This study aimed to explore the functional mechanism of the miRNA-20b-5p/cytoplasmic polyadenylation element binding protein 3 (miR-20b-5p/CPEB3) axis in hepatocellular carcinoma (HCC) so as to provide a new idea for targeted therapy of HCC.
Materials and MethodsBioinformatics analysis was employed to obtain markedly differentially expressed miRNAs and mRNAs in The Cancer Genome Atlas-Liver Hepatocellular Carcinoma (TCGA-LIHC) dataset, so as to find target miRNA and its target mRNA. Real-time quantitative PCR was conducted to detect miR-20b-5p and CPEB3 mRNA expression. Western blot was performed to determine CPEB3 protein expression. Dual-luciferase reporter assay was carried out to verify the targeting relationship between miR-20b-5p and CPEB3. Cell counting kit-8 assay, wound healing assay, Transwell invasion assay and flow cytometry were conducted to evaluate the proliferation, migration, invasion and apoptosis of HCC cells.
ResultsBioinformatics analysis suggested that miR-20b-5p and CPEB3 were markedly highly and lowly expressed, respectively, in HCC tissue in TCGA-LIHC dataset. Over-expressing miR-20b-5p facilitated the proliferation, migration and invasion, and suppressed the apoptosis of HCC cells. Dual-luciferase reporter assay validated that there was a targeting relationship between miR-20b-5p and CPEB3. The inhibitory effect of CPEB3 over-expression on HCC cell proliferation, migration and invasion was reversed by over-expressing miR-20b-5p.
ConclusionsThe present study proved that miR-20b-5p promotes HCC cell proliferation, migration and invasion by inhibiting CPEB3 expression, which may provide a theoretical basis for the prognosis and treatment of HCC patients.
Hepatocellular carcinoma (HCC) is the most common malignant tumor in liver [1]. Although early HCC patients have various effective therapeutic options, such as surgical resection, liver transplantation, chemotherapy [2], treatment for patients with advanced metastatic HCC remains rather unsatisfactory [3]. Besides, the molecular mechanisms underlying HCC recurrence and metastasis have not been fully clarified. Hence, exploring HCC pathogenesis is likely to help to identify effective molecules for treatment of HCC.
MicroRNAs (miRNAs) are small non-coding RNA molecules that negatively regulate gene expression by either inducing mRNA degradation or inhibiting translation. A considerable number of studies revealed that certain miRNAs are able to affect HCC progression by altering tumor cell physiological processes [4–6]. For instance, miR-20b-5p is abnormally expressed in breast cancer [7], gastric cancer [8], cervical cancer [9], colorectal cancer [10], etc. In view of this, investigating the effect of miRNAs on human cancers like HCC may help to provide a novel theoretical basis for the diagnosis and treatment of HCC.
The cytoplasmic polyadenylation element binding protein (CPEB) family of vertebrates consists of four paralogues: CPEB1, CPEB2, CPEB3 and CPEB4 [11]. Several studies uncovered the aberrant expression of CPEB3 in multiple cancers, such as cervical cancer [12], HCC [11], high-grade ovarian serous tumor [13], etc. Additionally, miRNA was found to regulate cancer progression by directly targeting the CPEB3/epidermal growth factor receptor (EGFR) axis. For instance, studies demonstrated that miR-107 and miR-452-3p are capable of repressing HCC cell proliferation by targeting the CPEB3/EGFR axis [11,14]. Nonetheless, the effect of the miR-20b-5p/CPEB3 axis on the occurrence and progression of HCC has not been elucidated by any research up to now.
This study aimed to clarify the effect of miR-20b-5p and CPEB3 on the occurrence and progression of HCC, and to discuss the underlying interaction between these two genes. Our study may provide a novel theoretical basis for exploring targeted therapies for HCC patients.
2Materials and methods2.1Bioinformatics analysisHCC-related miRNA mature data (normal: n = 50, tumor: n = 375) and mRNA Counts data (normal: n = 50, tumor: n = 374) were downloaded from The Cancer Genome Atlas-Liver Hepatocellular Carcinoma (TCGA-LIHC) dataset (https://portal.gdc.cancer.gov/). Differential expression analysis was conducted to screen differentially expressed mRNAs (DEmRNAs) using the “edgeR” package (|logFC| > 2.0, padj < 0.01). Bioinformatics databases including miRDB (http://mirdb.org/), mirDIP (http://ophid.utoronto.ca/mirDIP/index.jsp#r), TargetScan (http://www.targetscan.org/vert_71/), and starBase (http://starbase.sysu.edu.cn/) were employed to predict target genes of miR-20b-5p. The predicted target genes were then intersected with the DEmRNAs in TCGA-LIHC dataset to confirm the ultimate target gene.
2.2Cell cultureNormal human liver cell line L-02 (GNHu 6), human HCC cell lines Huh-7 (TCHu182), SK-Hep-1 (TCHu109), HepG2 (TCHu72) and MHCC97H (SCSP-528) were all purchased from Type Culture Collection Center, Chinese Academy of Sciences and cultured in Dulbecco’s Modified Eagle’s Medium (DMEM) (GibcoBRL, Gaithersburg, Maryland, USA) supplemented with 10% fetal bovine serum (FBS). All the cells were incubated in 5% CO2 at 37 °C for 2 d.
