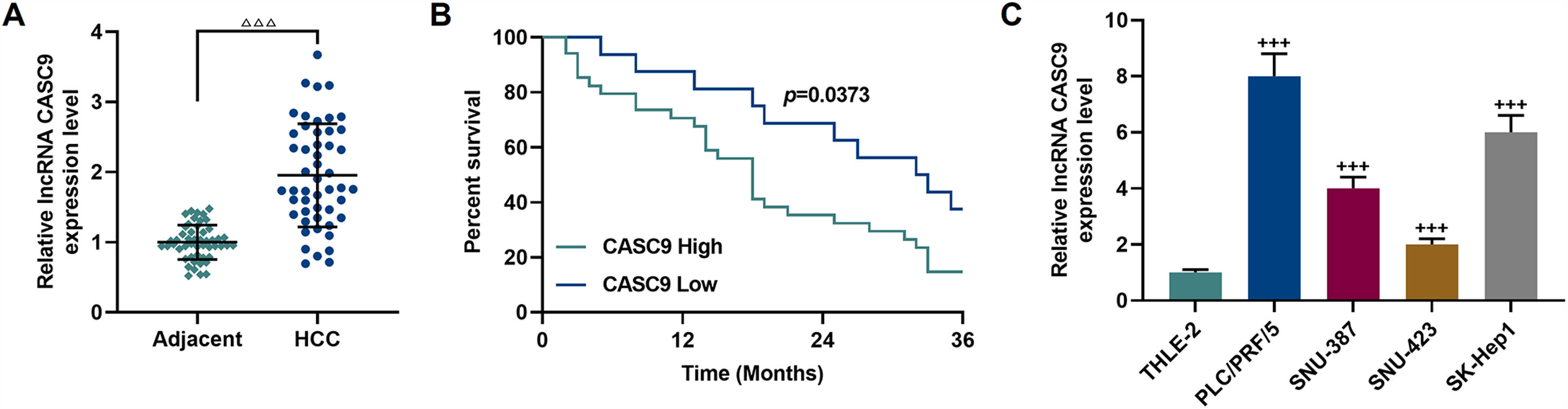
3Results3.1CASC9 was highly expressed in HCC tissues and cells, which was closely related to poor prognosis of HCC patientsCASC9 expression in HCC tissues and adjacent normal tissues from 50 patients was detected by qRT-PCR. The results demonstrated that CASC9 was highly expressed in HCC tissues relative to adjacent normal tissues (p < 0.001; Fig. 1A). Meanwhile, we analyzed the correlations between CASC9 expression and the clinicopathological data of HCC patients. It was observed that CASC9 was positively associated with TNM stage (p = 0.003), lymph node metastasis (p = 0.028), tumor size (p = 0.004), differentiation degree (p = 0.018) and alpha fetoprotein (AFP) (p = 0.010) (Table 1). Besides, highly expressed CASC9 was strongly related with poor prognosis of HCC (p < 0.05; Fig. 1B). Furthermore, we also measured the expression of CASC9 in human liver epithelial cell lines (THLE-2) and HCC cell lines (PLC/PRF/5, SNU-387, SNU-423 and SK-Hep1). As detailed in Fig. 1C, the expression of CASC9 in HCC cells was higher than that in human liver epithelial cells (p < 0.001). It must be mentioned that the expression of CASC9 in PLC/PRF/5 and SK-Hep1 cells was the highest among the 4 HCC cell lines, and therefore these two cell lines were used as experimental objects in subsequent experiments.
CASC9 was highly expressed in HCC tissues and cells, which was closely related to poor prognosis of HCC patients. (A): The expression of CASC9 in HCC tissues and adjacent normal tissues was detected by qRT-PCR. n = 50. △ vs. Adjacent. △△△ p < 0.001. (B): Kaplan-Meier was used to analyze the relationship between CASC9 and the survival rate of HCC patients. (C): The expression of CASC9 in HCC cells (PLC/PRF/5, SNU-387, SNU-423 and SK-Hep1) and normal epithelial cells of the liver (THLE-2) was detected by qRT-PCR. n = 3. + vs. THLE-2. +++ p < 0.001.
Abbreviation: qRT-PCR, quantitative reverse transcription polymerase chain reaction; HCC, Hepatocellular carcinoma.
The relationship between lncRNA CASC9 expression and clinical characteristics.
