Occult hepatitis B virus (HBV) infection (OBI) is characterized by presence of HBVDNA in blood or liver tissue without detectable HBV surface antigen (HBsAg), with or without antibodies to hepatitis B core antigen (anti-HBc) or antibodies against HBsAg (anti-HBs). A molecular and serological characterization was done of OBI in blood donors from Yucatan, Mexico. HBV DNA was found in 24 (6.4%) of the 372 evaluated samples. Anti-HBs was present in 15/24 samples (62.5%), and no significant difference was observed between HBV DNA positivity and anti-HBs levels. HBV genotype H was detected in 66.7% of samples, followed by genotypes D (20.8%) and F (8.3%). Amino acid substitutions were identified in the core region of nine samples, and most of these changes were located in immunodominant epitopes. No precore stop codon 28 mutant (W28Stop) was identified among the analyzed HBV isolates. In conclusion, genotype H is the main circulating HBV strain among OBI blood donors from Yucatan, Mexico. Mutations in the core region may contribute to viral persistence.
Serological evidence of acute or chronic hepatitis B is commonly generated by hepatitis B surface antigen (HBsAg) assays. This sensitive serological test for HBsAg detection is widely applied but transmission of hepatitis B virus (HBV) infections by transfusion continues to occur.1,2 Occult HBV infection (OBI) is characterized by presence of HBV DNA in blood or liver tissue with no detectable HBsAg, and with or without antibodies to hepatitis B core antigen (anti-HBc) or antibodies against HBsAg (anti-HBs).3,4 Clinical observations suggest that OBI carriers may be a HBV transmission source via blood transfusion or orthotopic liver transplantation. It is also considered a possible risk factor for developing hepatocarcinoma.4 Carrier rates for HBsAg vary geographically and Mexico, like other Latin American countries, is considered a low endemicity area < 2% of population are carriers).5 However, high anti-HBc prevalence (91.1%) has been reported in populations on Mexico’s southern border.6 Very little data has been collected on OBI prevalence among blood donors in Mexico.7 The present study objective was to molecularly and serolo-gically characterize OBI in a population of blood donors from Mexico.
MethodsSample selectionHBsAg-negative plasma samples from 20,328 blood donors were collected between January 2007 and July 2008 at the Central Blood Bank of the Ignacio Garcia Tellez National Medical Center, in Merida, Yucatan, Mexico, and screened for anti-HBc using a microparticle enzyme immunoassay (MEIA) AxSYM Core (Abbott Laboratories, Abbott Park, IL, USA), in addition to routine viral screening. Three hundred and seventy-two consecutive plasma samples found to be HBsAg-negative but anti-HBc-positive in at least two independent tests were selected for further analysis. These were stored at -20 °C until analysis. Donors provided written informed consent at the time of blood donation and this study was approved by the Institution Ethics Committee.
Serological and Biochemical assaysAll anti-HBc-positive plasmas were further tested for anti-HBs by MEIA AxSYM AUSAB (Abbott Laboratories, Abbott Park, IL, USA). DNA-positive samples were also tested for hepatitis B e antigen (HBeAg) and antibodies to HBeAg (anti-HBe) using commercial kits (ARCHITECT, Abbott Laboratories, Wiesbaden, Germany). Alanine aminotransferase (ALT) levels were identified with an automated Dimension RXL (Dade Behring Inc., Newark, USA) following manufacturer instructions.