2.3Cell transfectionmiR-20b-5p mimic (miR-mimic), mimic NC (miR-NC), pcDNA3.1-CPEB3 over-expression plasmid (oe-CPEB3) and pcDNA3.1 plasmid (oe-NC) were obtained from GenePharma (Shanghai, China). They were transiently transfected into Huh-7 cells using Lipofectamine 2000 (Invitrogen, Carlsbad, CA) and cultured in corresponding medium with 5% CO2 at 37 °C. All cells were kept in medium for at least 24 h and washed with phosphate-buffered saline (PBS, pH 7.4) before transfection. Subsequently, real-time quantitative PCR (qRT-PCR) were employed to verify the transfection efficiency.
2.4qRT-PCRTotal RNA was extracted from cells using TRIzol reagent (Life Technologies, Grand Island, NY, USA) according to the manufacturer’s instructions. The concentration of total RNA was measured using NanoDrop 2000 system (Thermo Fisher Scientific, Inc., Waltham, MA, USA). Then, miRNA and mRNA were transcribed into complementary DNA (cDNA) by using the miScript II RT Kit (Qiagen, USA) and PrimeScript RT Master Kit (TaKaRa, Dalian, P.R. China), respectively, following the manufacturer’s protocol. miRNA and mRNA expression levels were detected by miScript SYBR Green PCR Kit (Qiagen, Germany) and SYBR® Premix Ex Taq TM II (Takara Bio Inc., Shiga, Japan), respectively. qRT-PCR was performed on Applied Biosystems® 7500 Real-Time PCR Systems (Thermo Fisher Scientific, Waltham, MA). β-Actin and U6 were used as internal references for CPEB3 and miR-20b-5p, respectively. The primers were listed in Table 1.
Primer sequences for qRT-PCR.
| Gene | Sequences (5’→3’) | |
|---|---|---|
| miR-20b-5p | F: TGTCAACGATACGCTACGA | R: GCTCATAGTGCAGGTAGA |
| U6 | F: CTCGCTTCGGCAGCACA | R: AACGCTTCACGAATTTGCGT |
| CPEB3 | F: GAAAGGTAAACACTACCCTCCCA | R: CCAGGAAGGCATTGTTAAGTGC |
| β-Actin | F: TACCTCATGAAGATCCTCACC | R: TTTCGTGGATGCCACAGGAC |
The quantitative value was expressed using the 2−ΔΔCt method to compare the relative expression of miR-20b-5p and CPEB3 in the control group and the test group.
Huh-7 cells were suspended in DMEM supplemented with 10% FBS and were then seeded in 96-well plates at a density of 2 × 103 cells/well. Subsequently, at 24 h, 48 h, 72 h and 96 h, each well was added with 10 μL CCK-8 solution (CK04; Dojindo Laboratories, Kumamoto, Japan) and cells were incubated with 5% CO2 at 37 °C for additional 2 h. At last, the absorbance of each well at 490 nm was identified at designated time points, with the absorbance in the first day as control.
2.6Wound healing assayWound healing assay was performed to evaluate cell migratory ability. Huh-7 cells were inoculated into 6-well plates. After cells were grown to 80% in confluence, a scratch was made on cells using the tip of a 200 μL pipette. After the wells were washed twice to remove the floating cells, a fresh medium was added and cells were cultured for another 24 h. The cell migration distance at 0 h and 24 h was measured under a microscope. Each experiment was repeated 3 times.
2.7Transwell invasion assayCell invasion assay was conducted using 24-well Transwell chambers (8 μm pore size; BD Biosciences). Approximately 2 × 104 cells were seeded into the upper chamber coated with Matrigel, while the lower chamber contained DMEM supplemented with 10% FBS (Thermo fisher, USA) to stimulate cell invasion. After cells were cultured at 37 °C for 48 h, a cotton swab was used to remove non-invasive cells in the upper chamber, while invasive cells in the lower chamber were stained with 0.1% crystal violet. The experiment was repeated 3 times. The number of cells that successfully invaded the Matrigel was counted and used as an index for assessing cell invasive ability. Four fields were randomly chosen and photographed under a microscope.
2.8Western blotCell lysate was extracted using radio-immunoprecipitation assay (RIPA) lysis buffer (Beyotime, China) and the concentration of proteins was measured by bicinchoninic acid (BCA) protein assay kit (Beyotime, China). Protein samples were separated on sodium dodecyl sulphate-polyacrylamide gel electrophoresis (SDS-PAGE) and transferred onto polyvinylidene fluoride (PVDF) (Millipore) membranes. After being blocked with 5% non-fat milk for 2 h, the membranes were incubated with primary rabbit antibodies overnight at 4 ℃, followed by hybridization with secondary antibody goat anti-rabbit IgG H&L (Abcam, China) at room temperature for 1 h. The membranes were washed with PBS buffer containing Tween-20 (PBST) three times with 10 min for each time before and after hybridization. All protein bands were visualized using an enhanced chemiluminescence assay kit (GE Healthcare, Chicago, IL, USA). Image Lab (Bio-Rad) was performed to measure the band density.
