LINC01006 has been verified to be correlated with several cancer types, whereas its biological function in hepatocellular carcinoma (HCC) is still elusive. This study aimed to elucidate the specific regulatory mechanism of LINC01006 in the tumorigenesis of HCC.
Materials and methodsThe expression of LINC01006, miR-433-3p and CBX3 in HCC tissues and cells was assessed by qRT-PCR or Western blot. MTT, wound-healing, and transwell assays were used to evaluate the effects of LINC01006 on cell viability, migration, and invasion in vitro. A mouse xenograft model was established for in vivo assays. The relations among LINC01006, miR-433-3p, and CBX3 were analyzed by MS2-RNA immunoprecipitation (RIP) and Dual-luciferase reporter (DLR) assays.
ResultsThe expression of LINC01006 was up-regulated in HCC tissues and cells. LINC01006 knockdown inhibited the viability, wound healing rate, and invasive cell number of HeP3B and SK-HeP-1 cells, and decreased the tumor volume and weight in a mouse xenograft model. MiR-433-3p was a target of LINC01006, and LINC01006 overexpression inhibited the viability, wound healing rate, and invasive cell number of HeP3B and SK-HeP-1 cells. In addition, CBX3 was a target of miR-433-3p, which was negatively regulated by miR-433-3p. CBX3 overexpression and miR-433-3p inhibition reversed the inhibiting effects of LINC01006 knockdown on the viability, migration, and invasion of HeP3B cells.
ConclusionsSilencing of LINC01006 inhibited the viability, migration, and invasion of HCC cells through regulating miR-433-3p/CBX3 axis.
Hepatocellular carcinoma (HCC), ranking the sixth most common human cancers, is characterized by a low rate of early detection, poor prognosis, and high mortality [1,2]. Until now, surgery, chemo-radiotherapy, and transplantation are the main strategies for HCC treatment [3]. The 5-year survival rate of patients underwent transplantation is about 80%, while that of patients underwent resection is only about 50% [4]. In addition, most patients with HCC are diagnosed in the middle or late stage with poorer survival, and their therapeutic opportunity for amelioration is diminished [5]. Therefore, it is imperative to explore the pathogenesis of HCC and screen out effective diagnostic markers as well as therapeutic targets.
Increasing evidence has demonstrated that long non-coding RNAs (lncRNAs) are involved in various pathological and physiological processes, particularly in malignant tumors [6–8]. Many lncRNAs have been proved to play crucial roles in the occurence and development of HCC. For instance, KTN1-AS1 functions as an oncogene in HCC through regulating miR-23c/ERBB2IP axis [9]. TCONS00006195 suppresses the tumorigenesis of HCC through inhibiting the enzymatic activity of ENO1 [10]. Herein, we focused on the long intergenic non-protein coding RNA 1006 (LINC01006), which has been reported in gastric and pancreatic cancer. Zhu et al. have demonstrated that LINC01006 serves as a novel biomarker for gastric cancer [11]. LINC01006 contributes to the tumorigenesis of pancreatic cancer via regulating miR-2682-5p/HOXB8 axis [12]. However, the specific function of LINC01006 in HCC remains unclear.
MicroRNAs (miRNAs) have been reported in a variety of human cancers, including HCC [13–16]. For instance, miR-133a-3p acts as a tumor suppressor and a diagnostic marker in HCC [17]. MiR-221 contributes to the development of HCC by inhibiting PHF2 [18]. According to previous studies, miR-433-3p is identified as a tumor suppressor in many cancers, such as glioma [19], cervical cancer [20], and gastric cancer [21]. More importantly, Xue et al. [22] have demonstrated that miR-433-3p inhibits HCC progression by regulating p21 activated kinase. Recently, researches have demonstrated that lncRNAs usually act as competitive endogenous RNAs (ceRNAs) to sponge specific miRNAs, thus regulating the downstream mRNA targets [23–25]. For example, lncRNA SNHG14 functions as a ceRNA in regulating osteosarcoma progression by targeting miR-433-3p [26]. LINC00460 knockdown restrains the tumorigenesis of colon cancer through sponging miR-433-3p [27]. However, the specific interaction between LINC01006 and miR-433-3p in HCC has not been investigated.
Chromobox protein homolog 3 (CBX3) has been shown to play a critical role in a variety of human malignant tumors. For instance, CBX3 accelerates the proliferation of colorectal cancer cells both in vitro and in vivo [28]. CBX3 contributes to the tumorigenesis of osteosarcoma [29]. Moreover, Zhong et al. have revealed that CBX3 serves as a tumor-promoter in HCC progression [30]. Nevertheless, the specific relations among LINC01006, miR-433-3p, and CBX3 in HCC need to be further elucidated.
In this study, we measured LINC01006 expression in HCC tissues and cells, and investigated its role in HCC both in vitro and in vivo. Furthermore, we analyzed the action mechanisms of LINC01006, miR-433-3p, and CBX3 in HCC. Our findings disclosed that LINC01006/miR-433-3p/CBX3 may act as potential therapeutic targets for HCC.
2Material and methods2.1HCC tissue specimensForty-nine pairs of HCC tissues and corresponding adjacent normal tissues were obtained from 59 patients with HCC in our hospital by surgical resection. All patients had not received any chemotherapy or radiotherapy before surgery. This study was approved by the ethics committee of our hospital in accordance with the Declaration of Helsinki. Written informed consents were obtained from all patients.
2.2Cell culture and transfectionHuman fetal hepatocyte line L-02, and four HCC cell lines, including HeP3B, SK-HeP-1, HePG2, and Huh7 were obtained from American Type Culture Collection (ATCC, Manassas, VA, USA). Cells were cultured in DMEM containing 10% fetal bovine serum (FBS) and maintained in a humidified incubator supplied with 5% CO2 at 37 °C. ShRNAs against LINC01006 (sh-LINC01006-1/2), non-targeting shRNA (sh-NC), and pcDNA-CBX3 were purchased from Genepharma (Shanghai, China). MiR-433-3p mimics, miR-433-3p inhibitor, and corresponding negative control (miR-NC) were obtained from RiboBio (Guangzhou, China). Cell transfection was performed using Lipofectamine 3000 (Invitrogen, Carlsbad, CA, USA).
2.3qRT-PCRTotal RNAs were isolated using TRIzol reagent (Invitrogen), and the cDNA was reversely transcribed using miRNA reverse transcription kit (Takara, Dalian, China). qRT-PCR analysis was applied using SYBR® Premix Ex Taq™ II (Takara, Dalian, China) on ABI Prism 7500 Fast Real-Time PCR system (Applied Biosystems, Foster City, CA, USA). Specific primers were synthesized by Shenggong (Shanghai, China) and the sequences were shown as follows: LINC01006 forward 5′-TTTGTGGTGGTGAAGACGTG-3′, LINC01006 reverse 5′-TCCTCAAGAATAAGGAACATAGGC-3′; miR-433-3p forward 5′-GGAGAAGTACGGTGAGCCTGT-3′, miR-433-3p reverse 5′-GAACACCGAGGAGCCCATCAT-3′; CBX3 forward 5′- TGGCCTCCAACAAAACTACA-3′, CBX3 reverse 5′- TCCCATTCACTACACGTCGA-3′; GAPDH forward 5′-ACCCACTCCTCCACCTTTGAC-3′, LINC01006 reverse 5′-TGTTGCTGTAGCCAAATTCGTT-3′; U6 forward 5′-CATGCTTGTAGCTGCCCCAT-3′, U6 reverse 5′-GAGAGTACTGGGTGTCCGTTT-3′. The PCR program included 95 °C for 5 min, 40 cycles of 95 °C for 10 s, 56 °C for 20 s and 72 °C for 30 s. The relative expression level was calculated using the 2−ΔΔCt method. GAPDH and U6 were used as internal references.
