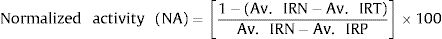Direct antiviral agents (DAAs) are very efficient in inhibiting hepatitis C virus and might be used to treat infections caused by other flaviviruses whose worldwide detection has recently increased. The aim of this study was to verify the efficacy of DAAs in inhibiting yellow fever virus (YFV) by using drug repositioning (a methodology applied in the pharmaceutical industry to identify new uses for approved drugs).
Materials and methodsThree DAAs were evaluated: daclatasvir, sofosbuvir and ledipasvir or their combinations. For in vitro assays, the drugs were diluted in 100% dimethyl sulfoxide. Vaccine strain 17D and a 17D strain expressing the reporter fluorescent protein were used in the assays. A fast and reliable cell-based screening assay using Vero cells or Huh-7 cells (a hepatocyte-derived carcinoma ell line) was carried out. Two patients who acquired yellow fever virus with acute liver failure were treated with sofosbuvir for one week as a compassionate use.
ResultsUsing a high-content screening assay, we verified that sofosbuvir presented the best antiviral activity against YFV. Moreover, after an off-label treatment with sofosbuvir, the two female patients diagnosed with yellow fever infection displayed a reduction in blood viremia and an improvement in the course of the disease, which was observed in the laboratory medical parameters related to disease evolution.
ConclusionsSofosbuvir may be used as an option for treatment against YFV until other drugs are identified and approved for human use. These results offer insights into the role of nonstructural protein 5 (NS5) in YFV inhibition and suggest that nonstructural proteins may be explored as drug targets for YFV treatment.
Flaviviruses represent some of the most important pathogenic arboviruses worldwide, comprising diseases caused by mosquito-borne viruses, such as dengue virus, West Nile virus, yellow fever virus (YFV) and Zika virus (ZKV). In the last three years, flaviviruses have received the attention of the international research community and have brought new concerns for world public health due to the global epidemic ZKV and recent YFV outbreaks [1–5].
Typically, flaviviruses are RNA viruses that share the same genomic structure, with a noncoding sequence flanking both extremities of an open reading frame encoding a polyprotein. Characteristically, the structural proteins for the capsid, prM/M and envelope are located in the amino terminus, and the nonstructural proteins NS1–NS5 are located in the carboxy terminus of the polyprotein [1]. Regarding the functions and differences among viruses, NS1 and NS2 proteins are involved in viral replication. NS3 and NS5 proteins have enzymatic functions in all Flavivirus members. NS3 contains a helicase and ATPase domain [2]. Hepatitis C virus (HCV) NS5 protein has a peculiar subdivision of NS5A and NS5B subproteins not observed for YFV and ZKV [3,4].
Yellow fever (YF) is an acute febrile illness with several clinical manifestations during a short period of time. In most cases, the disease begins with fever and myalgia and in 25–60% of cases eventually progresses to hepatorenal syndrome, with liver and kidney failure with a fulminant course [5,6]. The live YFV vaccine 17D is one of the most effective vaccines ever produced that, in a few cases, causes severe adverse neurologic and viscerotropic reactions [7]. Therefore, it is not recommended for individuals who are more prone to develop severe adverse events, such as pregnant and breastfeeding women, individuals older than 60 years and immunocompromised individuals [8]. Recently, there were many cases of YF in the southeast of Brazil, and some of these cases with fulminant evolution needed liver transplantation as a compassionate therapy with good results [9,10]. As YF occurs in Brazil and several fulminant cases develop a dramatic course, we believe that early intervention leading to viral replication interruption could avoid the worst scenarios.
There are no antivirals or other specific chemotherapies approved for the effective treatment of YF disease, and its clinical management involves only the mitigation of symptoms and intensive fluid replacement for seriously ill patients [11]. A specific treatment for these viral diseases has considerable value during outbreaks, when an antiviral could be administered as prophylaxis or at the onset of any symptoms that could be associated with the disease.
The NS5 protein is an interesting target for antiviral drug development, since it is the largest and most conserved flaviviral protein, performing a multifunctional role in viral replication. The N-terminal region of NS5 protein is a methyltransferase domain responsible for adding the Cap signature at the 5′ of the RNA, and the C-terminal region is an RNA-dependent-RNA-polymerase domain (RdRp) [3]. NS5B inhibitors are classified into nucleotide inhibitors (NIs) and nonnucleotide inhibitors (NNIs). NIs bind to NS5B active sites, mimicking natural polymerase substrates, causing chain termination when incorporated into a growing RNA chain. NNIs inhibit conformational changes in the polyprotein replication complex by binding to one of the four allosteric sites [12]. Sofosbuvir is a nucleotide prodrug of the active triphosphate GS-461203. This prodrug works as an inhibitor of HCV NS5B RdRp, acting as a chain terminator [13]. Daclatasvir and ledipasvir are inhibitors of the NS5A replication complex. Daclatasvir targets a specific function of NS5A, which involves the downregulation of the hyperphosphorylation of NS5A [14]. The exact mechanism of action of ledipasvir is unknown, but a suggested mechanism could be the inhibition of NS5A phosphorylation, which seems required for viral replication [15,16].