Primary antibodies included rabbit anti-CPEB3 (Abcam, China) and rabbit anti-β-Actin (Abcam, China).
2.9Dual-luciferase reporter assayIn order to verify the targeting relationship between miR-20b-5p and CPEB3, wild-type CPEB3 (CPEB3-WT) and mutant CPEB3 (CPEB3-MUT) vectors were generated by cloning CPEB3 3’-untranslated region (3’-UTR) -WT/MUT into psiCHECK luciferase vectors (Sangon Co., LTD, Shanghai, China). Then, Huh-7 cells were seeded into 48-well plates at 37 °C for 24 h. CPEB3-WT/CPEB3-MUT vectors and mimic NC/miR-20b-5p mimic were co-transfected into Huh-7 cells. Finally, luciferase activity was assessed using the Dual-Luciferase Reporter Assay Kit (Promega, Fitchburg, WI, USA) according to the manufacturer’s instructions. Each transfection was repeated in triplicate.
2.10Flow cytometryAfter being incubated to the logarithmic phase and digested with ethylene diamine tetraacetic acid (EDTA)-free trypsin, Huh-7 cells were washed with PBS twice, mixed evenly in 500 μL pre-cooled PBS for resuspension and were then set at a density of 1 × 106 cells/mL. Subsequently, 100 μL cell suspension was added with 5 μL Annexin-V-FITC (KeyGen Biotech, Nanjing, China) for 15 min of incubation in the dark at room temperature. Thereafter, 2.5 μL propidium iodide (PI) was added into the suspension to stain cells 5 min before flow cytometry. Cell apoptosis was detected using a flow cytometer (Thermo Fisher, NY, USA).
2.11Statistical analysisAll statistical analyses were performed using GraphPad Prism 6.0 (La Jolla, CA). All data were expressed as mean ± standard deviation (SD), and differences between two groups were analyzed by Student’s t-test. P < 0.05 was considered statistically significant.
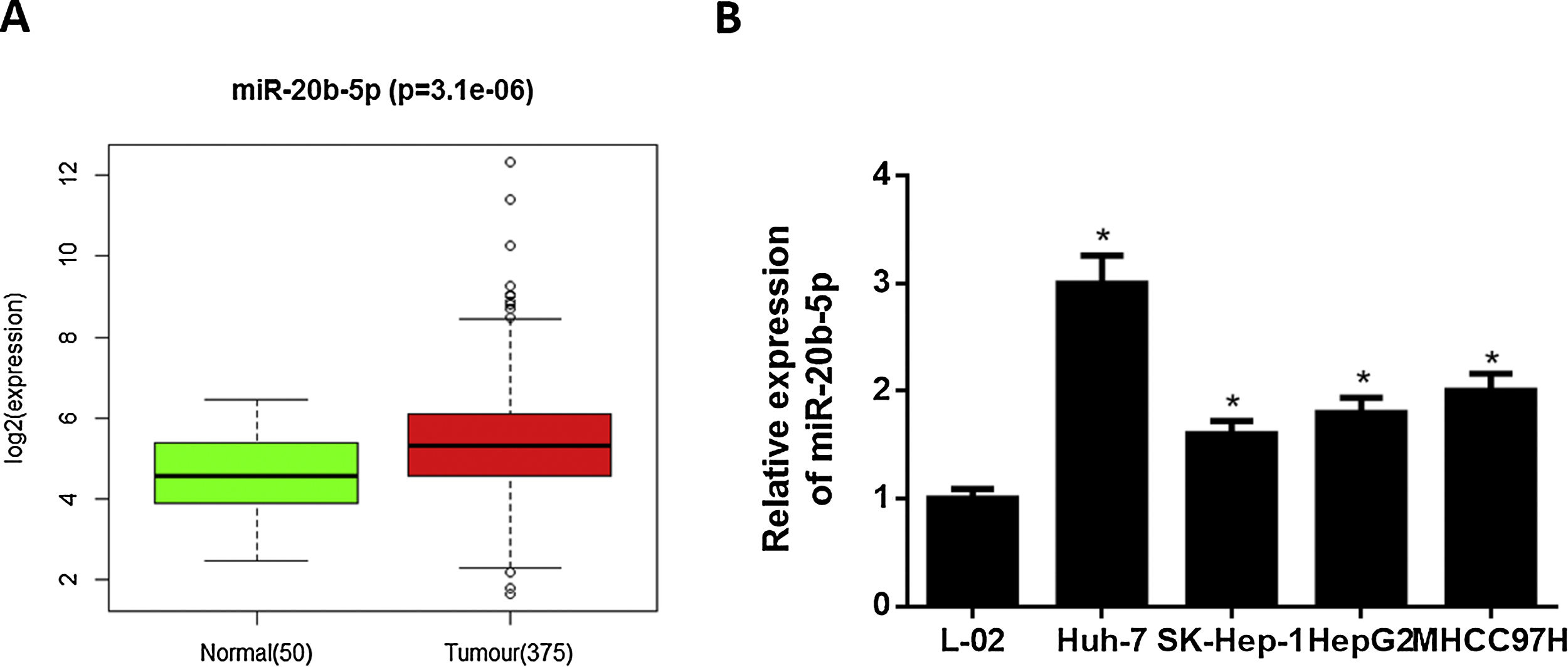
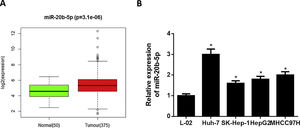
3Results3.1miR-20b-5p is up-regulated in HCC cellsNumerous studies indicated that miR-20b-5p can promote cancer cell proliferation and is expected to be a therapeutic marker for cancers. Thence, miR-20b-5p was chosen as the research object in this study. Relative expression of miR-20b-5p in the normal and tumor tissue samples in TCGA-LIHC dataset showed that miR-20b-5p was markedly up-regulated in HCC (Fig. 1A). Subsequently, we conducted qRT-PCR to determine miR-20b-5p expression in HCC cell lines (Huh-7, SK-Hep-1, HepG2 and MHCC97H) and normal human liver cell line L-02, the result of which suggested that miR-20b-5p was significantly highly expressed in HCC cell lines (Fig. 1B). Collectively, the above results unveiled that miR-20b-5p was up-regulated in HCC cells and was likely to act as an oncogene in cancer progression. In order to better investigate the functional mechanism of miR-20b-5p in HCC, Huh-7 cell line with the highest miR-20b-5p expression was chosen for follow-up experiments.