2.4MS2-RNA immunoprecipitation (MS2-RIP) assayHeP3B and SK-HeP-1 cells were co-transfected with pMS2-GFP and pcDNA-MS2/pcDNA-MS2-LINC01006 for 48 h. After transfection, RIP assay was performed using anti-GFP antibody (Roche, Germany) and MagnaRIP RNA-binding protein immunoprecipitation kit (Millipore, Bedford, MA, USA) according to the manufacturer’s protocol.
2.5Dual luciferase reporter (DLR) assayThe 3′UTR sequences of LINC01006 or CBX3 containing miR-433-3p binding sites were cloned into the psiCHECK-2 vector (Promega, Madison, WI, USA) to generate LINC01006-wt and CBX3-wt. The sequences containing mutant binding sites were used to generate LINC01006-mut and CBX3-mut. Then, HeP3B and SK-HeP-1 cells were co-transfected with miR-NC/miR-433-3p mimics and the above luciferase reporter vectors using Lipofectamine 3000 (Invitrogen). After 48 h of transfection, Dual-Luciferase Reporter Assay System (Promega) was applied to detect the luciferase activity.
2.6MTT assayHeP3B and SK-HeP-1 cells were seeded into 96-well plates and cultured for 24, 48, 72, and 96 h, respectively. Subsequently, cells were incubated with 5 mg/mL MTT (Sigma-Aldrich, St. Louis, MO, USA) for 4 h at 37 °C. Then, dimethyl sulfoxide (DMSO) was added into each well, and the absorbance of formazan crystals at 450 nm was measured by a microplate reader (Bio-Rad).
2.7Wound healing assayHeP3B and SK-HeP-1 cells (5 × 105/well) were seeded into 6-well plates. When cells cultured to 100% confluence, an artificial scratch was created using a 200 μL pipette tip. Wound closure images were taken at 0 and 24 h using an inverted microscope (Olympus, Japan). Wound healing rate = (initial scratch width – scratch width at 24 h post-culturing)/initial scratch width × 100%.
2.8Transwell assayCell invasion was detected using matrigel-coated transwell chambers (8 µm pore size; Millipore, Billerica, MA, USA). In brief, cells (5 × 105/well) suspended in serum free DMEM were added into the upper chamber. Correspondingly, the DMEM containing 10% FBS was added to the lower chamber. After incubated for 24 h, cells in the lower chamber were fixed in 4% paraformaldehyde for 20 min and stained with 0.1% crystal violet for 15 min. The invasion cells were counted at five randomly selected views under a microscope (Olympus).
2.9Western blotProtein samples were isolated by lysing cells in RIPA buffer (Beyotime, Shanghai, China). The protein concentration was determined using Pierce BCA protein assay kit (Pierce, Rockford, IL, USA). The proteins were then separated by 10% SDS-PAGE electrophoresis, transferred onto PVDF membranes, and blocked with 5% non-fat milk. The membranes were incubated with primary antibody (anti-CBX3, 1:1000, ab227478; anti-β-actin, ab11003, 1/500, Abcam, Cambridge, MA, USA) overnight at 4 °C, followed by horseradish peroxidase-conjugated secondary antibody (1/500, Abcam) for another 2 h at 25 °C. Protein bands were visualized using an ECL kit (Beyotime).
2.10Xenograft tumor assayMale nude mice (balb/c, 4-weeks-old) were purchased from HFK Bioscience Co., Ltd. (Beijing, China), and were randomly divided into sh-NC and sh-LINC01006-1 groups (5 mice in each group). HeP3B cells (1 × 106) stably transfected with sh-NC or sh-LINC01006-1 were injected into the right axilla of mice. The tumor volume was measured every 7 days using a vernier caliper. At the 4th week post-injection, mice were anesthetized with pentobarbital sodium (60 mg/kg) and sacrificed by cervical dislocation. The tumor xenograft was then removed and weighed. Animal experiments were approved by the ethics committee of our hospital in accordance with the Guide for the Care and Use of Laboratory Animals (eighth edition, 2011, National Institutes of Health, USA).
2.11Statistical analysisStatistical analysis was performed using SPSS 22.0 (SPSS, Chicago, USA). All experiments were repeated 3 times independently. Quantitative data were expressed as mean ± standard deviation (SD). The differences between two groups were analyzed by Student’s t-test. The differences among multi-groups were analyzed by One-way ANOVA, followed by Tukey’s multiple comparisons test for pairwise comparison. Qualitative data were expressed as number (data in Table 1), and the differences were analyzed by X2 test. The correlation between miR-433-3p and LINC01006/CBX3 was analyzed by Pearson correlation test. The difference was considered statistically significant at P < 0.05.
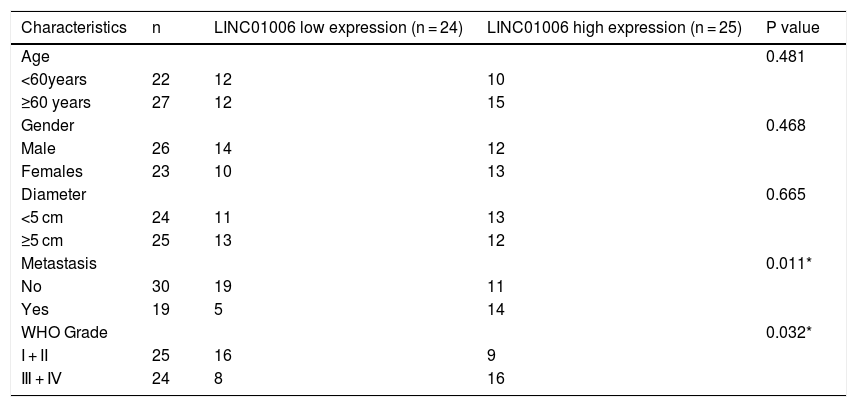
Correlation between the clinicopathological characteristics and LINC01006 expression in hepatocellular carcinoma (n = 49).