| Variable | n | lncRNA CASC9 | P value | |
|---|---|---|---|---|
| High | Low | |||
| Total | 50 | 34 | 16 | |
| Gender | ||||
| Male | 29 | 19 | 10 | 0.658 |
| Female | 21 | 15 | 6 | |
| Age | ||||
| <55 | 36 | 24 | 12 | 0.746 |
| ≥55 | 14 | 10 | 4 | |
| Tumor size | ||||
| ≥5 cm | 30 | 25 | 5 | 0.004 |
| <5 cm | 20 | 9 | 11 | |
| Combined hepatitis | ||||
| Yes | 22 | 14 | 8 | 0.558 |
| No | 28 | 20 | 8 | |
| Differentiation | ||||
| Well | 8 | 4 | 4 | 0.018 |
| Moderate | 20 | 11 | 9 | |
| Poor | 22 | 19 | 3 | |
| Lymph node | ||||
| Negative | 33 | 19 | 14 | 0.028 |
| Positive | 17 | 15 | 2 | |
| TNM stage | ||||
| I | 4 | 2 | 2 | 0.003 |
| II | 7 | 3 | 4 | |
| III | 17 | 9 | 8 | |
| IV | 22 | 20 | 2 | |
| AFP (ng/mL) | ||||
| ≥200 | 29 | 24 | 5 | 0.010 |
| <200 | 21 | 10 | 11 | |
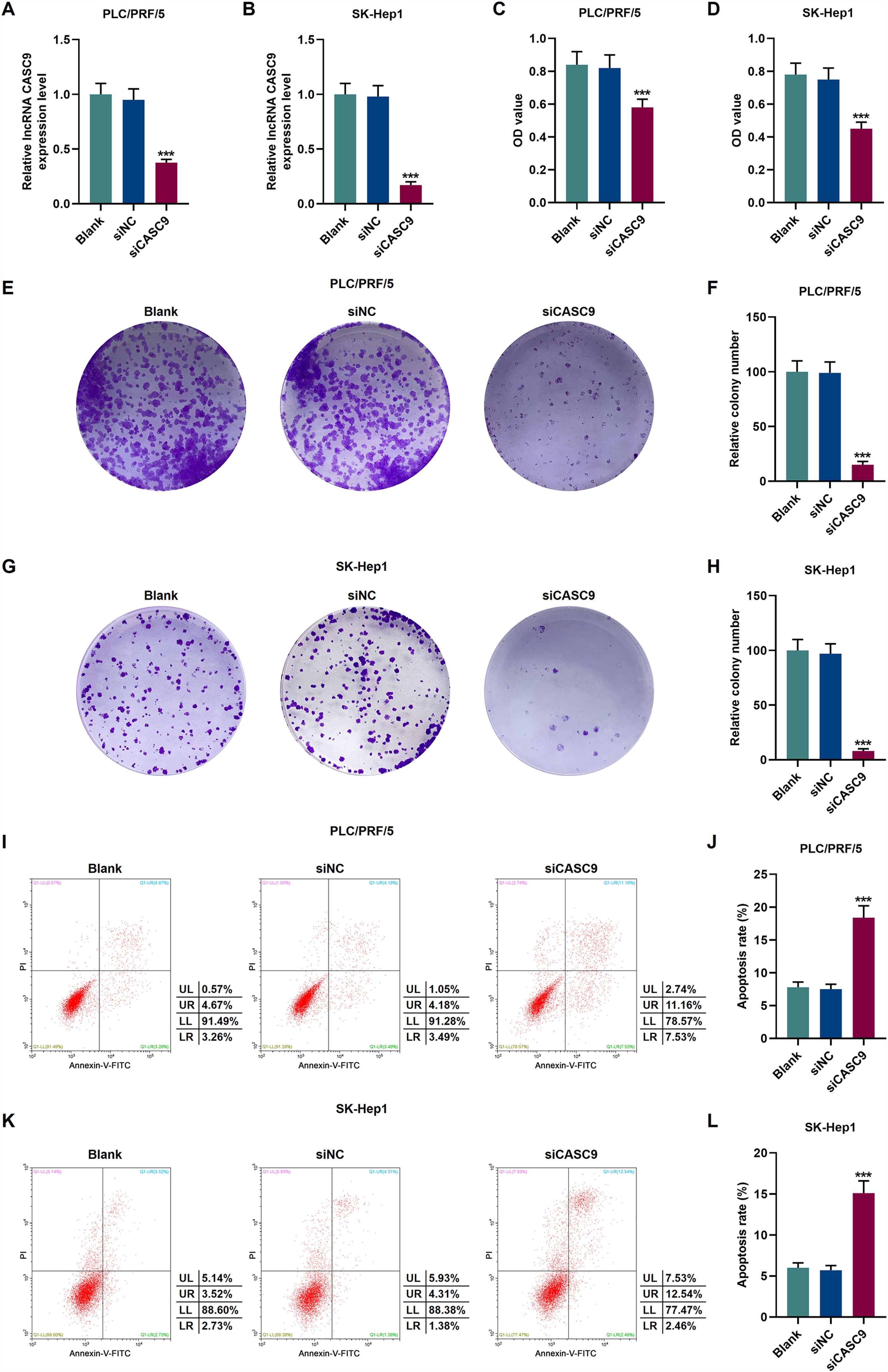
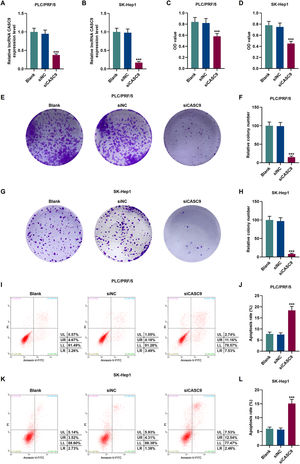
To verify the transfection efficiency of siCASC9, qRT-PCR was performed to detect its expression level. It is evident from Fig. 2A and 2B that the expression of siCASC9 could significantly silence CASC9 (p < 0.001). Afterwards, HCC cell proliferation was assessed with CCK-8 assay. As detailed in 2C and 2D, the down-regulated expression of CASC9 decreased the proliferation of HCC cells as compared to the siNC group (p < 0.001). Meanwhile, colony formation assay was performed to verify the role of CASC9 in HCC cell proliferation, and it was found that siCASC9 was able to reduce the colony formation ability of HCC cells in comparation to the siNC group (p < 0.001; Fig. 2E–H). In addition, down-regulated CASC9 promoted the apoptosis of HCC cells in comparation to the siNC group (p < 0.001; Fig. 2I–L).
Down-regulated CASC9 decreased the proliferation and promoted the apoptosis of HCC cells. (A): Transfection efficiency of siCASC9 in PLC/PRF/5 cells was detected by qRT-PCR. n = 3. (B): Transfection efficiency of siCASC9 in SK-Hep1 cells were detected by qRT-PCR. n = 3. (C): CCK-8 was used to detect the effect of CASC9 on the viability of PLC/PRF/5 cells. n = 3. (D): CCK-8 was used to detect the effect of CASC9 on the viability of SK-Hep1 cells. n = 3. (E): Colony formation assay was performed to measure the effect of CASC9 on the proliferation of PLC/PRF/5 cells. (F): The colonies containing more than 50 cells were counted. n = 3. (G): Colony formation assay was performed to measure the effect of CASC9 on the proliferation of SK-Hep1 cells. (H): The colonies containing more than 50 cells were counted. n = 3. (I): Flow cytometry was used to detect the effect of CASC9 on the apoptosis of PLC/PRF/5 cells. (J): The apoptosis rate was calculated. n = 3. (K): Flow cytometry was used to detect the effect of CASC9 on the apoptosis of SK-Hep1 cells. (L): The apoptosis rate was calculated. n = 3. * vs. siNC. *** p < 0.001.