HBV DNA detectionDNA was extracted from plasma samples using a commercial kit (QIAamp DNA Mini Kit, QIAGEN GmbH, Hilden, Germany) following manufacturer instructions. HBV DNA identification was done for all 372 plasma samples by PCR amplification of a 560-bp fragment of the C gene, followed by nested amplification of a 438-bp fragment. The outer primers were: sense (5‘-TTCAAGCCTCCAAGCTGT-GCCTTGG-3’, nt 1863 to 1887) and antisense (5‘-TCTGCGACGCGGCGATTGAGA-3’, nt 2402 to 2422). The inner primers were: sense (5’-CCTTGGG-TGGCTTTGGGGCA-3’, nt 1882 to 1901) and antisense (5’-AGGATAGGGGCATTTGGTGGTCTATA-3’, nt 2294 to 2319).8 First-round amplification was done in a volume of 50 μL, containing New England BioLabs 1 × PCR buffer, 0.5 μM of each primer, 0.2 mM dNTP mix; 2.5 U Taq polymerase (New England BioLabs) and 10 μL DNA. The reaction was run in a MyCycler Thermal Cycler (Bio-Rad) using the following specifications: initial denaturation at 94 °C for 5 min; followed by 40 cycles at 94 °C for 1 min; 63 °C for 1 min; and 72 °C for 2 min; and final extension at 72 °C for 10 min. The second PCR round was done under the same conditions except that annealing was done at 55 °C with 10 μL of the first-round product. PCR-amplified products (10 μL) were separated by electrophoresis on 2% agarose gel, stained with ethidium bromide and viewed under ultraviolet light. Recombinant HBV DNA samples were used as positive controls for PCR amplification and for establishing the lower detection limit of the assay as described previously.7 Detection limit for the nested PCR assay was approximately 30 copies per mL. Plasma samples from blood donors negative for all HBV serological markers and reagents without DNA were used as negative controls. Negative and positive controls were added to each amplification round. To prevent cross-contamination, DNA extraction and PCR reaction mixture preparation were done in rooms separate from that in which the amplified samples were handled. Furthermore, all instruments used in the procedures were ultraviolet-irradiated before use. A sample was considered positive when repeatedly found positive after amplification of newly extracted material.
DNA sequencingAfter 2% agarose gel electrophoresis, the second-round PCR products were purified using a commercial kit (QIAquick Gel Extraction Kit, QIAGEN GmbH, Hilden, Germany). Direct sequencing of the purified product was done with the dideoxy chain termination method using the same sense and antisense inner primers used in the PCR, with a BigDye Terminator Cycle Sequencing Kit and a 3730xl DNA Analyzer (Applied Biosystems, Foster City, CA, USA).
Sequence analysisNucleotide and amino acid sequences were edited, aligned and analyzed with the EditSeq, SeqMan II and MegAlign programs in the DNASTAR package. To determine the HBV genotypes, phylogenetic analysis was done by first aligning occult HBV infection sequences with 42 GenBank reference sequences corresponding to HBV genotypes A to H (V00866, X51970, AF121251, D00330, AB014362, AF411411, AF324131, AJ131956, AM422939, AY230114, AY233291, AY254515, DQ315776, EU155891, EU185779, U95551, X75657, X75664, AB064316, AB116552, AB116654, AF223964, AY090455, AY090458, AY090461, DQ899150, EU620070, EU670261, X69798, AB064312, AB375165, AF405706, AB059660, AB059661, AB179747, AB266536, AB375161, AB375163, AB375164, AY090457, EU498228, FJ356716). A phylogenetic tree was then generated using the neighbor-joining algorithm (1000 bootstrap replicates) based on Kimura two-parameter distance estimation (PHYLIP program package version 3.6). Woolly monkey hepatitis B virus (GenBank accession number AF046996) was utilized as an out group.
Statistical analysisStatistical differences were evaluated by applying a Chi-square test and a Fisher’s exact test. P values less than 0.05 were considered significant. All analyses were done using the SPSS software package (Version 14, SPSS Inc. Chicago, Illinois, USA).