miR-20b-5p is up-regulated in HCC cells.
(A) Relative expression of miR-20b-5p in TCGA-LIHC dataset. Green and red boxes stand for normal and tumor groups, respectively; (B) qRT-PCR was carried out to determine miR-20b-5p expression in HCC cell lines (Huh-7, SK-Hep-1, HepG2 and MHCC97H) and normal human liver cell line L-02. *p < 0.05.
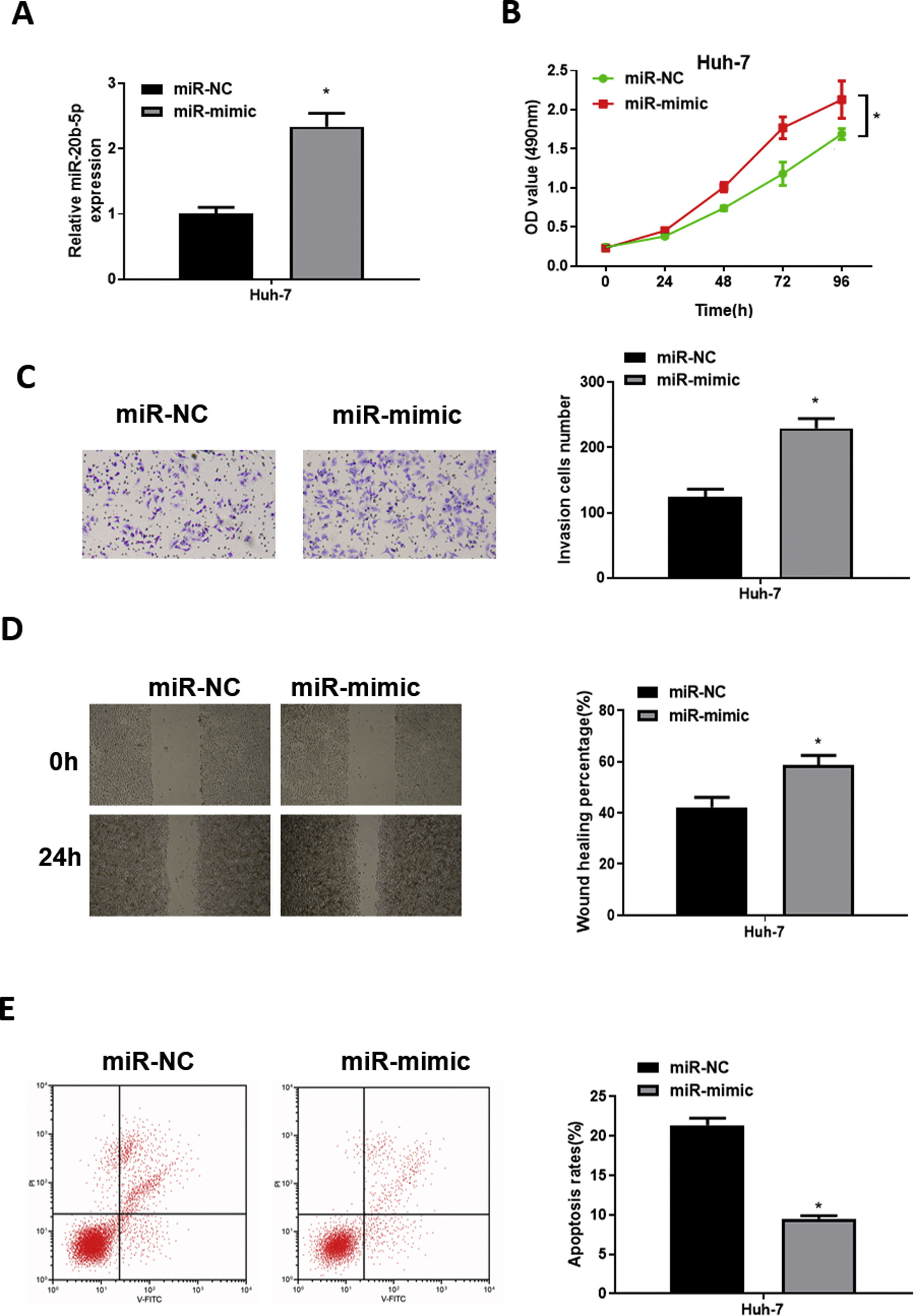
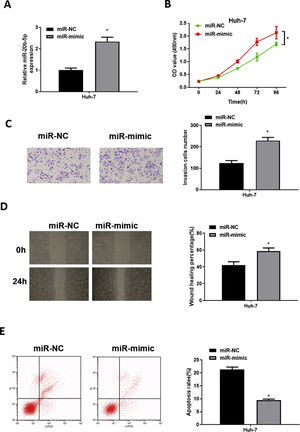
To explore the role of miR-20b-5p in HCC cells, we set up miR-NC and miR-mimic groups for examination. qRT-PCR was used to detect the transfection efficiency, uncovering that miR-20b-5p was noticeably up-regulated in the miR-mimic group (Fig. 2A). Then, CCK-8 assay, Transwell invasion assay and wound healing assay were performed to evaluate the effect of miR-20b-5p over-expression on cell activities. The results unveiled that the proliferative, invasive and migratory abilities of Huh-7 cells were prominently raised upon miR-20b-5p over-expression (Fig. 2B–D). Flow cytometry showed that Huh-7 cell apoptosis was markedly suppressed upon miR-20b-5p over-expression (Fig. 2E). Taken together, the above results demonstrated that over-expressing miR-20b-5p could facilitate HCC cell proliferation, migration and invasion, and inhibit cell apoptosis.