Abbreviation: HCC, Hepatocellular carcinoma; CCK-8, Cell Counting Kit-8; siNC, small interfering RNA of negative control.
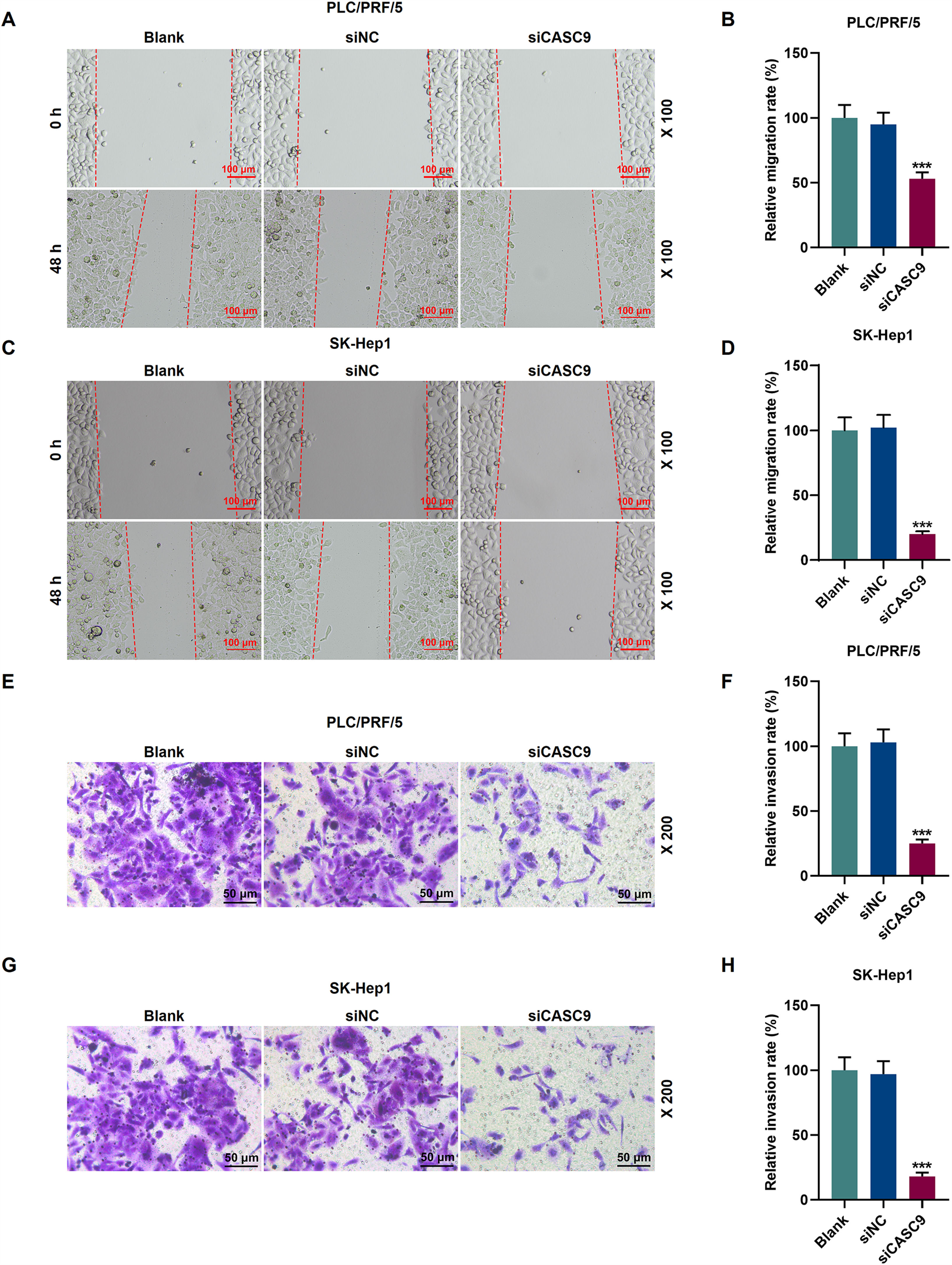
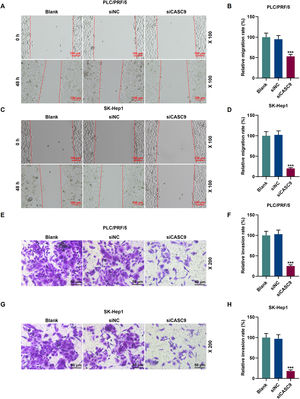
To examine the migratory property of HCC cells, we conducted scratch wound assay. According to Fig. 3A–D, the down-regulated expression of CASC9 attenuated the migration of HCC cells, compared with the siNC group (p < 0.001). Transwell assay results exhibited that siCASC9 could reduce the invasion of HCC cells as compared to the siNC group (p < 0.001; Fig. 3E–H).
Down-regulated CASC9 inhibited the migration and invasion of HCC cells. (A): Wound healing assay was used to detect the effect of CASC9 on the migration of PLC/PRF/5 cells. Scale: 100 μm; magnification: ×100. n = 3. (B): The migration rate of PLC/PRF/5 cells was calculated. (C): Wound healing assay was used to detect the effect of CASC9 on the migration of SK-Hep1 cells. Scale: 100 μm; magnification: ×100. n = 3. (D): The migration rate of SK-Hep1 cells was calculated. (E): Transwell assay was used to detect the effect of CASC9 on the invasion of PLC/PRF/5 cells. Scale: 50 μm; magnification: ×200. (F): The number of invasive PLC/PRF/5 cells was counted. n = 3. (G): Transwell assay was used to detect the effect of CASC9 on the invasion of SK-Hep1 cells. Scale: 50 μm; magnification: ×200. (H): The number of invasive SK-Hep1 cells was counted. n = 3. * vs. siNC. *** p < 0.001.
Abbreviation: HCC, Hepatocellular carcinoma; siNC, small interfering RNA of negative control.