These drugs belong to a new class of drugs called direct antiviral agents (DAAs), which are efficient inhibitors of HCV and might be useful to treat infections caused by other flaviviruses that have been widely detected worldwide in recent years [17]. In this context, the identification of new drugs to treat YF disease is a very relevant issue. The aim of this study was to verify the efficacy of HCV-approved drugs against YFV infections in vitro. The data generated here represent new starting points for the development of a treatment for YFV using broad spectrum antiflaviviral medicines.
2Materials and methodsCompound libraries and reference compoundsThree anti-HCV commercially available drugs, daclatasvir (BMS-790052), sofosbuvir (β-d-2′-deoxy-2′-α-fluoro-2′-β-C-methyluridine triphosphate; GS-7977) and ledipasvir (GS-5885), were used in the present work (MedChem Express, Monmouth Junction, USA). Human recombinant interferon-α-2A (IFNα2A) was used as a reference compound (Sigma–Aldrich, Saint Louis, USA). All drugs were diluted in 100% dimethyl sulfoxide (DMSO) (Sigma–Aldrich), except IFNα2A, which was directly dissolved in Dulbecco's phosphate-buffered saline (DPBS) (Sigma–Aldrich) containing 0.5% (W/V) bovine albumin (Sigma–Aldrich).
2.1VirusesA vaccinal YFV strain (17D) was provided by Professor Amadou Alpha Sall from Institute Pasteur, Dakar, Senegal. A YFV strain (17D) expressing the reporter yellow fluorescent protein (YFV-YFP) was provided by Laura H. V. G. Gil from the Research Center Aggeu Magalhaes, FIOCRUZ, Recife, Brazil.
2.2CellsAedes albopictus C6/36 cells and the human hepatoma cell line Huh7 were kindly provided by Dr. Amílcar Tanuri from Universidade Federal do Rio de Janeiro, Rio de Janeiro, Brazil. The C6/36 cells were cultivated in Leibovitz's L-15 medium (Sigma–Aldrich) at 28°C, and Huh7 cells were cultivated in Dulbecco's Modified Eagle's Medium F-12 (Sigma–Aldrich) at 37°C with 5% CO2. In both cases, the media were supplemented with 10% fetal bovine serum (FBS) (Thermo Fisher, Waltham, USA), 100U/mL penicillin and 100μg/mL streptomycin (Sigma–Aldrich). The Vero E6 cell line was cultured in Modified Eagle's Medium supplemented with 10% FBS, 1% nonessential amino acids (Thermo Fisher), 1% sodium pyruvate (Thermo Fisher), 100 units/mL penicillin and 100μg/mL streptomycin (Sigma–Aldrich), 0.05% Fungizone® Amphotericin B (Thermo Fisher) at 37°C in the presence of CO2.
2.3Viral expansionFor viral expansion, C6/36 cell culture was maintained in Leibovitz's L-15 medium and cultivated in 75cm3 flasks. After reaching approximately 70–80% confluence in the monolayer, the cells were infected at a multiplicity of infection (MOI) of 0.01–0.1 for 4–8 days with the YF vaccinal strain 17D or YFV-YFP [18]. After one hour (h) of adsorption, with gentle shaking every 10min to allow the homogeneous adsorption of the viruses, 15mL of culture media plus 2% FBS, 1% nonessential amino acids and 1% sodium pyruvate were added. After 8–12 days of incubation, the supernatant was collected and aliquoted in sterile conical tubes, frozen at −80°C and tested for the presence of virus by a quantitative reverse transcription polymerase chain reaction (q-RTPCR) and a plate assay using Vero cells, as described by Medina et al., 2012 [19].
2.4YFV-YFP titration by plate assayThe infectious viral titer was obtained by plate assay. Briefly, the virus samples were serially diluted 1:10, and 100μL of each dilution was transferred in duplicate to a 24-well plate with Vero or Huh7 cells prepared the day before titration. The plate was incubated for 1h at 37°C, and 500μL of a 1:1 mix of MEM 2% carboxymethylcellulose supplemented with 2% FBS was added to each well. The plate was further incubated at 37°C/5% CO2 for 4–5 days. For the development of plaques, 300μL of formaldehyde was added to each well to fix the cells, and 300μL/well of trypan blue dye was used to develop the plate, followed by 3 washes with phosphate-buffered saline. The plates were dried at room temperature before viral plate visualization and scoring.