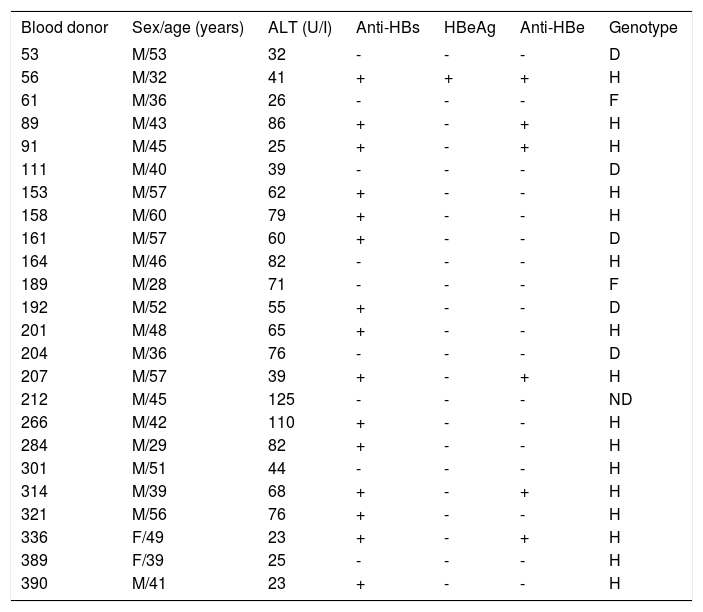
ResultsSerological characterizationOf the 372 blood donors positive for anti-HBc and negative for HBsAg, 202 (54.3%) were also anti-HBs-positive. Twenty-four (6.4%) of the 372 evaluated anti-HBc positive samples were also DNA-positive and thus categorized as OBI carriers. All the HBV DNA-positive samples were identified as such only after a second PCR amplification round. No HBV DNA was detected in any of the negative controls. In the positive controls, a visible 560-bp DNA band was detected after the first amplification round and a 438-bp DNA band after the second round. All donors were clinically asymptomatic and none of the viremic donors had previously received HBV immunization. Additional serological markers such as anti-HBe and HBeAg were evaluated using HBV DNA-positive samples. No significant difference (P = 0.53) was found between HBV DNA positivity and anti-HBs levels. OBI-positive donor serological, biochemical and molecular data are summarized in Table 1.
Serological, biochemical and molecular data of 24 donors with OBI.
| Blood donor | Sex/age (years) | ALT (U/I) | Anti-HBs | HBeAg | Anti-HBe | Genotype |
|---|---|---|---|---|---|---|
| 53 | M/53 | 32 | - | - | - | D |
| 56 | M/32 | 41 | + | + | + | H |
| 61 | M/36 | 26 | - | - | - | F |
| 89 | M/43 | 86 | + | - | + | H |
| 91 | M/45 | 25 | + | - | + | H |
| 111 | M/40 | 39 | - | - | - | D |
| 153 | M/57 | 62 | + | - | - | H |
| 158 | M/60 | 79 | + | - | - | H |
| 161 | M/57 | 60 | + | - | - | D |
| 164 | M/46 | 82 | - | - | - | H |
| 189 | M/28 | 71 | - | - | - | F |
| 192 | M/52 | 55 | + | - | - | D |
| 201 | M/48 | 65 | + | - | - | H |
| 204 | M/36 | 76 | - | - | - | D |
| 207 | M/57 | 39 | + | - | + | H |
| 212 | M/45 | 125 | - | - | - | ND |
| 266 | M/42 | 110 | + | - | - | H |
| 284 | M/29 | 82 | + | - | - | H |
| 301 | M/51 | 44 | - | - | - | H |
| 314 | M/39 | 68 | + | - | + | H |
| 321 | M/56 | 76 | + | - | - | H |
| 336 | F/49 | 23 | + | - | + | H |
| 389 | F/39 | 25 | - | - | - | H |
| 390 | M/41 | 23 | + | - | - | H |
ND: Not determined.
Phylogenetic analysis comparing donor sequences with those of various HBV genotype strains revealed that sixteen (66.7%) isolates belonged to genotype H, 5 (20.8%) to genotype D and 2 (8.3%) to genotype F (Figure 1). Genotype could not be determined in one (4.2%) donor, and therefore the results include only 23 donors.
Phylogenetic tree of 23 HBV DNA sequences from isolates from the study population. Genetic distanceswere estimated using the Kimura two-parameter matrix and the phylogenetic tree built using the neighbor-joining method. Bootstrap values produced with 1,000 replicates appear at main branch nodes. Reference strains for each hepatitis B virus genotype are included, and identified by their Gen-Bank accession numbers. Woolly monkey hepatitis B virus (GenBank accession number AF046996) was utilized as an out group.