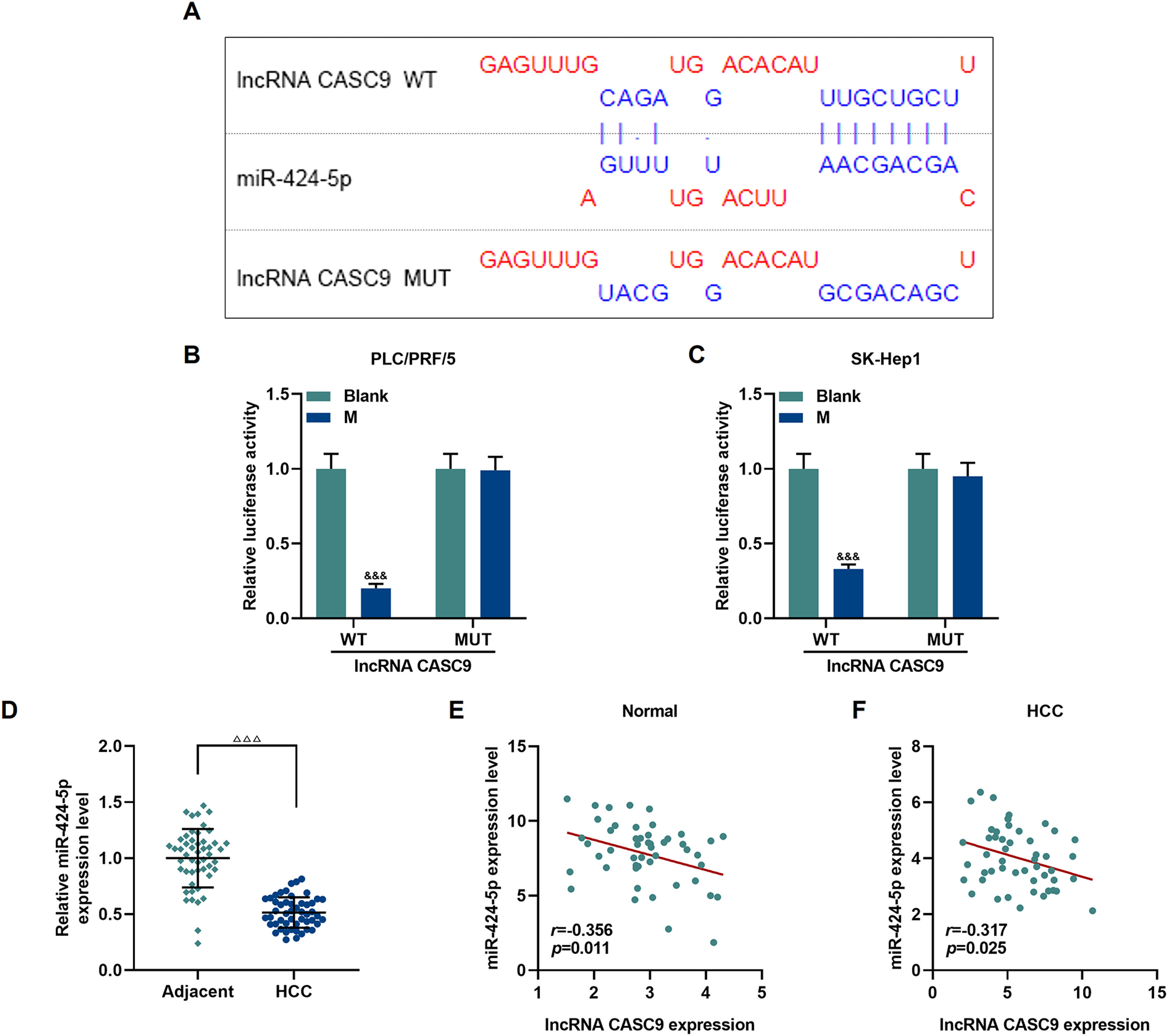
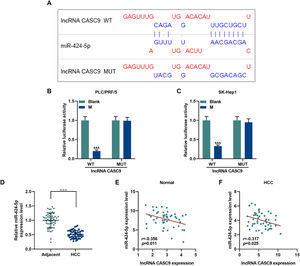
The relationship between miR-424-5p and CASC9 was predicted by Diana-lncbase V2, and then verified by dual-luciferase reporter gene assay (Fig. 4A). However, no significant change in fluorescence intensity was found after mutation in the binding sites of CASC9 and miR-424-5p in comparison to the control group (p < 0.001; Fig. 4B and C), while the fluorescence intensity in the wild-type CASC9 group was reduced in comparison to the control group. Thus, the results revealed that CASC9 was targeted by miR-424-5p. Afterwards, we found that the expression of miR-424-5p in HCC tissues was lower than that in adjacent normal tissues (p < 0.001; Fig. 4D). Finally, the relationship between CASC9 and miR-424-5p expressions in HCC tissues and adjacent normal tissues was analyzed by Pearson correlation, and we found that they were negatively correlated (Fig. 4E and F).
CASC9 specifically targeted miR-424-5p. (A): DIANA-LncBase V2 predicts the binding sites of miR-424-5p and CASC9. (B): Dual-luciferase reporter gene assay was performed to verify the targeted binding sites of miR-424-5p and CASC9 in PLC/PRF/5 cells. n = 3. (C): Dual-luciferase reporter gene assay verified the targeted binding sites of miR-424-5p and CASC9 in SK-Hep1 cells. n = 3. (D): The expression of miR-424-5p in HCC tissues and adjacent normal tissues was detected by qRT-PCR. n = 50. (E): Pearson correlation coefficient was used to analyze the relationship between CASC9 and miR-424-5p in adjacent normal tissues. (F): Pearson correlation coefficient was used to analyze the relationship between CASC9 and miR-424-5p in HCC tissues. & vs. Blank. &&& p < 0.001; vs. Adjacent. △△△ p < 0.001.
Abbreviation: HCC, Hepatocellular carcinoma; M: miR-424-5p mimic.