2.5Drug tests in Vero cellsNinety-six-well plates were seeded with 1×104 Vero cells/well and incubated overnight. The cells were then treated with different concentrations of sofosbuvir, ledipasvir, daclatasvir, DMSO, or plain media (used as negative controls). After 1h of incubation, YFV was added at 0.1–1 MOI, and the plates were further incubated for 72 hs. After this period, the cells were collected for the quantification of YFV growth by viral load quantitation.
2.6YFV viral loadA TaqMan qRTPCR assay for YFV-RNA was performed. RNA extracted using Trizol™ reagent (Thermo Fisher) was eluted in 40μL of water and stored at −80°C until use. All extracts were tested for the human RNase P gene by polymerase chain reaction (PCR) qRTPCR to confirm sample quality. The reaction was carried out with RNA from each sample on a 7500 Real-time PCR System (Applied Biosystems, Foster City, CA, USA) with a set of primers and probes with FAM as a dye reporter for the probe, as previously described [20,21]. The presence of viral RNA in Vero cells was calculated using the RNase P gene as a normalization parameter and considering the amount of virus contained in the negative control (considered as 1) compared to the treatments.
2.7YFV high-content screening assay.The assay was performed as described by Pascoalino et al., 2016 [22], with minor differences. The drugs were tested in a 16-point dose–response curve (serially diluted by a factor of 2) starting at 100μM for daclatasvir, sofosbuvir and ledipasvir and 5.2nM for IFNα2A, reaching a final DMSO concentration of 1.0%. Mock-infected Huh-7 cells and IFNα2A were used as positive controls, and 1% DMSO vehicle-treated cells were used as negative controls (Sigma–Aldrich). Following the addition of compound, Huh-7 cells at a density of 2000 cells/well and YFV-YFP at an MOI of 2.5 were added onto μClear Black 384-well plates (Greiner Bio-One, Frickenhause, Germany). After 72h of incubation, the plates were fixed, and infection was then quantified by YFP signal as described below.
2.8Detection of YFV-YFP infection by fluorescenceYFV-YFP-infected cells were fixed with 4% (w/v) paraformaldehyde (PFA) for 20min at room temperature and then incubated with 5μg/mL of DAPI (4′,6-diamidino-2-phenylindole) (Sigma–Aldrich) in 1× PBS for 20min. The plates were washed twice with 1× phosphate-buffered saline (PBS). Subsequently, four digital images of different fields from each well were acquired at 20× magnification by the high-throughput confocal fluorescence imaging system Operetta (Perkin Elmer, Waltham, USA). The acquired images were analyzed as described by Pascoalino et al., 2016 [22].
2.9Data normalization and assay quality controlThe acquired images were analyzed with the High Content Analysis (HCA) software Harmony (Perkin Elmer) for the identification, segmentation and quantification of the host cell nucleus, cytoplasm and intracellular virus. The HCA provides output data for all images from one well of the total number of cells, the total number of infected cells and the intensity of the YFV-YFP signal. For the purpose of this study, the infection ratio (IR) was defined as the ratio between (i) the total number of infected cells in all images from one well and (ii) the total number of cells in all images from the same well. The raw data for IR values were normalized to negative (infected cells, DMSO-mock treated) and positive (non-infected cells) controls to determine the normalized antiviral activity, according to the equations below:
where:
Av. IRN: average infection ratio of negative control wells,
Av. IRP: average infection ratio of positive control wells,
Av. IRT: average infection ratio of test compound wells (in a given concentration).
Normalized activity values of the reference compound dose–response curve were processed with GraphPad Prism software, version 6, for sigmoidal dose–response (variable slope) nonlinear curve fitting and the determination of EC50 values by interpolation. The statistical validity of the high-throughput screening was determined by calculating the Z′-factor [23] using normal-infected and non-infected cells as negative and positive controls, respectively.
2.10Structural analysisThe amino acid sequences of hepatitis C virus (HCV) and YFV (accession codes CAB_10747.1 and NP_041726.1, respectively) were aligned based on structural information using Expresso from the T-COFFEE server (http://tcoffee.crg.cat/apps/tcoffee/do:expresso). The three-dimensional model of the YFV NS5 RdRp domain was built using the I-TASSER program [24]. The structural coordinates of HCV NS5B genotype 2A in complex with sofosbuvir (PDB code 4WTGhttps://www.rcsb.org) were used for comparisons with the YFV model [25]. The figures were prepared using Pymol 2.2 by Schrodinger (https://pymol.org/2/).