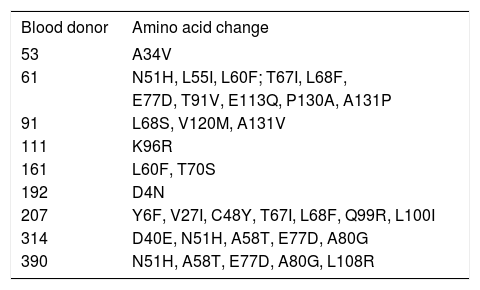
In an effort to identify mutations in the precore and core regions, an analysis was done of the 23 sequences with an identified genotype by comparing each genomic fragment sequence with a consensus sequence constructed from representative strains of each isolate’s respective genotype. The GenBank accession numbers for the H, D and F genotypes used to build the consensus sequence were the same as those used in the phylogenetic analysis (Figure 1). Amino acid changes were detected in HBV isolates from nine donors (Table 2,Figures 2 and 3), while the remaining fourteen HBV isolates had wild type amino acid sequences. Of the nine samples with amino acid mutations in the core protein, six (91, 161, 192, 207, 314, and 390) were anti-HBs positive and three (53, 61 and 111) were anti-HBs negative. Four samples contained anti-HBe and all were HBeAg negative (Table 1). No statistically significant association was found between the presence of core protein mutations and anti-HBe (P = 0.363) or anti-HBs (P = 1.00) positivity. The precore stop codon 28 mutant (W28Stop) can interfere with secreted HBeAg production, but none was observed among the 23 analyzed HBV isolates (Figures 2 and 3). Neither insertions nor deletions were recorded within the C gene.
Amino acid substitutions in the C region.
| Blood donor | Amino acid change |
|---|---|
| 53 | A34V |
| 61 | N51H, L55I, L60F; T67I, L68F, |
| E77D, T91V, E113Q, P130A, A131P | |
| 91 | L68S, V120M, A131V |
| 111 | K96R |
| 161 | L60F, T70S |
| 192 | D4N |
| 207 | Y6F, V27I, C48Y, T67I, L68F, Q99R, L100I |
| 314 | D40E, N51H, A58T, E77D, A80G |
| 390 | N51H, A58T, E77D, A80G, L108R |
Deduced amino acid sequences encoded by a portion of the hepatitis B virus (HBV) C gene from blood donors with occult hepatitis B infection. Each donor’s sequence was aligned with reference sequences of HBV genotype H (consensus sequence). Numbers indicate amino acid position within the protein and each dot corresponds to a non-variable amino acid.
Deduced amino acid sequences encoded by a portion of the hepatitis B virus (HBV) C gene from blood donors with occult hepatitis B infection. Each donor’s sequence was aligned with reference sequences for the same genotype. Consensus sequences: A) HBV genotype D; and B) HBV genotype F. Numbers indicate amino acid position within the protein and each dot corresponds to a nonvariable amino acid.
Blood donor screening for HBV infection in Mexico involves only HBsAg. However, this marker may not be detected for a number of reasons, such as HBV infection during the window period, OBI and presence of HBsAg escape mutants.9 Studies in other countries have shown that HBsAg seronegative donors can transmit HBV to these units’ receptors.1,10,11 In Mexico, very little research has been done on OBI occurrence,7,12,13 and no data is available on post-transfusion HBV incidence.
Previous reports have shown that OBI is characterized by a very low viral load.3,14 In the present study, all HBV DNA-positive samples were negative in the first PCR round but positive in the second (nested) PCR round, suggesting low viral DNA levels in these blood donors. Blood HBV DNA copy number in these donors was not determined.
Prevalence of OBI individuals (with or without detectable anti-HBs) varies depending on the studied population. For example, the 7.4% observed in the present study is notably higher than reported for Venezuela in HBsAg-negative donors with HBV DNA who are also anti-HBc and anti-HBs positive (1.42, 4.8 and 4.3%),15-17 and higher than reported for Germany (1.59%).18 The 62.5% of donors with OBI as well as anti-HBs in the present study is comparable to the 42.86% reported in Greece19 and the 63.63% reported for Venezuela.17 No association was identified here between HBV DNA positivity and anti-HBs levels, which coincides with other reports.17 This is a vital aspect since transmission via blood containing anti-HBc and anti-HBs has been reported recently.2 The present finding of high HBV DNA prevalence among HBsAg-negative blood donors from Mexico is particularly significant because Mexico is considered a low HBV prevalence region. Roman, et al.13 have suggested that the high anti-HBc prevalence versus the low HBsAg prevalence observed in Mexico may be an artifact of the limited sensitivity immunoassays used for HBsAg detection in the country and/or the presence of as yet unidentified immunogenetic characteristics within the Mexican population.