| Characteristics | n | LINC01006 low expression (n = 24) | LINC01006 high expression (n = 25) | P value |
|---|---|---|---|---|
| Age | 0.481 | |||
| <60years | 22 | 12 | 10 | |
| ≥60 years | 27 | 12 | 15 | |
| Gender | 0.468 | |||
| Male | 26 | 14 | 12 | |
| Females | 23 | 10 | 13 | |
| Diameter | 0.665 | |||
| <5 cm | 24 | 11 | 13 | |
| ≥5 cm | 25 | 13 | 12 | |
| Metastasis | 0.011* | |||
| No | 30 | 19 | 11 | |
| Yes | 19 | 5 | 14 | |
| WHO Grade | 0.032* | |||
| I + II | 25 | 16 | 9 | |
| Ⅲ + Ⅳ | 24 | 8 | 16 |
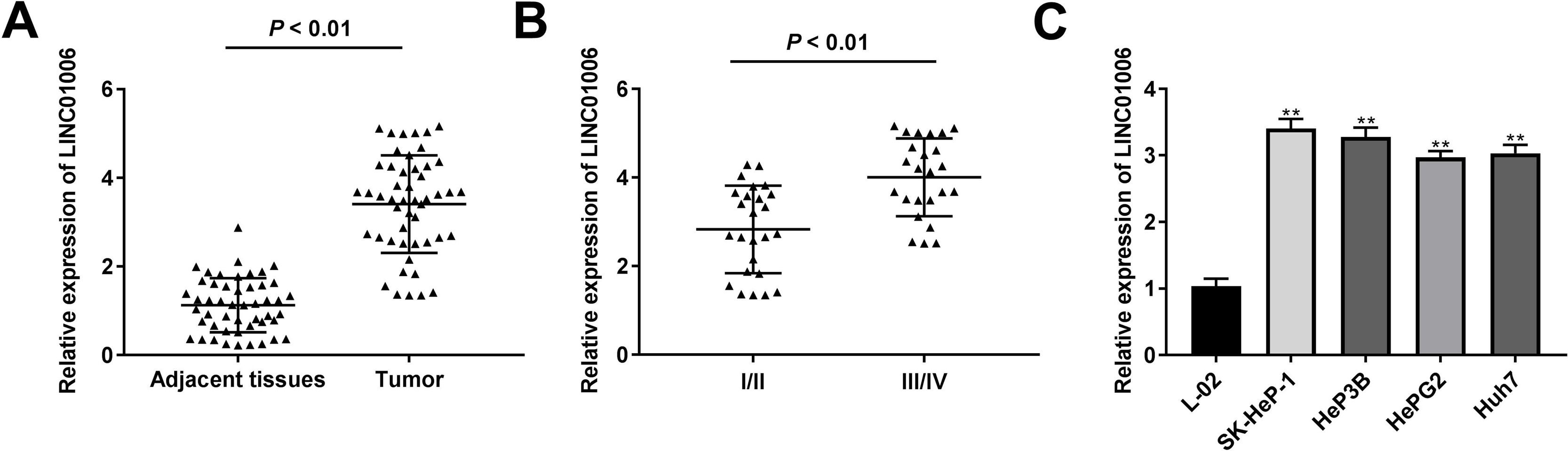
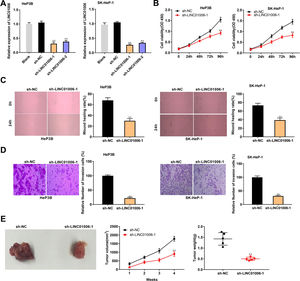
To determine the functional role of LINC01006 in HCC, we primarily detected LINC01006 expression in 49 pairs of HCC tissues and adjacent tissues. qRT-PCR showed that LINC01006 was significantly up-regulated in tumor tissues in contrast to adjacent normal tissues (3.41 ± 1.09 vs. 1.13 ± 0.61; P < 0.01, Fig. 1A). Then, 49 pairs of HCC samples were classified into high (n = 25) and low (n = 24) expression groups based on the mean expression of LINC01006 (3.41). As shown in Table 1, high expression of LINC01006 was significantly associated with metastasis (P = 0.011) and advanced WHO grade (P = 0.032) in HCC patients. As shown in Fig. 1B, LINC01006 expression was significantly higher in WHO grade Ⅲ/Ⅳ than that in grade I/II (4.01 ± 0.87 vs. 2.83 ± 0.99; P < 0.01). Subsequently, we detected the expression of LINC01006 in HCC cell lines (SK-HeP-1, HeP3B, HepG2 and Huh7) and human fetal hepatocyte line L-02. LINC01006 expression was significantly increased in HCC cell lines compared to that in L-02 cells (3.41 ± 0.41, 3.28 ± 0.14, 2.97 ± 0.09, 3.03 ± 0.13 vs. 1.00 ± 0.12; P < 0.01, Fig. 1C). SK-HeP-1 and HeP3B cells with relatively high expression of LINC01006 were selected for the subsequent experiments.
LINC01006 expression was up-regulated in HCC tissues and cells. A, The expression of LINC01006 in 49 pairs of HCC specimens and adjacent tissues was detected by qRT-PCR. B, The expression of LINC01006 was detected in HCC tissues at grade I/II and III/IV by qRT-PCR. C, The expression of LINC01006 was detected in HCC cell lines (SK-HeP-1, HeP3B, HepG2 and Huh7) and normal human fetal hepatocyte line L-02 by qRT-PCR. ** P < 0.01 vs. L-02.
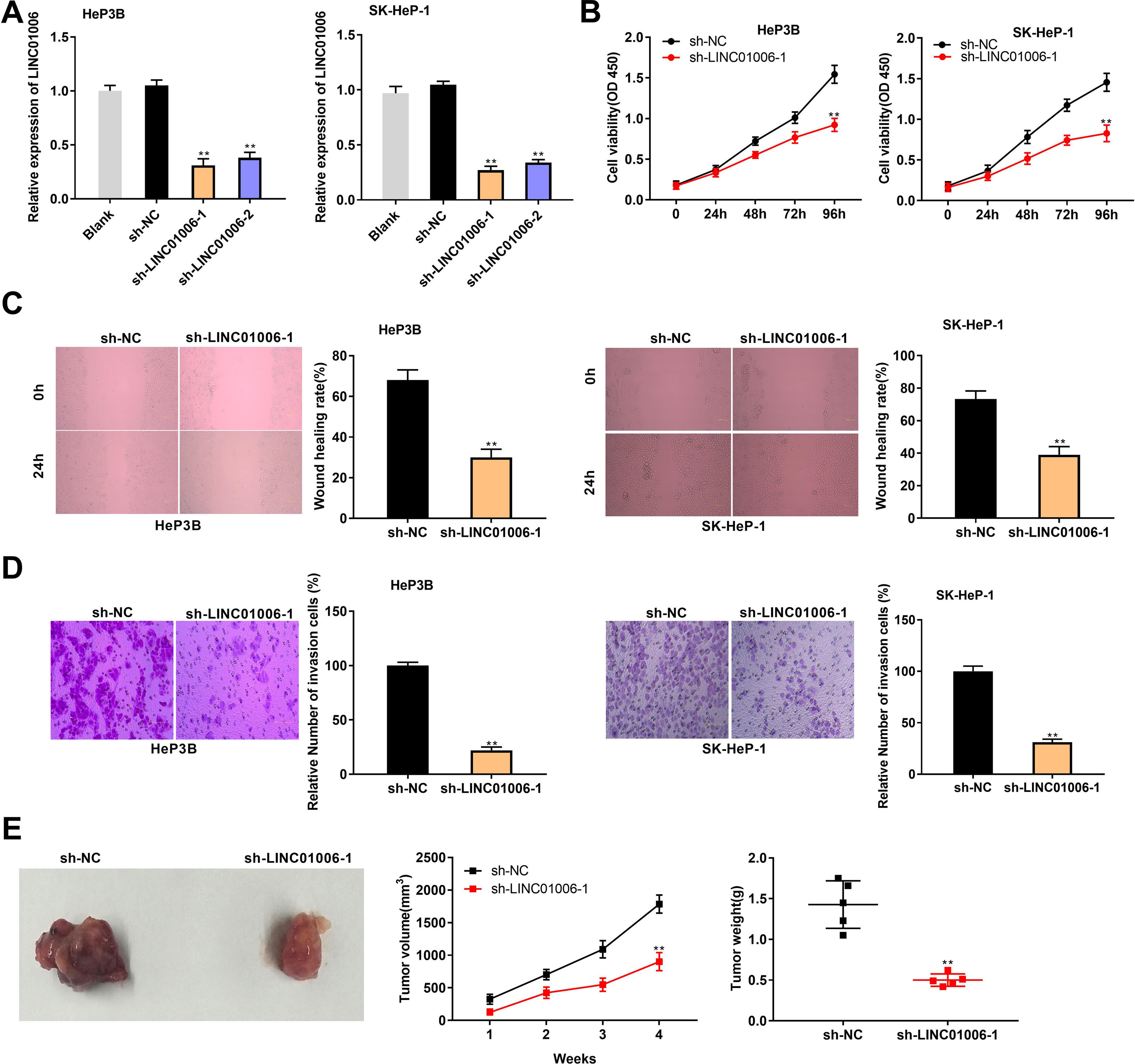
ShRNAs against LINC01006 were transfected into HeP3B and SK-HeP-1 cells. qRT-PCR demonstrated that the transfection of sh-LINC01006-1/2 dramatically decreased the LINC01006 expression compared to Blank group (P < 0.01, Fig. 2A). Sh-LINC01006-1 with relatively high silencing efficiency was selected for subsequent assays. As illustrated in Fig. 2B–D, knockdown of LINC01006 markedly reduced the cell viability (HeP3B: 0.92 ± 0.08 vs. 1.54 ± 0.11; SK-HeP-1: 0.83 ± 0.10 vs. 1.46 ± 0.11; P < 0.01), wound healing rate (HeP3B: 30.12 ± 4.05 vs. 68.05 ± 5.03; SK-HeP-1: 39.45 ± 5.13 vs. 73.37 ± 5.22; P < 0.01), and relative number of invasion cells (HeP3B: 22.18 ± 3.08 vs. 100.00 ± 3.33; SK-HeP-1: 31.15 ± 3.08 vs. 100.00 ± 5.16; P < 0.01) in HeP3B and SK-HeP-1 cells. To further illustrate the functional role of LINC01006 in vivo, a mouse xenograft model was established by subcutaneous injection of sh-LINC01006-1-transfected HeP3B cells. The results of tumor volume (902 ± 139 vs. 1785 ± 138; P < 0.01) and weight (0.50 ± 0.08 vs. 1.43 ± 0.29; P < 0.01) demonstrated that silencing of LINC01006 suppressed the tumor growth in mice (Fig. 2E). Taken together, LINC01006 knockdown inhibited the viability, migration, and invasion of HCC cells in vitro, and inhibited the tumor growth in vivo.