2.11Statistical analysisThe means and standard deviations were calculated for all data points from at least two independent experiments in triplicate. Statistical significance was determined using one-way ANOVA or the Wilcoxon test, in which P values less than 0.05 were considered significant.
2.12Compassionate use of sofosbuvir in two YFV-infected patientsSofosbuvir (Gilead, Foster City, CA, USA) in a 400mg single daily dose per os was used in an off label treatment and as a compassionate use in two patients diagnosed with YF. The patients included in this study were included in a research protocol approved by HC-FMUSP Ethical Committee (Process number: 2.669.963). Informant consent was waived as anonymity of participants was preserved. The patients were analyzed daily for the following laboratory parameters: complete blood count (CBC), aspartate aminotransferase (AST), alanine aminotransferase (ALT), gamma-glutamyl transferase (GGT), lactate, creatinine, ammonia, lipase, factor V, bilirubin, international normalized ratio (INR), albumin, globulin and bicarbonate. Other concomitant viral infections were also investigated by using serological and molecular assays (anti-HAV IgM, HAV RNA, anti-HBc, HBsAg, HBV DNA, anti-HCV, HCV RNA, anti-HIV, and HIV RNA). YF viremia was quantified utilizing the methodology described by Casadio et al., 2019 [26]. Samples for YF viral load were collected just before the daily sofosbuvir dose intake.
3Results3.1In vitro activity of approved antivirals against YFVSince YFV has several aspects in common with HCV, we decided to evaluate whether the preapproved commercial drugs, sofosbuvir, daclatasvir and ledispavir, were able to control YFV infection in vitro. We also tested the combination of sofosbuvir and ledipasvir, mimicking another approved HCV drug called Harvoni (Gilead, Foster City, CA, USA) and sofosbuvir and daclatasvir, which is a drug regimen widely utilized in Brazil for HCV.
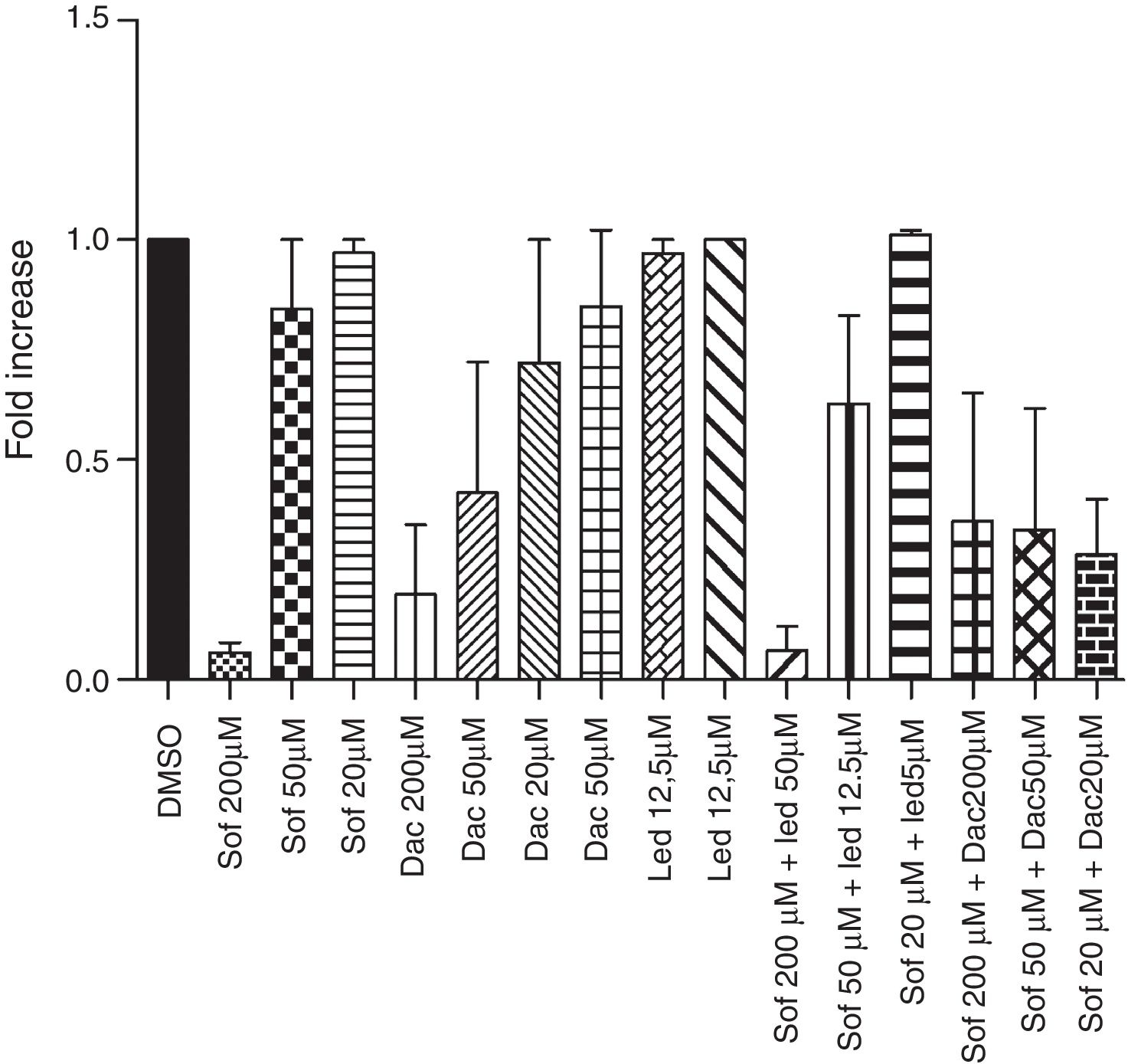