miR-20b-5p promotes HCC cell proliferation, migration and invasion, and inhibits cell apoptosis.
(A) qRT-PCR was performed to test miR-20b-5p expression in Huh-7 cells. The proliferative ability, invasive ability and migratory ability of Huh-7 cells were detected via (B) CCK-8 assay, (C) Transwell invasion assay (100×) and (D) wound healing assay (40×); (E) Flow cytometry was conducted to assess the apoptotic rate of Huh-7 cells. *p < 0.05.
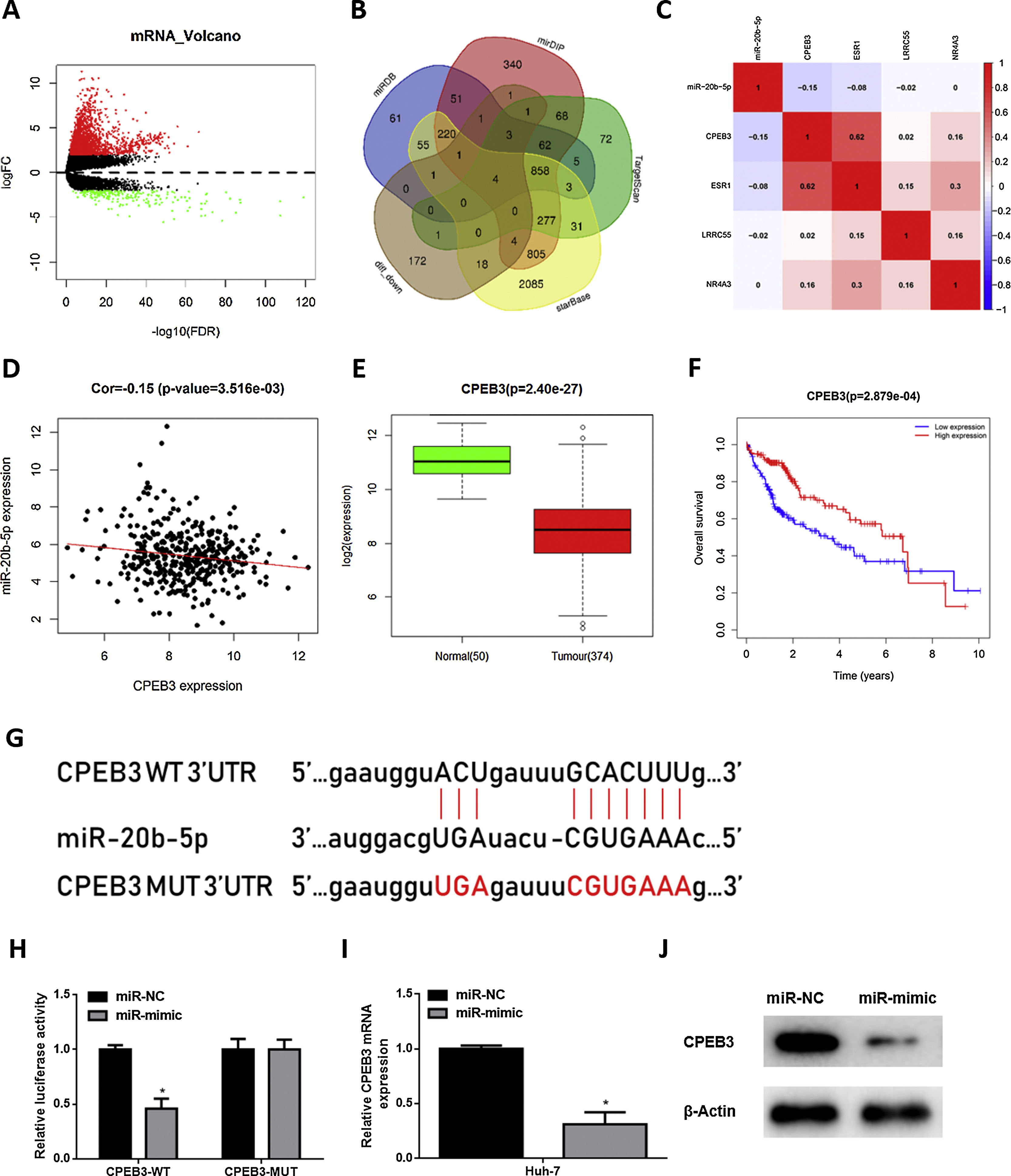
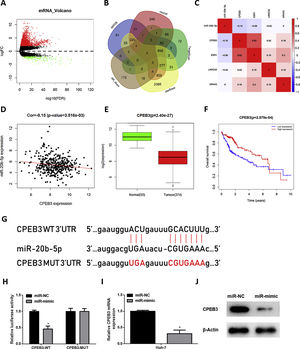
In order to research the regulatory mechanism of miR-20b-5p in HCC cells, we further analyzed downstream target genes of miR-20b-5p. Firstly, we integrated GTEx data and FPKM data of mRNA in TCGA-LIHC dataset, and performed differential gene expression analysis. A total of 1981 DEmRNAs were obtained, including 1774 up-regulated DEmRNAs and 207 down-regulated DEmRNAs (Fig. 3A). Meanwhile, starBase, TargetScan, mirDIP and miRDB databases were employed to predict target genes of miR-20b-5p, which were then intersected with the 207 down-regulated DEmRNAs, and finally 4 mRNAs were overlapped (Fig. 3B). Pearson correlation analysis indicated that among the 4 genes (CPEB3, ESR1, LRRC55 and NR4A3), miR-20b-5p had the highest negative correlation with CPEB3. Hence, CPEB3 was identified as