LINC01006 knockdown inhibited the viability, migration, and invasion of HCC cells in vitro, and the tumor growth in vivo. A, The expression of LINC01006 was decreased by transfection of sh-LINC01006-1/2 in HeP3B and SK-HeP-1 cells. ** P < 0.01 vs. Blank. B, Cell viability was determined by MTT analysis. C, Wound healing assay was carried out to detect the migration of HeP3B and SK-HeP-1 cells. D, Transwell assay was performed to detect the invasion of HeP3B and SK-HeP-1 cells. E, Tumor volume and weight were measured in a mouse xenograft model. ** P < 0.01 vs. sh-NC (B–E).
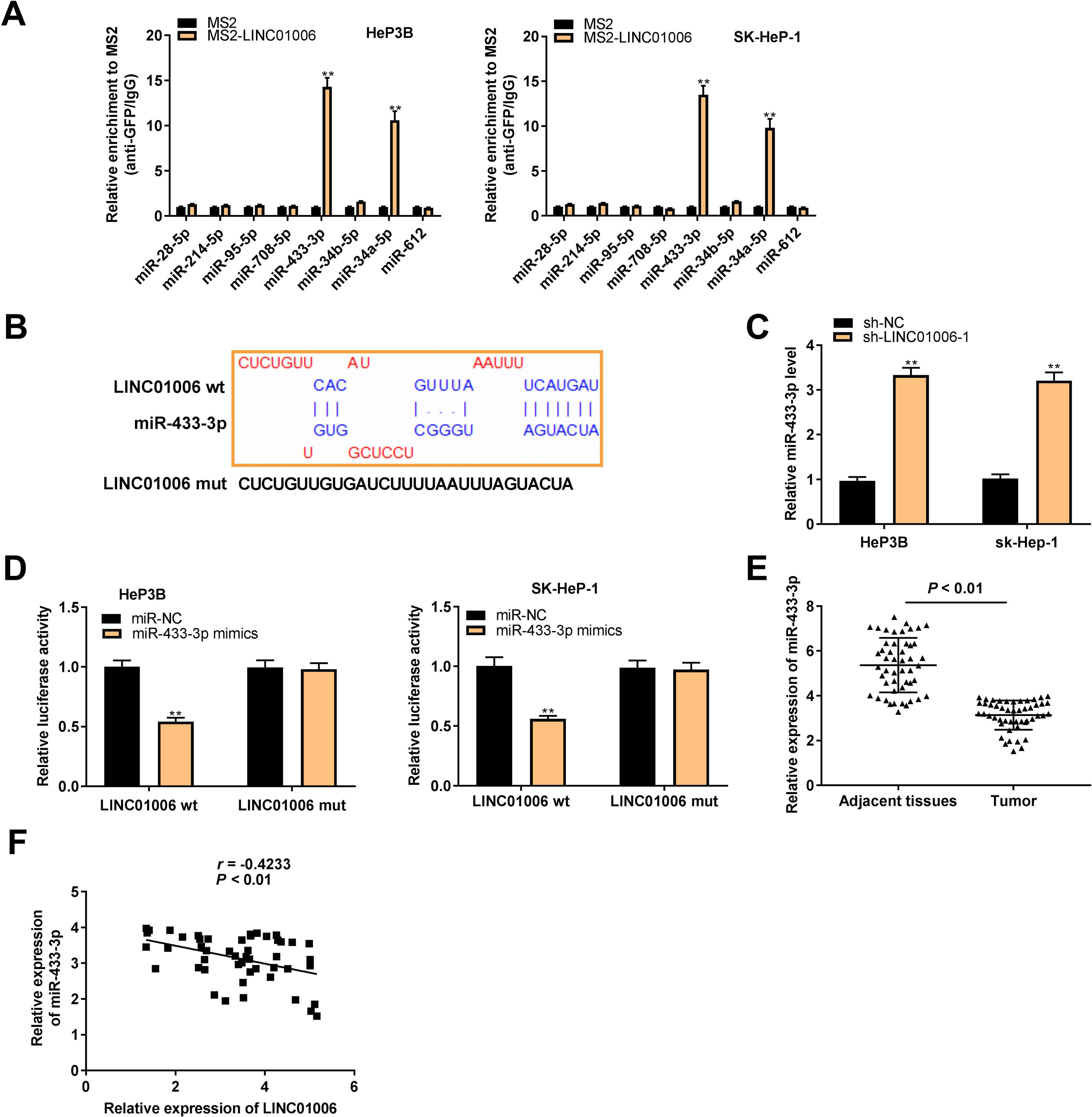
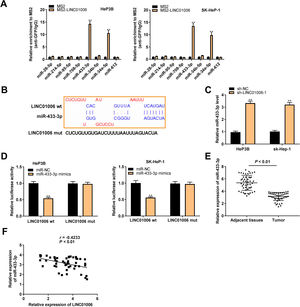
To explore the action mechanism of LINC01006 in HCC, eight target miRNAs containing LINC01006 binding sites were predicted by lncbase and starbase. Subsequently, we carried out MS2-RIP assay to further identify the interactions between LINC01006 and eight candidate miRNAs. The results showed that the relative enrichments of miR-433-3p and miR-34a-5p were prominently high in MS2-LINC01006 group compared with those in MS2 group, suggesting that both miR-433-3p and miR-34a-5p were targets of LINC01006 (P < 0.01, Fig. 3A). MiR-433-3p with relatively higher enrichment was applied to the follow-up experiments. The binding site of miR-433-3p with LINC01006 was shown in Fig. 3B. As presented in Fig. 3C, LINC01006 knockdown significantly elevated miR-433-3p expression in HeP3B and SK-HeP-1 cells (HeP3B: 3.33 ± 0.17 vs. 1.00 ± 0.09; SK-HeP-1: 3.21 ± 0.19 vs. 1.00 ± 0.10; P < 0.01). DLR analysis disclosed that miR-433-3p mimics significantly reduced the luciferase activity of LINC01006 wt (HeP3B: 0.54 ± 0.03 vs. 1.00 ± 0.05; SK-HeP-1: 0.56 ± 0.02 vs. 1.00 ± 0.07; P < 0.01, Fig. 3D). In addition, miR-433-3p expression was significantly decreased in HCC tissues in comparison with that in adjacent tissues (3.14 ± 0.64 vs. 5.36 ± 1.20; P < 0.01, Fig. 3E). A negative association was detected between miR-433-3p expression and LINC01006 expression (r = −0.4233, P < 0.01, Fig. 3F). Collectively, LINC01006 directly targeted miR-433-3p, and negatively regulated miR-433-3 in HCC cells.