Very little research has been done on HBV genotype distribution in Mexico and most reports have been done on HBsAg-positive individuals.20-23 The HBV genotype distribution pattern observed here is similar to that reported by Sánchez et al.21 in a group of patients with chronic and acute hepatitis; the exception is genotype A, which was not observed in the present study. As reported previously for HBV in Mexico,13,21-23 the H genotype was found to be dominant in the present study. To our knowledge, no previous reports exist of HBV genotypes in OBI blood donors from Mexico.
The hepatitis B core protein (HBc) is highly immunogenetic whereas HBV envelope proteins have comparatively low immunogeneticity.24 This means that HBc is an important target for immune-mediated viral clearance via inducement of B cell, T helper cell and cytotoxic T limphocyte (CTL) responses.25 Important B cell epitopes of the HBc protein are located near amino acid sequences 74-89 and 126-135,25,26 while important HBc protein immune recognition sites for T helper epitopes are at sequences 1-20, 28-47, 50-69, 72-105 and 108-165, and those for CTL epitopes are at 18-27, 88-96 and 141-151.25-27 In the present results, mutations in immunodominant epitopes were identified in 9/23 (39.1%) of the analyzed OBI donors.
Changes in amino acids were observed in positions 40, 51, 58, 60, 77, 80 and 91, which coincides with reports for OBI donors in Venezuela.17 Amino acid substitution also occurred at position 27 in one of the studied OBI donors. In a study of patients administered interferon therapy, Naoumov, et al.28 reported that HBeAg-positive patients did not respond initially or subsequently developed amino acid changes in the CTL epitope (residues 21 and/or 27), whereas these mutations did not occur in patients who responded. Mutant HBV strains resistant to antiviral therapy have also been reported in chronic asymptomatic HBV carriers who have not received treatment.29 Substitution of amino acids at position 27 may influence immune modulation, which may in turn contribute to development of viral persistence.30
Amino acid substitution was also observed here in positions 77 and 80, which is known to reduce HBe and HBc antigenicity.31 Substitution of glutamine for arginine at position 99 within the CTL epitope was identified in the present study in a donor negative for HBeAg but positive for anti-HBe, a phenomenon reported elsewhere in a chronic liver disease patient who was also HBeAg-negative and anti-HBe-positive.32 In addition, six OBI donors exhibited amino acid changes in the hepatitis B core antigen hot-spot mutational domain (residues 80120) reported to be associated with severe forms of liver disease.32,33 Amino acid changes in positions 27, 40, 48, 60, 67, 77, 80, 100, 108, 113 and 130 observed here have also been reported in hepatocarcinoma, exacerbated chronic hepatitis B and liver fibrosis patients.33-36
Mutations in the HBV core are also reported in HBeAg-negative and anti-HBe positive chronic liver patients with wild type precore sequences, suggesting that absence of HBeAg is never solely due to a high proportion of precore mutants.30,32 Core region mutations have frequently been identified in patients with chronic HBV infections and these mutations are even more common in HBeAg-negative patients,30 which coincides with the present results. Amino acid changes in HBc protein immune recognition sites are probably influenced by the host immune system and core mutation frequency may be associated with hepatitis severity.26,32,33
ConclusionThe H genotype is the main circulating HBV strain in blood donors with OBI from Yucatan, Mexico. Mutations in the core region may contribute to viral persistence.
Abbreviations- •
HBV: Hepatitis B virus
- •
HBsAg: Hepatitis B surface antigen
- •
OBI: Occult hepatitis B virus infection
- •
anti-HBc: Antibodies to hepatitis B core antigen
- •
anti-HBs: Antibodies against HBsAg
- •
MEIA: Microparticle enzyme immunoassay
- •
HBeAg: Hepatitis B e antigen
- •
anti-HBe: Antibodies to HBeAg
- •
ALT: Alanine aminotransferase
- •
PCR: Polymerase chain reaction
- •
HBc: Hepatitis B core protein
- •
CTL: Cytotoxic T lymphocyte
The authors thank the blood bank staff of the Ignacio Garcia Tellez National Medical Center, Merida, Yucatan, Mexico for their assistance. This research was financed by the FOMIX CONACYT-Gobierno del Estado de Yucatán (grant No. YUC-2006-C05-66115).