the ultimate target gene (Fig. 3C,D). Relative expression of CPEB3 in TCGA-LIHC dataset suggested that CPEB3 was markedly down-regulated in HCC tissue (Fig. 3E), and low CPEB3 expression was associated with poor prognosis of HCC sufferers (Fig. 3F). Next, to verify the regulatory effect of miR-20b-5p on CPEB3, we firstly predicted the binding site sequence of miR-20b-5p on CPEB3 3’-UTR by a bioinformatics database, finding that miR-20b-5p had binding sites on CPEB3 3’-UTR (Fig. 3G). Then, dual-luciferase reporter assay was carried out to validate the targeting relationship between miR-20b-5p and CPEB3. The result unveiled that miR-20b-5p over-expression repressed the luciferase activity of CPEB3-WT but showed no effect on that of CPEB3-MUT (Fig. 3H). Thereafter, qRT-PCR was performed to determine CPEB3 mRNA expression in Huh-7 cells in different treatment groups, suggesting that CPEB3 mRNA expression was significantly knocked down in the miR-mimic group (Fig. 3I). Similarly, western blot also indicated that CPEB3 protein expression was remarkably down-regulated upon miR-20b-5p over-expression (Fig. 3J). Collectively, the above results illustrated that miR-20b-5p could target and inhibit the expression of CPEB3 in HCC cells.
miR-20b-5p targets and silences CPEB3 in HCC cells.