MiR-433-3p was a direct target of LINC01006. A, Eight candidate genes that potentially interacted with LINC01006 were predicated by lncbase and starbase, and MS2-RNA immunoprecipitation (MS2-RIP) assay was performed to determine the interaction between LINC01006 and these candidates. ** P < 0.01 vs. MS2. B, The binding site between LINC01006 and miR-433-3p was predicted by starbase. C, MiR-433-3p expression in sh-LINC01006-1-transfected HeP3B and SK-HeP-1 cells was measured by qRT-PCR. ** P < 0.01 vs. sh-NC. D, Dual luciferase reporter (DLR) assay was performed to verify the relationship between LINC01006 and miR-433-3p. ** P < 0.01 vs. miR-NC. E, MiR-433-3p expression in HCC tissues and adjacent tissues was detected by qRT-PCR. F, The correlation between LINC01006 and miR-433-3p expression was analyzed by spearman's correlation analysis.
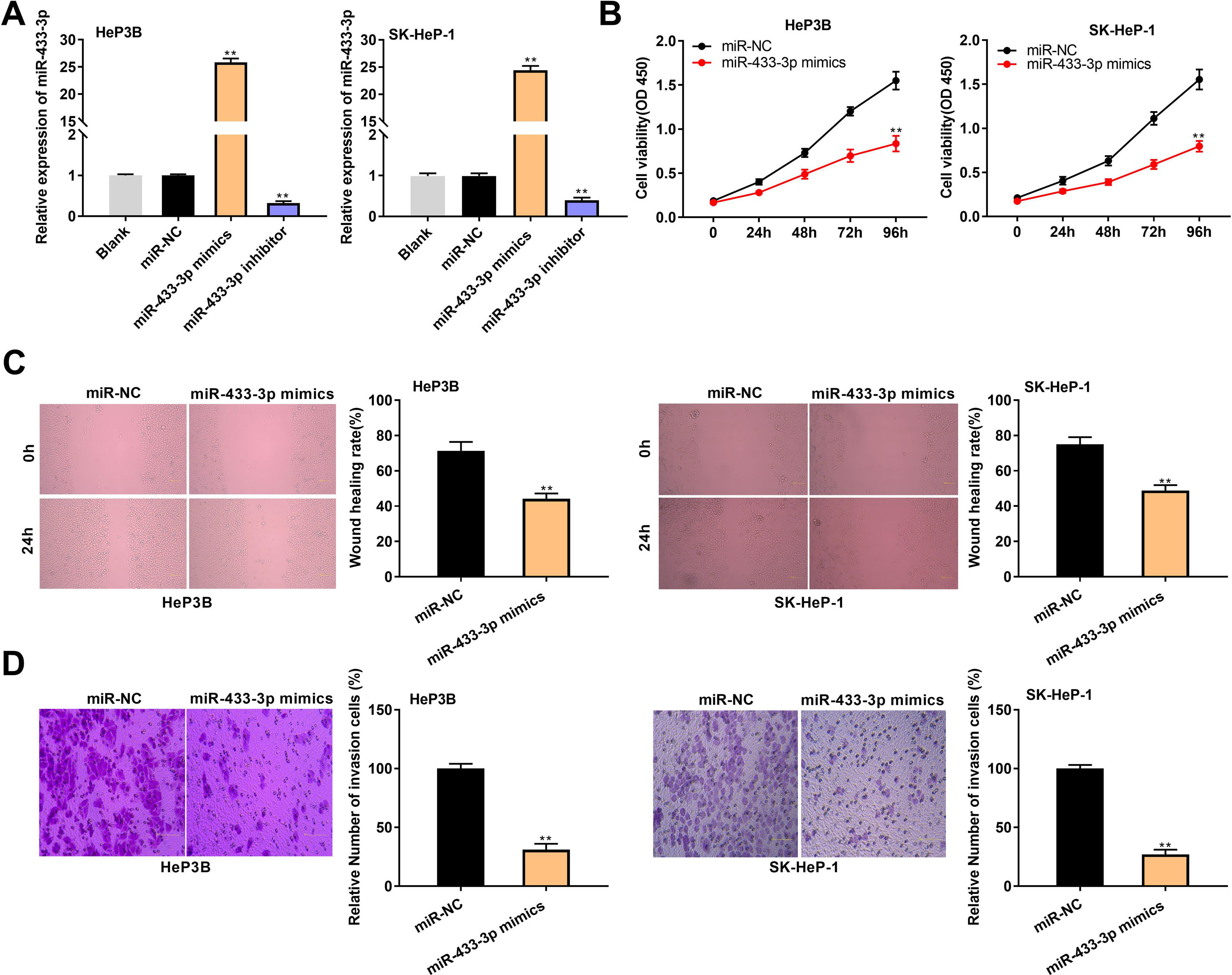
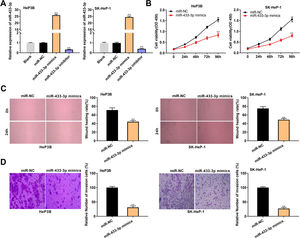
MiR-433-3p expression was overexpressed or silenced in HeP3B and SK-Hep-1 cells to reveal the biological role of miR-433-3p in HCC. qRT-PCR showed that miR-433-3p expression was dramatically increased in miR-433-3p mimics group, while was decreased in miR-433-3p inhibitor group in comparison to that in Blank group (P < 0.01, Fig. 4A). As shown in Fig. 4B–D, miR-433-3p overexpression significantly inhibited the viability (HeP3B: 0.84 ± 0.09 vs. 1.55 ± 0.10; SK-HeP-1: 0.80 ± 0.06 vs. 1.55 ± 0.11; P < 0.01), wound healing rate (HeP3B: 44.21 ± 3.04 vs. 71.44 ± 5.03; SK-HeP-1: 48.90 ± 3.11 vs. 75.07 ± 4.13; P < 0.01), and relative number of invasion cells (HeP3B: 31.26 ± 5.03 vs. 100.00 ± 4.11; SK-HeP-1: 27.08 ± 4.01 vs. 100.00 ± 3.06; P < 0.01) in HeP3B and SK-Hep-1 cells. Collectively, overexpression of miR-433-3p inhibited the viability, migration, and invasion of HCC cells.
MiR-433-3p overexpression inhibited the viability, migration, and invasion of HCC cells. A, MiR-433-3p expression in miR-433-3p mimics or inhibitor-transfected HeP3B and SK-HeP-1 cells was measured by qRT-PCR. ** P < 0.01 vs. Blank. B, Cell viability of miR-433-3p mimics-transfected HeP3B and SK-HeP-1 cells was determined by MTT assay. C, Wound healing assay was performed to detect the migration of miR-433-3p mimics-transfected HeP3B and SK-HeP-1 cells. D, Transwell assay was perfromed to detect the invasion of miR-433-3p mimics-transfected HeP3B and SK-HeP-1 cells. ** P < 0.01 vs. miR-NC (B–D).