In the Vero cell model, sofosbuvir was the most effective drug in inhibiting YFV replication, particularly when associated with daclatasvir, but no statistically significant differences were detected with other treatment regimens (Fig. 1). The compound activities were also tested using the Huh-7 cell model infected with YFV-YFP to double check our results. All drugs were screened in a 16-point dose–response assay, and the experimental plates were considered approved (in terms of quality control), with a mean Z′-factor of 0.88±0.06. The drugs reached maximum activities comparable to the reference compound interferon-α 2A, showing 100.1% for sofosbuvir, 100.0% for daclatasvir and 53.3% for ledipasvir (Table 1a). The addition of ledispavir to sofosbuvir did not increase the activity against YFV in culture (Table 1b). Sofosbuvir and daclatasvir presented selectivity for YF, as demonstrated in the obtained images (Fig. 2). The most potent and efficacious compound in Huh-7 cells was sofosbuvir with a high selectivity index (Table 1, Fig. 3).
Inhibition of Yellow fever virus growth in vitro. (a) Vero cells were treated with different concentrations of sofosbuvir (Sof), daclatasvir (Dac), ledispavir (Led), Sof+Led, Sof+Dac and then infected with YFV. 72h after infection, YFV presence was analyzed by qRTPCR. Data obtained from three independent experiments. p>0.05 by Wilcoxon test.
(a) Summary of the results obtained in the YFV high-content screening of the commercial drugs and reference drug Interferon-α-2a (IFN α2a). (b) Combined treatment Sofosbuvir+Ledipasvir.
| (a) Compound name | EC50 (μM) | CC50 (μM) | Selectivity index | Maximum activity % | Z′ factor (mean) |
|---|---|---|---|---|---|
| Daclatasvir | 9.3±3.2 | 11.3±5.6 | 1.2±0.2 | 100.0 | 0.88±0.06 |
| Sofosbuvir | 0.4±0.1 | ND | >265.6 | 100.1 | 0.88±0.06 |
| Ledipasvir | ND | ND | ND | 53.3 | 0.88±0.06 |
| IFNα 2a | 25.9±20.2 | ND | >288.7 | 100.5 | 0.88±0.06 |
| (b) Compound name | EC50 (μM) | CC50 (μM) | Maximum activity % | Z′ factor (mean) |
|---|---|---|---|---|
| Sofosbuvir 75μM | 0.26±0.01 | ND | 100.1 | 0.93 |
| Sofosbuvir 75μM+Ledipasvir 25μM | 0.41±0.03 | ND | 100.1 | |
| Sofosbuvir 75μM+Ledipasvir 20μM | 0.52±0.04 | ND | 100.1 | |
| Sofosbuvir 75μM+Ledipasvir 15μM | 0.44 | ND | 100.1 | |
| Sofosbuvir 75μM+Ledipasvir 10μM | 0.41±0.01 | ND | 100.1 | |
| Sofosbuvir 75μM+Ledipasvir 5μM | 0.36±0.03 | ND | 100.1 | |
| Ledipasvir 25μM | ND | ND | 23.7 |
Where:
YFV=yellow fever virus.
CC50=concentration corresponding to 50% of cell toxicity.
EC50=concentration corresponding to 50% viral inhibition (of activity).