(A) Volcano plot of DEmRNAs in TCGA-LIHC dataset; (B) Venn diagram of down-regulated DEmRNAs in TCGA-LIHC dataset and predicted target mRNAs of miR-20b-5p; (C) Pearson correlation analysis of miR-20b-5p and 4 predicted target genes; (D) Pearson correlation analysis of miR-20b-5p and CPEB3 in tissue samples; (E) Relative expression of CPEB3 in TCGA-LIHC dataset; (F) Survival analysis of CPEB3 in TCGA-LIHC dataset. Abscissa and ordinate stand for time (years) and overall survival, respectively. Red and blue lines represent high- and low-expression groups, respectively; (G) The binding sites between miR-20b-5p and CPEB3 3’UTR predicted by a bioinformatics database; (H) The luciferase activity of CPEB3-WT and CPEB3-MUT detected via dual-luciferase reporter assay; (I) CPEB3 mRNA expression in Huh-7 cells (miR-NC and miR-mimic) determined through qRT-PCR; (J) CPEB3 protein expression in Huh-7 cells (miR-NC and miR-mimic) detected by western blot. *p < 0.05
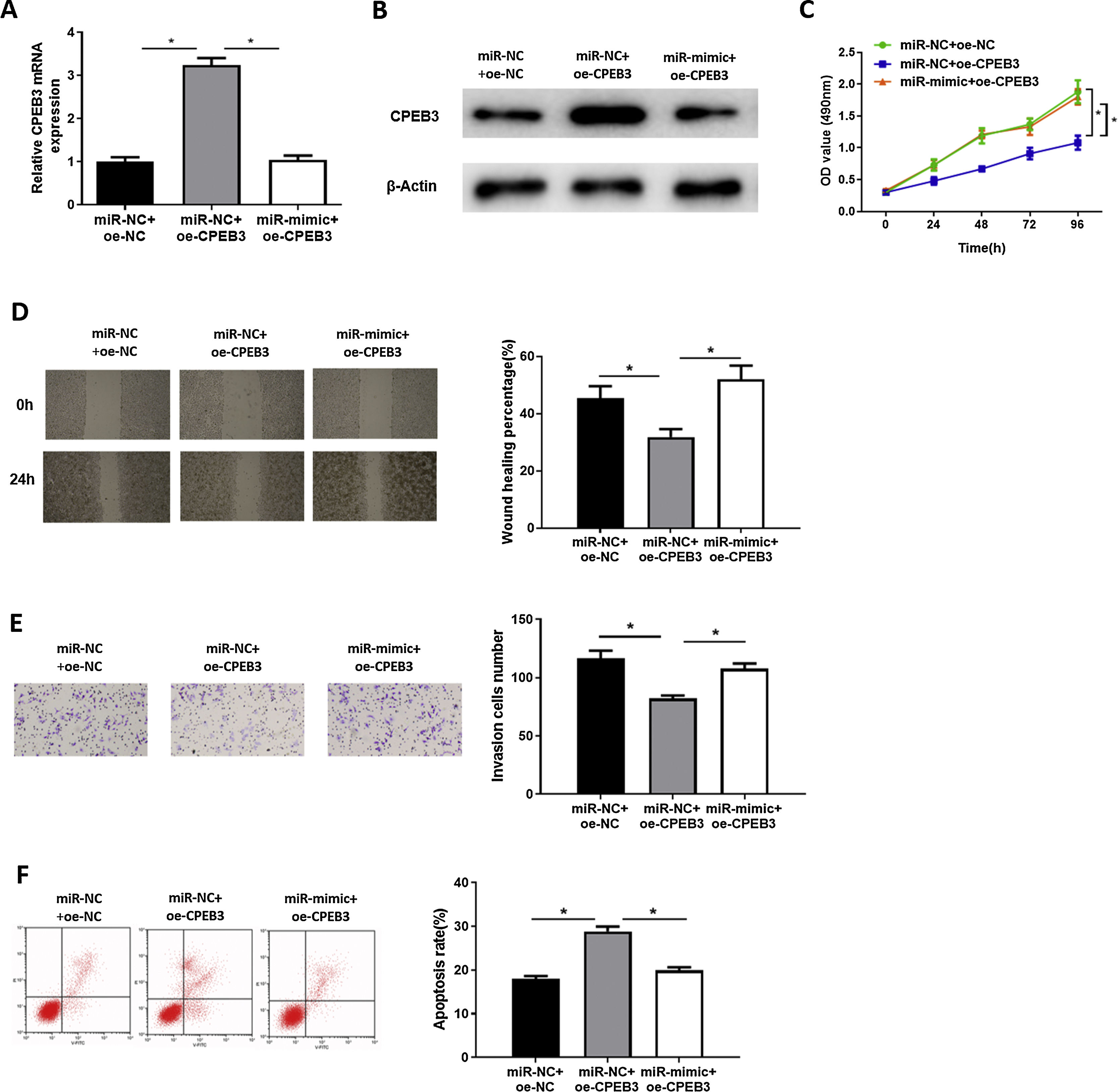
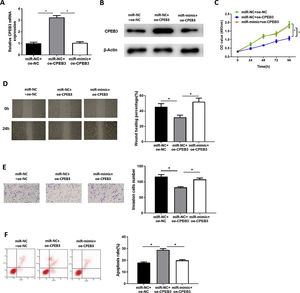
In order to explore whether miR-20b-5p regulates HCC cells through targeting CPEB3, we set miR-NC + oe-NC, miR-NC + oe-CPEB3 and miR-mimic + oe-CPEB3 groups. The results of qRT-PCR and western blot suggested that CPEB3 expression was prominently increased in the miR-NC + oe-CPEB3 group but was reduced in the miR-mimic + oe-CPEB3 group near to that in the miR-NC + oe-NC group (Fig. 4A,B), which further proved that CPEB3 expression could be down-regulated by miR-20b-5p. CCK-8 indicated that the proliferative ability of HCC cells was weakened upon CPEB3 over-expression, while such inhibitory effect was attenuated by further over-expressing miR-20b-5p (Fig. 4C). Thereafter, wound healing assay and Transwell invasion assay revealed that HCC cell migratory and invasive abilities were markedly decreased upon CPEB3 over-expression, whereas such inhibitory effect was significantly reduced in the miR-mimic + oe-CPEB3 group (Fig. 4D,E). Additionally, the result of flow cytometry demonstrated that Huh-7 cell apoptosis was noticeably fostered by transfecting CPEB3, while such promoting effect was reversed upon miR-20b-5p over-expression (Fig. 4F). Collectively, the present study confirmed that miR-20b-5p was able to facilitate HCC cell proliferation, migration and invasion, and suppress cell apoptosis by targeting CPEB3.
miR-20b-5p promotes HCC cell proliferation, migration and invasion, and inhibits cell apoptosis by targeting CPEB3.