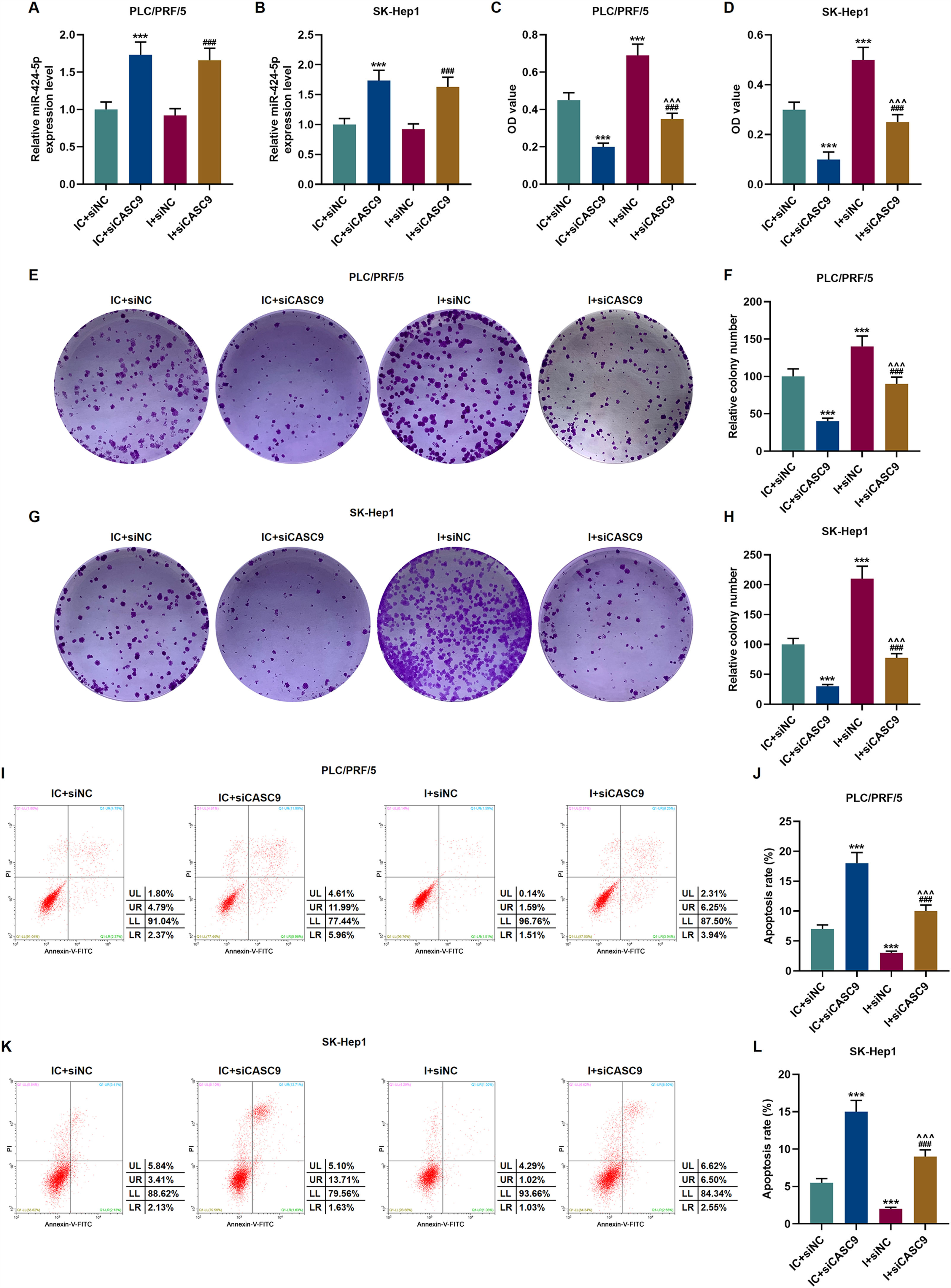
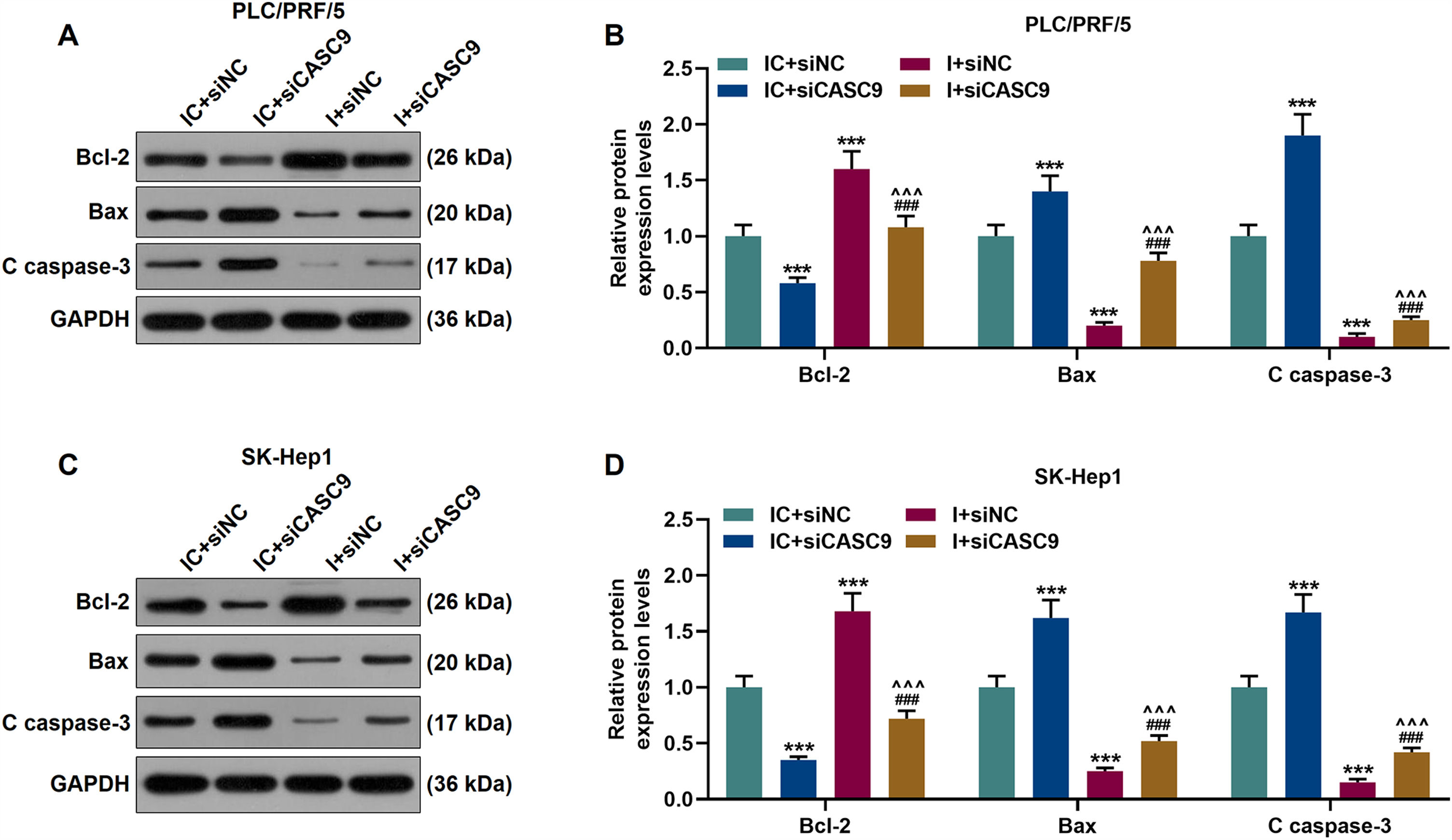
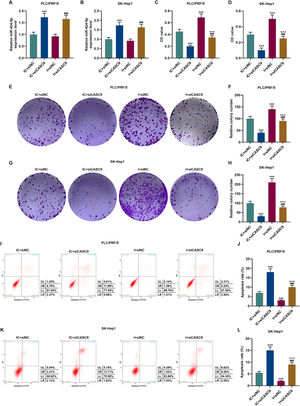
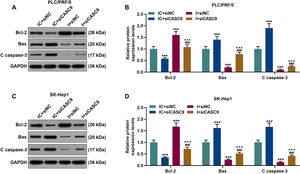
According to qRT-PCR results, siCASC9 promoted miR-424-5p expression as compared to the IC + siNC group (p < 0.001; Fig. 5A and B). Then, we identified the effects of miR-424-5p on the proliferation of HCC cells. The results from CCK-8 and colony formation assays both displayed that miR-424-5p inhibitor promoted the proliferation of HCC cells as compared to the IC + siNC group (p < 0.001; Fig. 5C–H). In addition, down-regulated CASC9 partially offset the effect of miR-424-5p inhibitor (p < 0.001; Fig. 5C–H). Afterwards, we also found down-regulated miR-424-5p decreased HCC cell apoptosis, compared with the IC + siNC group (p < 0.001). Similarly, siCASC9 partially reversed the effect of down-regulated miR-424-5p on the apoptosis of HCC cells. Furthermore, we detected the expressions of apoptosis-related proteins (Bcl-2, Bax and C caspase-3). As can be seen in Fig. 6A–D, down-regulated CASC9 inhibited the expression of Bcl-2 and promoted the expressions of Bax and C caspade-3 as compared to the IC + siNC group (p < 0.001), while miR-424-5p inhibitor produced an opposite effect. However, down-regulated CASC9 could partly reverse the changes of apoptosis-related proteins caused by miR-424-5p inhibitor (p < 0.001; Fig. 6A–D). Consistent with the above results, this finding also revealed that CASC9 promoted the apoptosis of HCC cells by inhibiting miR-424-5p expression.
Down-regulated CASC9 inhibited the proliferation and promoted the apoptosis of HCC cells by negatively regulating miR-424-5p. (A): The expression of miR-424-5p in PLC/PRF/5 cells treated with siCASC9 or/ and miR-424-5p inhibitor was detected by qRT-PCR. n = 3. (B): The expression of miR-424-5p in SK-Hep1 cells treated with siCASC9 or/ and miR-424-5p inhibitor was detected by qRT-PCR. n = 3. (C): CCK-8 was used to detect the effect of CASC9 and miR-424-5p on the viability of PLC/PRF/5 cells. n = 3. (D): CCK-8 was used to detect the effect of CASC9 and miR-424-5p on the viability of SK-Hep1 cells. n = 3. (E): Colony formation assay was performed to evaluate the effect of CASC9 and miR-424-5p on the proliferation of PLC/PRF/5 cells. (F): The colonies containing more than 50 cells were counted. n = 3. (G): Colony formation assay was performed to measure the effect of CASC9 and miR-424-5p on the proliferation of SK-Hep1 cells. (H): The colonies containing more than 50 cells were counted. n = 3. (I): Flow cytometry was used to detect the effect of CASC9 and miR-424-5p on the apoptosis of PLC/PRF/5 cells. (J): The apoptosis rate was calculated. n = 3. (K): Flow cytometry was used to detect the effect of CASC9 and miR-424-5p on the apoptosis of SK-Hep1 cells. (L): The apoptosis rate was calculated. n = 3. * vs. IC + siNC; ^ vs. IC + siCASC9; # vs. I + siNC. *** p < 0.001; ^^^ p < 0.001; ### p < 0.001.
Abbreviation: HCC, Hepatocellular carcinoma; CCK-8, Cell Counting Kit-8; siNC, small interfering RNA of negative control; IC, inhibitor control; I, miR-424-5p inhibitor.
Down-regulated CASC9 promoted the apoptosis of HCC cells by negatively regulating miR-424-5p. (A): Western blot was used to detect the effects of CASC9 and miR-424-5p on the expressions of Bcl-2, Bax and C caspase-3 in PLC/PRF/5 cells. (B): The signal intensities of Bcl-2, Bax and C caspase-3 were quantified. n = 3. (C): Western blot was used to detect the effects of CASC9 and miR-424-5p on the expressions of Bcl-2, Bax and C caspase-3 in SK-Hep1 cells. (D): The signal intensities of Bcl-2, Bax and C caspase-3 were quantified. n = 3. * vs. IC + siNC; ^ vs. IC + siCASC9; # vs. I + siNC. *** p < 0.001; ^^^ p < 0.001; ### p < 0.001.