SI (selectivity index)=ratio between CC50 value and EC50 value (CC50/EC50). Whenever CC50 is not generated through curve fitting, the highest concentration tested is used to estimate the SI (Max [] tested/EC50).
Maximum activity was obtained from the highest value spotted by the non-linear regressed curve.
Fluorescent images of Huh7 cells infected with YFV-YFP treated with tested drugs in three different concentrations. Representative images taken at 20× magnification. Cells were nuclear-stained with DAPI (blue) and YFV was detected through YFP signal (yellow). (A) First dilution point; (B) fourth dilution point; (C) ninth dilution point. Dilutions of Interferon-α-2A were: 5.2nM, 0.65nM and 0.02nM; dilutions of all other compounds: 100μM, 12.5μM and 0.39μM; + Control: non-infected cells treated with vehicle (1% DMSO); − Control: Infected cells treated with vehicle (1% DMSO). Scale bar: 50μm.
Anti-YFV activity of tested drugs and reference compound Interferon-α-2A (IFNα2A). Left-Y-axis: normalized activity (%) (black squares and curves); right-Y-axis: cell ratio (red dots and curves); X-axis: Log of compound concentration (in molar). Mean values (dots and squares) and standard deviation (bars) are shown. Data obtained from two independent experiments. Cell ratio=number of cells/well divided by the mean of cells in the negative control DMSO 1%; Cpd=compound (drug) utilized in each assay.
Sofosbuvir performance results were not surprising, since HCV and YFV present high levels of amino acid sequence overlap in the RdRp domain, leading to a similar tridimensional protein structure (Fig. 4A, r.m.s.d. 3.5Å). When we compared the residues that coordinated the interaction of sofosbuvir in the three-dimensional structure of HCV-NS5 protein (PDB 4WTGhttps://www.rcsb.org), high conservation was observed in both NS5 ligand-binding pockets, with the unique substitution of residue F224 in HCV by the similar W539 in YFV protein (Fig. 4B and C). Moreover, this substitution would not affect the binding, since the drug interaction is performed with nitrogen from the main chain. Hence, the ligand-binding regions in YFV and HCV conserve the charge profile as evidenced by the calculation of the electrostatic potential (Fig. 4D, red for negative, blue for positive and gray for neutral). Altogether, these structural analyses corroborate the previous results and reinforce the activity of sofosbuvir in patients diagnosed with YF. Additionally, the cavity present in the NS5 protein from YFV has a high volume compared to the HCV-NS5 cavity (155 and 86 Angstroms, respectively), which may result in a YFV ligand binding pocket able to bind to more molecules at once.
Structural comparison between HCV and YFV NS5 protein. (A) Structural superimposition of the HCV-NS5-RNA-dependent-RNA-polymerase domain (RdRp) (green cartoon) bound to sofosbuvir (showed in stick) and the model of YFV-NS5 (gray cartoon). (B) Detail of the residues (in stick) that interact with sofosbuvir in HCV and their conservation in YFV protein (showed in green and gray, respectively). Residues from YFV are named by one letter code and number and followed by HCV ones. Sofosbuvir is shown in lime green sticks. (C) Amino acid sequence alignment of HCV (residues 140–383) and YFV (residues 456–727) NS5-domains performed by T-coffee server. All residues of the pocket that interact with sofosbuvir in HCV protein are shown in red bold and compared in the YFV sequence. (D) Comparison of the electrostatic potential of the sofosbuvir interaction region in the two proteins reveals conservation of the charges, which explain the inhibitory results (red for negative, blue for positive and gray for neutral). r.m.s.d.=root mean square deviation of atomic positions.
These promising results in our in vitro screening model indicated that these drugs may be translated to in vivo situations and are useful for eliminating YFV infection. To understand its efficacy in patients, sofosbuvir was used in an off-label treatment and as a compassionate use in two patients diagnosed with YF. The patients were negative for all markers related to HIV and hepatitis virus. The results of the clinical laboratory assays are shown in Table 2. Patient 01, a 42-year-old female that arrived consciously at the hospital after presenting symptoms for 6 days. The patient was hospitalized in January 29, 2018, treated with 400mg/day per os of sofosbuvir for 7 days starting on January 30, 2018, and she remained in the Intensive Care Unit (ICU) until February 5, 2018, when she presented improvement in her parameters. On February 9, 2018, the patient was discharged from the hospital, with normal INR (0.95) but still with increased liver enzymes.