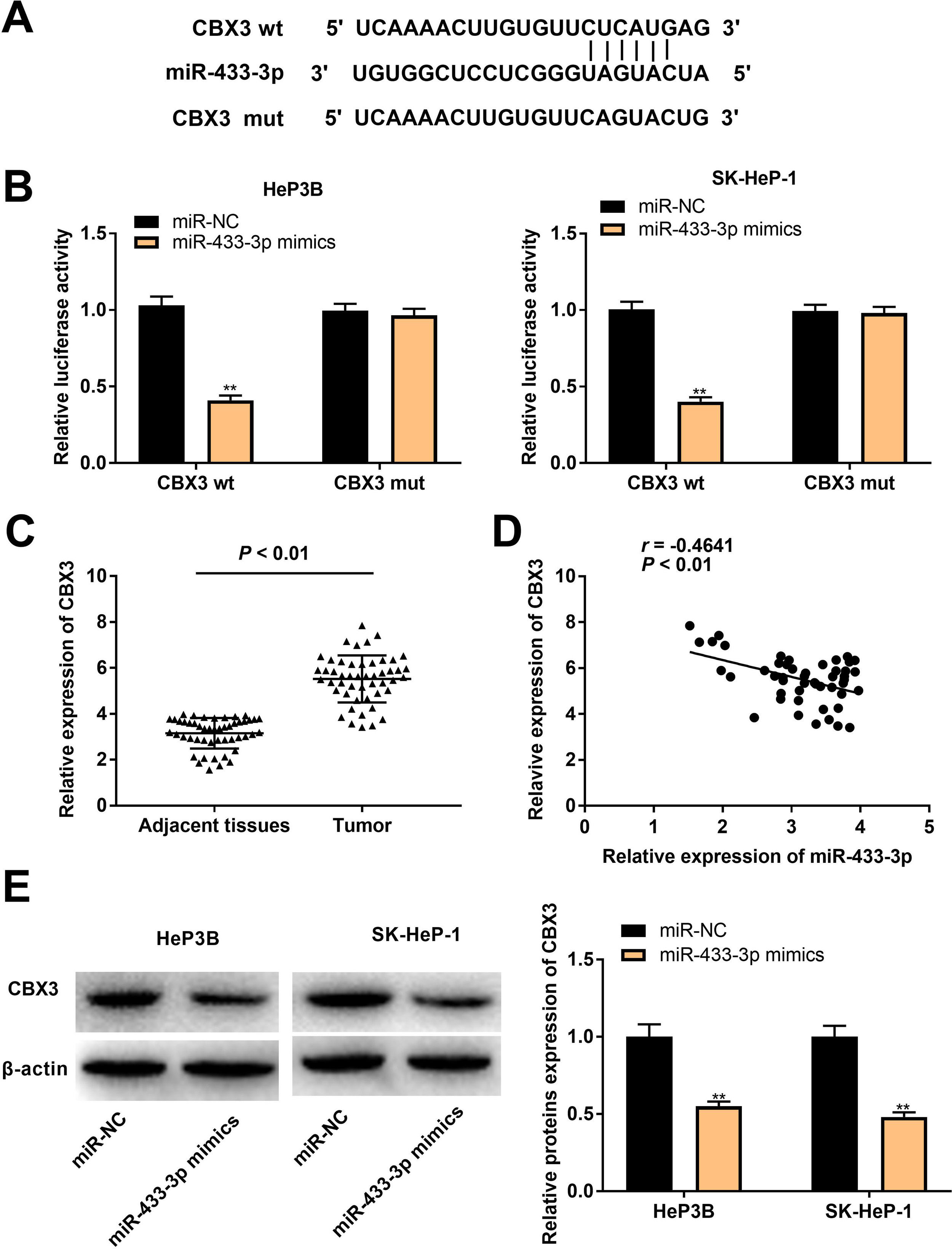
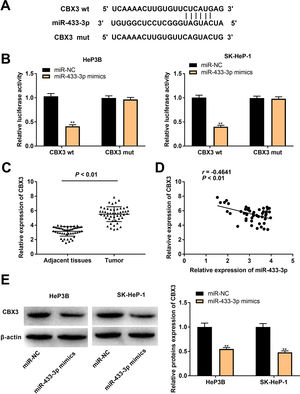
In order to further reveal the action mechanism of miR-433-3p in HCC, we predicted the downstream targets of miR-433-3p. A binding site of miR-433-3p on the 3′-UTR of CBX3 was predicted by TargetScan (Fig. 5A). DLR assay was performed to verify the target relation between miR-433-3p and CBX3. As shown in Fig. 5B, miR-433-3p mimics significantly decreased the luciferase activity of CBX3 wt (HeP3B: 0.41 ± 0.03 vs. 1.00 ± 0.06; SK-HeP-1: 0.40 ± 0.02 vs. 1.00 ± 0.05; P < 0.01). CBX3 expression was significantly higher in HCC tissues than that in adjacent tissues (5.52 ± 1.01 vs. 3.15 ± 0.66; P < 0.01, Fig. 5C). Moreover, a negative correlation was found in the expression of miR-433-3p and CBX3 (P < 0.01, Fig. 5D). The protein expression of CBX3 was significantly reduced by miR-433-3p overexpression (HeP3B: 0.55 ± 0.03 vs. 1.00 ± 0.12; SK-HeP-1: 0.48 ± 0.03 vs. 1.00 ± 0.10; P < 0.01, Fig. 5E). The above data illustrated that miR-433-3p directly targeted CBX3 and decreased CBX3 expression.
MiR-433-3p directly targeted CBX3. A, TargetScan predicted the binding site of miR-433-3p on 3′-UTR of CBX3. B, Dual luciferase reporter (DLR) gene assay was performed to verify the relationship between CBX3 and miR-433-3p. ** P < 0.01 vs. miR-NC. C, CBX3 expression in HCC tissues and adjacent tissues was detected by qRT-PCR. D, The correlation between CBX3 and miR-433-3p expression was analyzed by spearman’s correlation analysis. E, Relative protein expression of CBX3 was measured by Western blot. ** P < 0.01 vs. miR-NC.