Abbreviation: HCC, Hepatocellular carcinoma; Bcl-2, B-cell lymphoma-2; Bax, BCL2-Associated X; siNC, small interfering RNA of negative control; IC, inhibitor control; I, miR-424-5p inhibitor.
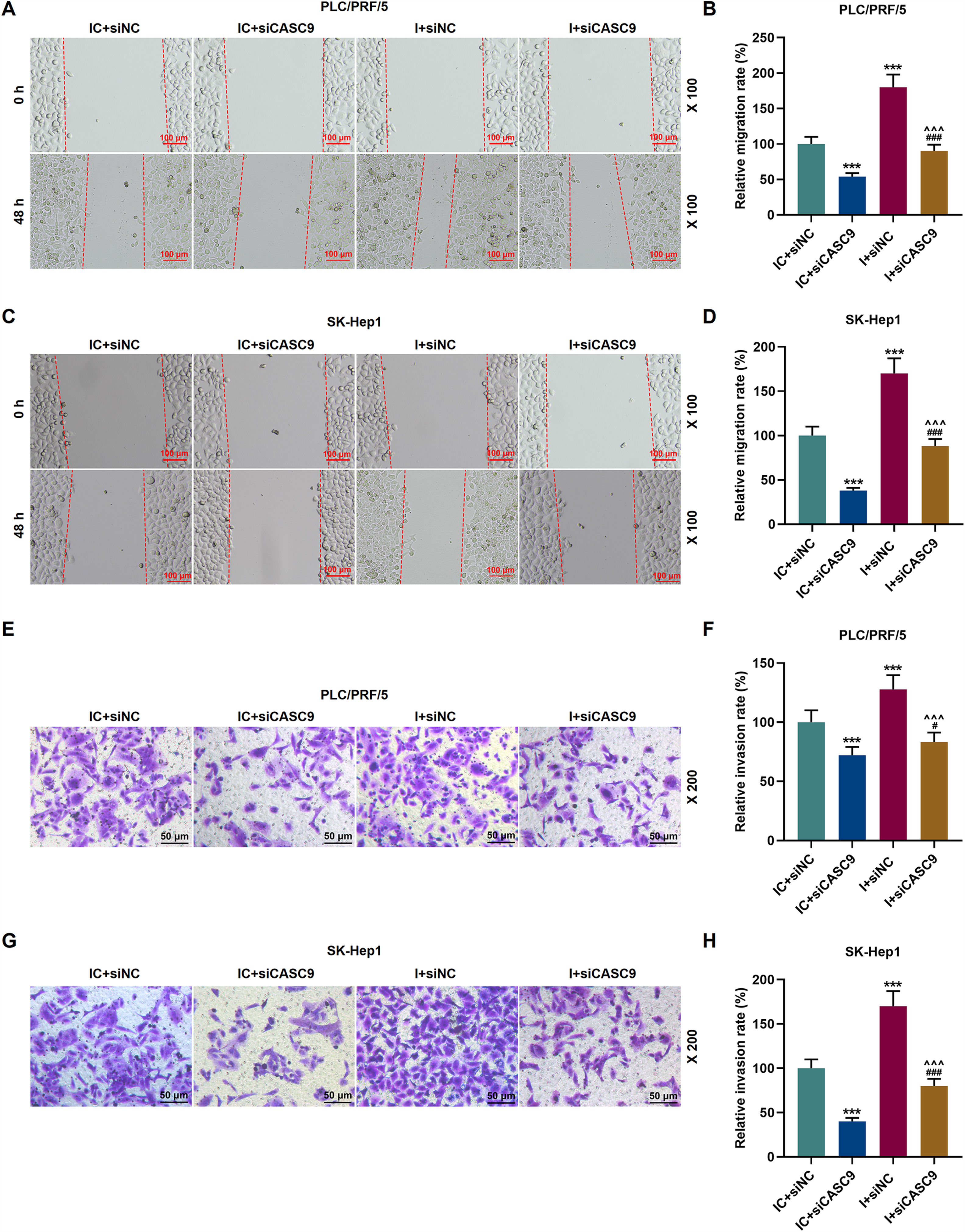
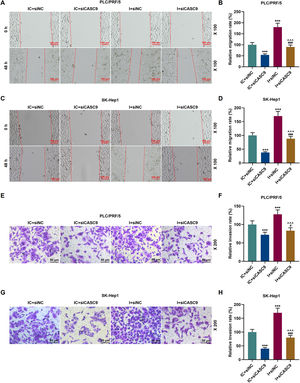
As shown in Fig. 7A–D, down-regulated miR-424-5p promoted the migration of HCC cells in comparison to the control group (p < 0.001). Besides, down-regulated CASC9 could partially offset the effect of down-regulated miR-424-5p on HCC cell migration. Transwell assay was performed to detect the effect of miR-424-5p on the invasion ability of HCC cells. As can be seen in Fig. 7E–H, miR-424-5p inhibitor enhanced the invasion of HCC cells in comparison to the control group. Furthermore, down-regulated CASC9 could partially reverse the effect of down-regulated miR-424-5p on HCC cell invasion (p < 0.001).
Down-regulated CASC9 inhibited the migration and invasion of HCC cells by negatively regulating miR-424-5p. (A): Wound healing assay was used to detect the effects of CASC9 and miR-424-5p on the migration of PLC/PRF/5 cells. Scale: 100 μm; magnification: ×100. (B): The migration rate of PLC/PRF/5 cells was calculated. n = 3. (C): Wound healing assay was used to detect the effects of CASC9 and miR-424-5p on the migration of SK-Hep1 cells. Scale: 100 μm; magnification: ×100. (D): The migration rate of SK-Hep1 cells was calculated. n = 3. (E): Transwell assay was used to detect the effects of CASC9 and miR-424-5p on the invasion of PLC/PRF/5 cells. Scale: 50 μm; magnification: ×200. (F): The number of invasive PLC/PRF/5 cells was counted. n = 3. (G): Transwell assay was used to detect the effects of CASC9 and miR-424-5p on the invasion of SK-Hep1 cells. Scale: 50 μm; magnification: ×200. (H):The number of invasive SK-Hep1 cells was counted. n = 3.