Clinical evolution of two patients submitted to Sofosbuvir therapy.
| Patient 1 (female, 42 yrs old) | Patient 2 (female, 68 yrs old) | Reference value | |||||
|---|---|---|---|---|---|---|---|
| Jan 29 Sofosbuvir | Feb 5 Sofosbuvir | Feb 9 Discharge | Jan 27 Sofosbuvir | Feb 7 Sofosbuvir | Feb 9 Discharge | ||
| AST (U/L) | 5789 | 379 | 114 | 7312 | 155 | 179 | <31U/L |
| ALT (U/L) | 3960 | 690 | 356 | 3587 | 192 | 178 | <31U/L |
| GGT (U/L) | 99 | 1.046 | 832 | 317 | 382 | 664 | 5–36U/L |
| Lactate (mg/dL) | 15 | NA | NA | 33 | 539 | NA | 4.5–14.4mg/dL |
| Total bilirubin (mg/dL) | 0.82 | 0.72 | 0.42 | 3.38 | 11.65 | 13.96 | 0.2–1mg/dL |
| Direct bilirubin (mg/dL) | 0.75 | 0.73 | 0.32 | 3.32 | 10.75 | 12.44 | <0.30mg/dL |
| Creatinine (mg/dL) | 0.96 | 0.59 | 0.50 | 2.69 | 0.94 | 1.12 | 0.5–0.9mg/dL |
| Ammonia (μmol/L) | 60 | NA | NA | 50 | 59 | NA | 11–32μmol/L |
| Lipase (U/L) | 42 | 51 | NA | 167 | 189 | 189 | 13–60U/L |
| Factor V (%) | 96 | >150 | NA | 46 | >150 | >150 | 70–150% |
| INR | 1.83 | 0.95 | 0.95 | 1.63 | 1.01 | 0.95 | 0.95–1.2 |
| Hb/Ht (g/dL; %) | 14.8/42.9 | 12.6/37.2 | 12/36.6 | 12.6/35.5 | 7.2/20.2 | 8.3/24.6 | 12–16g/dL/35–47% |
| Leucocytes (×103/mm3) | 2.69 | 7.15 | 6.88 | 4.48 | 4.3 | 5.29 | 4–11×103/mm3 |
| Platelets (×103/mm3) | 156 | 233 | 407 | 91 | 222 | 361 | 140–450×103L/mm3 |
| Albumin (g/dL) | 3.6 | NA | NA | 2.7 | 2.1 | 3 | 3.4–4.8g/dL |
| Globulin (g/dL) | 3.3 | NA | NA | 2.2 | 2.5 | 3.5 | 1.7–3.5g/dL |
| Bicarbonate (mmol/L) | 18.3 | NA | NA | 18.3 | 23.1 | NA | 21–28mmol/L |
| Viral load(log copies/mL) | 6.1 | 3.3 | ND | 5.8 | 4.6 | 3.3 | Not detected (ND) |
AST – aspartate aminotransferase.
ALT – alanine aminotransferase.
GGT – gamma-glutamyl transferase.
INR – international normalized ratio calculated from prothrombin time.
Hb/Ht – hemoglobin/hematocrit.
NA indicates that the value was not available.
ND – not detected.
Patient 02, a 68-year-old female that arrived conscious after 5 days, presenting with symptoms and receiving 400mg/day per os of sofosbuvir for one week from January 27, 2018 and remained in the ICU on February 7, 2018 for ambulatory care. She was discharged from the hospital on February 9, 2018.
YFV viral loads in these two patients are shown in Fig. 5. Both patients exhibited an important reduction in their viral loads. Patient 01 displayed a viral reduction from 1,265,839 (6.10log) to 1464 (3.16log) RNA viral copies/mL during treatment that was accompanied by a relevant improvement in other laboratorial markers. Patient 02 had a viral reduction from 639,333 (5.80log) to 792 (2.89log) RNA viral copies/mL. Both analyzed patients showed a decrease in the viral load slightly higher than −2.9log (the mean decrease in viral load each day was −0.41log).
YF viral load in two patents submitted to sofosbuvir therapy. Y axis – viral load expressed in log10RNA copies/mL. X axis – days. Viral load of patient 1 is show as a dashed curve while for patient 2 it was utilized a black curve. The initial day for sofosbuvir therapy is shown as a gray arrow while the black arrow shows the last day of therapy. Both patients showed a decrease in their viral loads.
These initial results suggest that sofosbuvir is active against YFV infection, confirming the obtained in vitro results, which should be further assessed utilizing a larger number of patients.
4DiscussionThe recent increase in YF cases in the southeast of Brazil reinforces the importance of the identification of different antiviral compounds able to inhibit Flavivirus. The current available treatment is driven to manage viral cytopathic effects that might lead the patient to a fatal course due to the failure of multiple organs. DAAs might be effective in controlling viral infection by inhibiting its cytopathic effects or at least controlling these effects before liver transplantation. Therefore, the objective of this study was to rapidly identify safe and potent molecules capable of eliminating the infection of YFV in vitro due to the urgency and absence of specific treatments. This matter increases in importance every year, as evidenced by the recent increase in YF cases in Brazil.