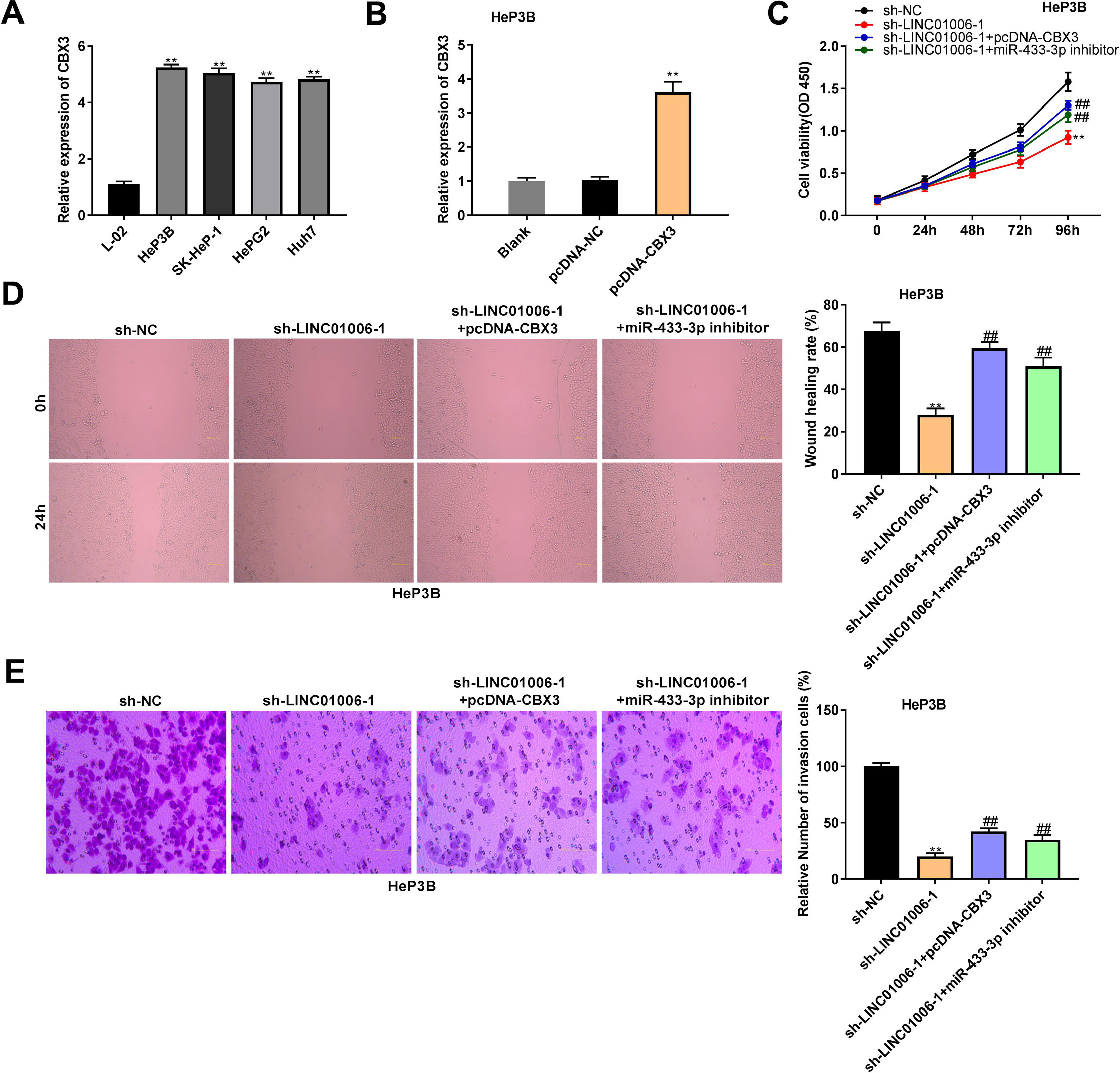
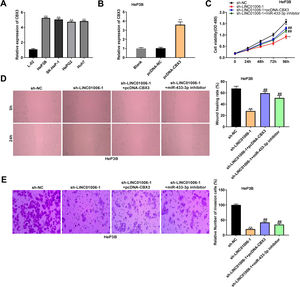
qRT-PCR showed a significantly higher expression of CBX3 in HeP3B, SK-HeP-1, HepG2 and Huh7 cells than that in L-02 cells (5.25 ± 0.08, 5.06 ± 0.13, 4.74 ± 0.10, 4.83 ± 0.07 vs. 1.00 ± 0.08; P < 0.01, Fig. 6A). In order to clarify the interactions among LINC01006, miR-433-3p, and CBX3, HeP3B cells were co-transfected with sh-NC/sh-LINC01006-1 and pcDNA-CBX3 or miR-433-3p inhibitor. As shown in Fig. 6B, pcDNA-CBX3 significantly enhanced the expression of CBX3 in HeP3B cells (3.61 ± 0.31 vs. 1.00 ± 0.10; P < 0.01). The cell viability (0.92 ± 0.08 vs. 1.58 ± 0.11; P < 0.01), wound healing rate (28.03 ± 3.08 vs. 67.70 ± 4.23; P < 0.01), and number of invasion cells (20.04 ± 2.87 vs. 100.00 ± 3.32; P < 0.01) were significantly inhibited by LINC01006 knockdown, while these inhibitory effects were partially reversed by CBX3 overexpression (cell viability: 1.30 ± 0.05 vs. 0.92 ± 0.08; wound healing rate: 59.42 ± 3.06 vs. 28.03 ± 3.08; number of invasion cells: 42.13 ± 3.11 vs. 20.04 ± 2.87; P < 0.01) or miR-433-3p inhibition (cell viability: 1.19 ± 0.09 vs. 0.92 ± 0.08; wound healing rate: 51.13 ± 4.06 vs. 28.03 ± 3.08; number of invasion cells: 35.21 ± 4.21 vs. 20.04 ± 2.87; P < 0.01) (Fig. 6C–E). Above all, LINC01006 knockdown inhibited the viability, migration, and invasion of HCC cells through regulating miR-433-3p/CBX3 axis.
The anti-tumor effect of LINC01006 knockdown on HCC cells was partially reversed by miR-433-3p inhibition or CBX3 overexpression. A, CBX3 expression in HCC cell lines was measured by qRT-PCR. ** P < 0.01 vs. L-02. B, CBX3 expression in pcDNA-CBX3-transfected HeP3B cells was detected by qRT-PCR. ** P < 0.01 vs. Blank. C, Cell viability of co-transfected HeP3B cells was determined by MTT assay. D, Wound healing assay was carried out to detect the migration of co-transfected HeP3B cells. E, Transwell assay was performed to detect the invasion of co-transfected HeP3B cells. ** P < 0.01 vs. sh-NC; ## P < 0.01 vs. sh-LINC01006-1.
More and more studies have demonstrated that lncRNAs participate in the tumorigenesis and development of malignant tumors, including HCC [31]. Different lncRNAs are identified to function as either tumor-promoter or tumor-suppressor in HCC progression. For instance, lncRNA AGAP2-AS1 is highly expressed in HCC, whcih accelerates the development of HCC [32]. lncRNA EPB41L4A-AS2 is down-regulated in HCC, which exerts an inhibitory effect on the progression of HCC [33]. LINC01006 is a novel identified lncRNA that involved in cancer progression. Previous studies have proved that LINC01006 is up-regulated in pancreatic cancer [34], prostate cancer [35], but is down-regulated in gastric cancer [11]. In this study, we discovered that LINC01006 expression was up-regulated in HCC, which is consistent with its expression in pancreatic and prostate cancer. In addition, we also found that high expression of LINC01006 was correlated with metastasis and advanced WHO grade, suggesting that LINC01006 may be a potential biological marker in HCC. However, LINC01006 may not be able to distinguish various cancers, because its expression is not specific in different tissues. Although this defect limits the predictive value of LINC01006 in clinical practice to some degrees, the help of clinical characteristics can greatly improve the diagnostic efficiency of HCC. Therefore, LINC01006 may be an auxiliary diagnostic factor for HCC.
Cell proliferation and metastasis are the main cellular processes involved in cancers. Zhang et al. [34] have illustrated that knockdown of LINC01006 inhibits the proliferation, invasion and migration of pancreatic cancer cells, and up-regulation of LINC01006 led to the opposite results. Ma et al. [35] have shown that silencing of LINC01006 suppresses cell proliferative, migratory, invasive capacities while accelerated apoptosis in prostate cancer. In this study, we found that LINC01006 knockdown inhibited the viability, migration and invasion of HCC cells in vitro. Our findings are consistent with previous studies and illustrated that silencing of LINC01006 can inhibit the malignant characteristics of HCC. Furthermore, we also determined that LINC01006 knockdown inhibited the tumor growth in vivo, which further indicated that LINC01006 is a potential therapeutic target for HCC. We speculated that silencing of LINC01006 may be a promising therapeutic strategy for HCC.
Accumulating evidences have suggested that lncRNAs exert their functions via acting ceRNAs [36,37]. For instance, XIST silencing impedes the tumorigenesis of HCC through serving as a sponge of miR-194-5p [38]. Overexpression of CRNDE contributes to HCC progression by sponging miR-217 [39]. Herein, we identified that LINC01006 acted as a molecular sponge of miR-433-3p, which negatively regulated miR-433-3p in HCC cells. Previous studies have indicated that miR-433-3p serves as an anti-tumor gene that is down-regulated in a variety of human malignancies, including gastric cancer [21], glioma [19], and HCC [22,40]. Xue et al. [22] have demonstrated that miR-433 is lowly expressed in HCC, and overexpression of miR-433 inhibits the proliferation of HCC cells. Yang et al. [40] have found that miR-433 dramatically restrains the migration and invasion of HCC cells. Consistent with previous studies, we found that miR-433-3p overexpression inhibited the viability, migration, and invasion of HCC cells. Our results illustrated that miR-433-3p is a tumor suppressor in HCC. Combined with the regulatory relationship between LINC01006 and miR-433-3p, we speculated that silencing of LINC01006 may inhibit the progression of HCC through up-regulating miR-433-3p. Encouragingly, our following feedback experiments confirmed this speculation.