(A-B) CPEB3 mRNA and protein expression in the miR-NC + oe-NC, miR-NC + oe-CPEB3 and miR-mimic + oe-CPEB3 groups. The proliferative ability, migratory ability and invasive ability of Huh-7 cells were detected via (C) CCK-8 assay, (D) wound healing assay (40×) and (E) Transwell invasion assay (100×); (F) Flow cytometry was conducted to evaluate the apoptotic rate of Huh-7 cells. *p < 0.05.
The annual death toll resulting from liver cancer is as high as 200,000 globally, and there are approximately 500,000 newly developed cases [15], among which HCC makes up over 90% [16]. The major challenges facing the treatment of liver cancer include intrahepatic recurrence and metastasis, which lead to the poor prognosis of cancer sufferers [17]. Hence, finding novel therapeutic methods for HCC has become an urgent issue. Previous studies indicated that miRNAs play significant roles in the occurrence and progression of HCC. For instance, miR-17 [18], miR-92b [19], miR-93-5p [20], miR-616 [21], etc. are remarkably up-regulated in HCC cells and can serve as biomarkers and potential therapeutic targets. In view of this, we explored therapeutic method on the molecular level, and endeavored to find underlying biomarkers and therapeutic targets for HCC.
In recent years, accumulating research uncovered that miRNA plays a crucial role in multiple cancers. For example, miRNAs are related to cell differentiation, proliferation, apoptosis and migration of colorectal cancer [22]. Nevertheless, the biological function and molecular mechanism of miRNAs in HCC have not been fully researched currently. Previous studies unveiled that miR-20b-5p is highly expressed in multiple cancers and is likely to act as an oncogene to promote cancer cell progression. For example, miR-20b-5p fosters the proliferation and migration of non-small cell lung cancer cells by targeting BTG3 [23]. miR-20b-5p is up-regulated in breast cancer, nasopharyngeal carcinoma, etc., and can be used as a biomarker for the diagnosis of breast cancer [24,25]. In the present study, we found that miR-20b-5p was noticeably up-regulated in HCC cell lines. It is reported that miR-20b-5p acts as an oncogene in various cancers. Similarly, this study also proved that over-expressing miR-20b-5p could drive HCC progression. Taken together, by conducting bioinformatics analysis and a series of in vitro experiments, this research confirmed that miR-20b-5p was prominently up-regulated and played a carcinogenic role in HCC. In other words, the present study clarified that miR-20b-5p functioned as a cancer-promoting factor in HCC.
Next, in order to further explore the molecular mechanism of miR-20b-5p influencing HCC, we excavated its target genes by bioinformatics methods. Results showed that CPEB3 was abnormally lowly expressed in HCC, and there was a binding site between CPEB3 3’UTR and miR-20b-5p. Current studies revealed that CPEB3serves as a target of various miRNAs in cancers. For instance, miR-301b-3p accelerates the invasion and migration of high-grade ovarian serous tumor by targeting the CPEB3/EGFR axis [13]. CPEB1 and CPEB3 are able to serve as biomarkers for the carcinogenesis of cervical and ovarian tissues [12]. miR-496 inhibits glioma progression via regulating CPEB3 [26]. In the present research, we speculated that CPEB3 might be a target gene of miR-20b-5p in HCC cells via bioinformatics analysis. Subsequently, we verified the targeting relationship between miR-20b-5p and CPEB3 by Pearson correlation analysis, binding sites prediction and dual-luciferase reporter assay. Additionally, the results of qRT-PCR and western blot also revealed that miR-20b-5p could modulate CPEB3 mRNA and protein expression in Huh-7 cell line. Cell functional analysis showed that over-expressing CPEB3 markedly repressed HCC cell proliferation, migration and invasion, and induced cell apoptosis, while such effect was attenuated after miR-20b-5p was overexpressed. Collectively, these results deepened our understanding of the downstream regulatory mechanism of miR-20b-5p in HCC.
In conclusion, this study demonstrated that miR-20b-5p was up-regulated in HCC and could promote HCC cell proliferation, migration and invasion. CPEB3 was a target gene of miR-20b-5p and was down-regulated in HCC cells. miR-20b-5p could foster HCC progression by silencing CPEB3. The present research unveiled the functional mechanism of the miR-20b-5p/CPEB3 axis in HCC cell progression, and explored the function of miR-20b-5p in HCC, which will provide a new idea for finding novel therapeutic approaches to HCC.AbbreviationsHCC hepatocellular carcinoma MicroRNAs the 3’-untranslational region cytoplasmic polyadenylation element binding protein RNA recognition motifs epidermal growth factor receptor real-time quantitative PCR Cell Counting Kit-8 radio-immunoprecipitation assay sulphate-polyacrylamide gel electrophoresis wild type mutant ethylene diamine tetraacetic acid standard deviation
This study was supported by the funds from Projects of Lishui Key Research and Development Plan in Zhejiang Province, Grant Number: 2017ZDYF12.
Conflict of interestThe authors declare no conflicts of interest.