* vs. IC + siNC; ^ vs. IC + siCASC9; # vs. I + siNC. *** p < 0.001; ^^^ p < 0.001; ### p < 0.001.
Abbreviation: HCC, Hepatocellular carcinoma; siNC, small interfering RNA of negative control; IC, inhibitor control; I, miR-424-5p inhibitor.
Liver cancer is one of the most common cancers in the world and causes ∼782,000 deaths annually [17]. It is the second most deadly cancer and there are very limited treatment options. The most common subtype of liver cancer (90%) is HCC [18]. HCC is characterized by insidious onset and lack of obvious early clinical symptoms, which makes early detection and diagnosis difficult. HCC is usually diagnosed in the middle and late stages, and its mortality rate remains high due to the high invasion and metastasis rates. Recent studies have found that the 5-year survival of HCC is incredibly low [19–21]. Therefore, it is urgently needed to find valuable molecule markers for early diagnosis and therapeutic targets in the treatment of HCC.
Previous studies have suggested that long non-coding RNAs are functionless in humans, even though they make up 98.5% of the total human genome [22]. However, after the restart of the ENCODE research plan, a large number of studies proved that lncRNA could regulate the expression of protein-coding genes at the post-transcriptional level, thus affecting various biological functions of cells [23]. In recent years, lncRNA has become a hot spot in HCC research. Some researches have demonstrated that lncRNAs are involved in the HCC process [24,25]. CASC9 is an important member of the lncRNA family. Evidence has begun to emerge that CASC9 participates in the process of tumor occurrence and development and affects tumor progression [26–29]. The proliferation of healthy cells is strictly regulated by growth signals and cell cycle regulators to maintain homeostasis, while the abnormally high expression of CASC9 enhances the uncontrolled proliferation of HCC cells [30]. Klingenberg et al. demonstrated that the CASC9 gene is associated with the proliferation and apoptosis of HCC cells. Migration and invasion are also phenotypes of tumor progression [31]. However, there is no evidence that CASC9 is involved in the invasion and migration of HCC cells. Our study found that silenced CASC9 was found to decrease the proliferation, invasion and migration of HCC cells while promoting apoptosis, which enriches the previous studies [30]. At the same time, we also determined the correlation between CASC9 expression and the clinical pathological data of HCC patients, and it was found that the expression of CASC9 was strongly associated with tumor size, lymph node metastasis, TNM stage, differentiation degree and AFP. According to prior study, these indicators are closely related to the prognosis of HCC [10]. Using the kaplan-Meier method, we further concluded that high expression of CASC9 was strongly associated to the poor prognosis of HCC patients, thereby verifying the above results. All the aforementioned studies have demonstrated the involvement of CASC9 in the HCC process, but there are few examples of the effect of CASC9 protein complexes on the survival rate of HCC cells.
MiR-424-5p is also a molecule that plays a central role in HCC procession [32–34]. miR-424-5p could inhibit the proliferation and migration of HCC cells by targeting TRIM29 [35]. Similarly, Piao et al. also found that miR-424-5p inhibited the proliferation and promoted the apoptosis of HCC cells by inhibiting the expression of YAP1 [36]. Therefore, we hypothesized a targeting relationship between miR-424-5p and CASC9. Then, the binding site between miR-424-5p and CASC9 was predicted by the Diana-LncBase V2 website, and subsequently verified by dual-luciferase reporter gene assay. In Paraskevopoulou et al. study, the miRNA targets on lncRNA were predicted by the LncBase V2 host, which provides information on the regulation of cell-type specific miRNA and lncRNA [37]. Then, we found that miR-424-5p expression was low in HCC tissues. Pearson correlation coefficient analysis showed that CASC9 was inversely related with miR-424-5p. However, siCASC9 could partially reverse the effects of miR-424-5p inhibitor on the proliferation, migration, invasion and apoptosis of HCC cells. This suggests that CASC9 affects the proliferation, migration, invasion and apoptosis of HCC cells by negatively regulating miR-424-5p. However, this study also has limitations. For example, the expression of CASC9 in cancer stem cell population from HCC is yet to be tested; and this study only analyzed the effect of CASC9 on HCC in cells in vitro. Hence, in vivo animal experiments need to be conducted in the future for a comprehensive analysis of our results.
The data from this study showed that the down-regulation of CASC9 expression inhibited the malignant behavior of HCC, which further verified the role of CASC9 in liver cancer. Besides, miR-424-5p was identified as the downstream target molecule of CASC9. In conclusion, our results confirm that CASC9 affects HCC progression by negatively regulating miR-424-5p, suggesting that CASC9 may be a potential prognosis predictor and therapeutic target of HCC.
Declarations of interestNone.
FundingThis research did not receive any specific grant from funding agencies in the public, commercial, or not-for-profit sectors.
Ethics statementThe current study was reviewed and approved from Ethics committee of the People’s Hospital of Rizhao (RZ201912072), and all patients had signed informed consent. There were 50 patients (including 29 male patients and 21 female patients) diagnosed with HCC between December 2019 and June 2020 in the People’s Hospital of Rizhao. A total of 100 tissue samples (50 pairs of paracancerous tissues and tumor samples) were collected and agreed by these patients.
Not applicable.
LncRNAs, long non-coding RNAs; HCC, hepatocellular carcinoma; AFP, alpha-fetoprotein; CASC9, cancer sensitivity 9; ESCC, esophageal squamous cell carcinoma; qRT-PCR, quantitative reverse transcription polymerase chain reaction; ATCC, American Type Culture Collection; DMEM, Dulbecco's Modified Eagle Medium; CCK-8, cell counting kit-8; RIPA, radio-immunoprecipitation assay; SDS-PAGE, sodium dodecylsulphate polyacrylamide gel electrophoresis; PVDF, polyvinylidene fluoride; GAPDH, glyceraldehyde-3-phosphate dehydrogenase; siNC, small interfering RNA of negative control; M: miR-424-5p mimic; IC, inhibitor control; I, miR-424-5p inhibitor.