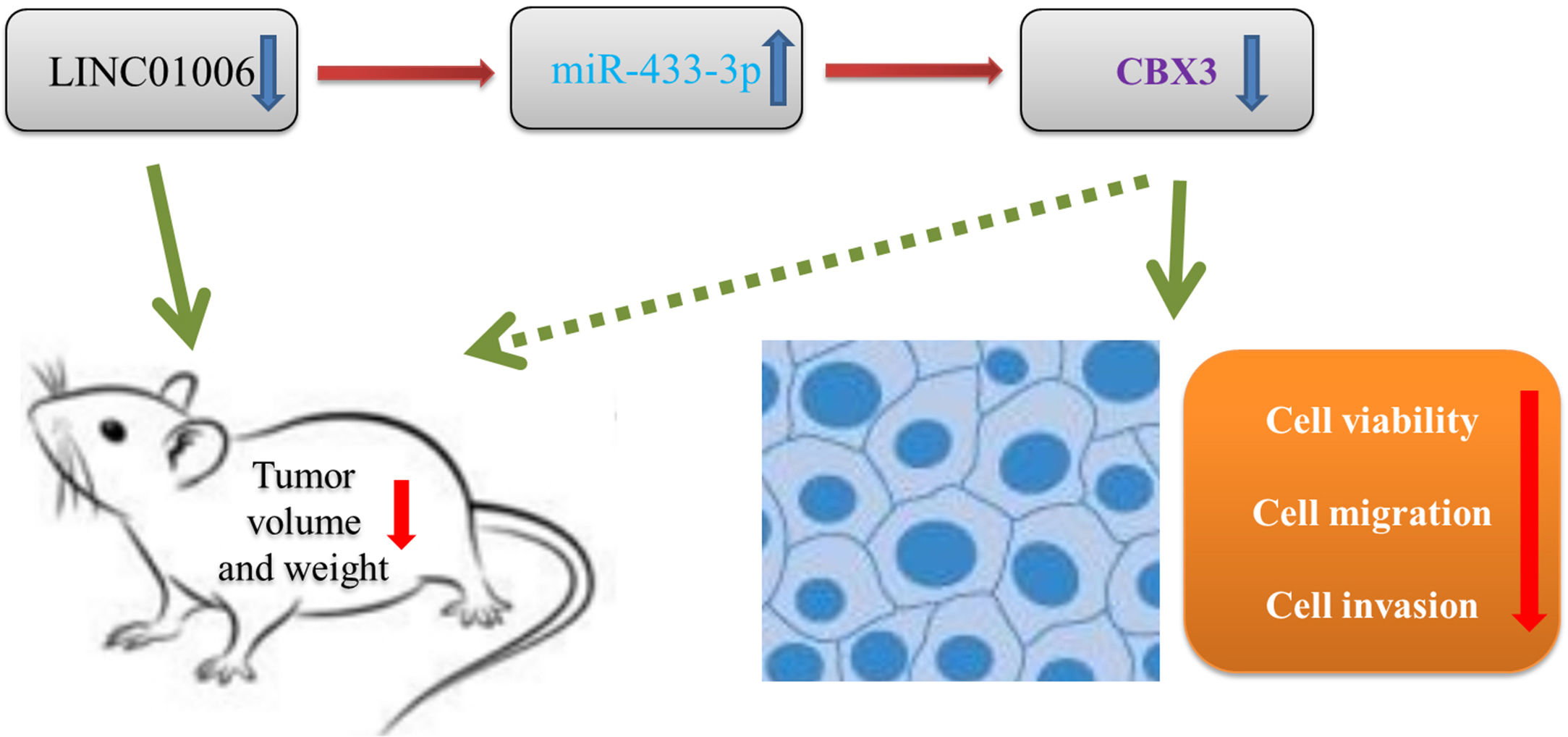
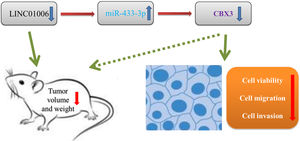
In order to further evaluate the downstream mechanisms of LINC01006, we further screened out the downstream target of miR-433-3p. We discovered that CBX3 is a target of miR-433-3p. CBX3 serves as an oncogene that is up-regulated in diverse human cancers [41–44]. For instances, CBX3 accelerates cell proliferation in colorectal cancer [45]. CBX3 promotes the tumor growth of osteosarcoma. More importantly, CBX3 is overexpressed in HCC tissues, and over-expression of CBX3 promotes the proliferation of HCC cells [30]. In line with previous studies, we also found that CBX3 expression was significantly increased in HCC tissues. Because CBX3 was negatively regulated by miR-433-3p, miR-433-3p may exert a tumor-suppressor role in HCC by regulating CBX3. Nowadays, the axis of lncRNA/miRNA/mRNA has been widely recognized in cancer. Zhang et al. [34] have found that LINC01006 promotes cell proliferation and metastasis in pancreatic cancer via regulating miR-2682-5p/HOXB8 axis. Ma et al. [35] have shown that LINC01006 promotes the progression of prostate cancer by regulating miR-34a-5p/DAAM1 axis. In this study, the feedback experiments showed that the anti-tumor effect of LINC01006 knockdown on HCC cells was partially reversed by miR-433-3p inhibition or CBX3 overexpression. Our findings illustrated that LINC01006 knockdown may inhibit the progression of HCC cells through regulating miR-433-3p/CBX3 axis (Fig. 7). The LINC01006/miR-433-3p/CBX3 axis provides new insights into the development of novel gene-target therapy of HCC.
5ConclusionsIn conclusion, LINC01006 was up-regulated in HCC tissues and cells, and high LINC01006 expression was notably related to metastasis and advanced WHO grade of HCC patients. LINC01006 silencing inhibited the viability, migration, and invasion of HCC cells in vitro, and inhibited the tumor growth in vivo. Notably, LINC01006 mediated miR-433-3p/CBX3 axis in HCC cells. Our findings provide deeper insight into the progression of HCC, and implicate that LINC01006/miR-433-3p/CBX3 may be promising therapeutic targets for HCC. AbbreviationsHCC Hepatocellular carcinoma Dual-luciferase reporter Long non-coding RNAs Long intergenic non-protein coding RNA 1006 MicroRNAs Competitive endogenous RNAs Chromobox protein homolog 3 MS2-RNA immunoprecipitation
This study was approved by the ethics committee of Yantai Mountain Hospital in accordance with the Declaration of Helsinki (No. 2020030). Written informed consents were obtained from all patients. Animal experiments were approved by the ethics committee of Yantai Mountain Hospital in accordance with the Guide for the Care and Use of Laboratory Animals (eighth edition, 2011, National Institutes of Health, USA) (No. 2020030).
Data availabilityUpon reasonable request, relevant data for this study may be obtained from the corresponding author.
FundingThis research did not receive any specific grant from funding agencies in the public, commercial, or not-for-profit sectors.
Conflict of interestThe authors have no conflicts of interest to declare.
Authors’ contributionsAll authors made substantial contributions to conception and design, acquisition of data, or analysis and interpretation of data; took part in drafting the article or revising it critically for important intellectual content; agreed to submit to the current journal; gave final approval of the version to be published; and agree to be accountable for all aspects of the work.