We verified by drug repositioning the activity of anti-HCV commercially available drugs against YFV. Since both viruses belong to the same family of Flaviviridae have some degree of homology in their genome organization, and infect the liver as the main target, DAAs developed against HCV may also inhibit the activity of YFV. The results showed that sofosbuvir presented activity and selectivity against YFV in Huh-7 cells. The potency of sofosbuvir against YFV was not enhanced when combined with ledipasvir. The high selectivity of sofosbuvir against another Flavivirus was already reported in cell lines and in animal models and was even indicated for the treatment of Brazilian ZKV cases, although this virus preferentially targets tissues from the nervous system [22,27]. In cell lines, such as BHK-21, and in the human neuroblastoma SH-Sy5y, for example, sofosbuvir was able to inhibit ZKV replication in a dose-dependent manner, which was not observed when Vero cells were used [28]. Interestingly, we also observed the same tendency of YFV inhibition after sofosbuvir treatment in Vero cells. Vero cells do not express high levels of carboxyl esterase, which is important for the intracellular activation of sofosbuvir and could be responsible for the observed differences in the potency of sofosbuvir between Vero cells and Huh-7 cells [29].
Ribavirin, a compound with broad-spectrum antiviral activity that has been used to treat different viral infections, including HCV and syncytial respiratory virus, also induced viral replication inhibition against YFV. Ribavirin efficacy in the treatment of YF in humans is still not deeply clarified, and some studies have emphasized its ability to reduce viremia in vitro and in hamster models. In hamster models, the effect of ribavirin was only achieved when the drug was administered before the establishment of infection, which brings difficulties for human use, since patients usually look for treatment when viremia is already present [29–32]. Furthermore, the use of ribavirin is restricted in specific groups, mainly because this drug seems to be teratogenic and able to induce rash and anemia in patients under therapy [32]. Because of these concerns, the identification of alternative treatments against YF as proposed is still required. Thus, two patients received sofosbuvir in an off label and compassionate treatment against YF to test its efficacy in humans based on the observed in vitro results and due to the need to provide some treatment for patients and the absence of a specific therapy. After one week of treatment, reduced viremia as well clinical parameters related to improvement of the liver conditions associated with YF infection were observed in both patients. Similar effects were observed in a neonatal Swiss mouse model, indicating that sofosbuvir is a feasible compound to control YF viremia in vivo[8].
Sofosbuvir showed a relevant effect on viral replication in the two in vitro YFV cultivation systems analyzed using two different cell culture systems. When we analyzed the interaction between sofosbuvir and the active site of NS5 YFV RdRp, the NS5 ligand binding pocket was highly conserved, with only one residue substitution (phenylalanine in HCV to tryptophan in YFV). The structural comparisons between HCV bound to sofosbuvir and YFV showed that both NS5-RdRp domains share conserved residues and charges for the sofosbuvir interaction. In addition, the model of the YFV-RdRp domain showed a higher volume of the cavity when compared to the HCV-NS5 cavity (155 and 86Å, respectively), suggesting that there are other possibilities for YFV-NS5 inhibitor development. In addition, the NS5 protein of ZKV and chikungunya has already been demonstrated as a potential drug target, and both viruses had NS5 proteins sharing high similarity with YFV and dengue virus crystallized protein structures [3].
Both analyzed patients showed a decrease in the YFV load slightly higher than −2.9log during the 7-day treatment (mean decrease in viral load each day was −0.41log). This level of reduction in HCV load can be achieved only in two days. The effect of sofosbuvir on HCV is very rapid, reducing viral production equal to 99.96%, on average, and may be detected on the first two days of infection, likely representing the time needed to accumulate intracellular triphosphates [33].
These results are consistent with a recent published paper that also showed that sofosbuvir inhibits YFV replication in liver cell lines and animal models [8], but to our knowledge, these results are the first to show the use of this drug in human patients infected with YFV as a compassionate use.
The differences in the efficacy of the drug in vitro and in vivo might be related to the severe clinical picture of YF patients, with a generalized failure of multiple organs involving not only the liver but also the brain, lungs, kidneys, pancreas, testis and heart in a recent study of the autopsy findings from cases in São Paulo City [34]. This severe picture likely adds difficulty in achieving therapeutic levels at sites of virus replication.
In conclusion, using a high-content screening assay, sofosbuvir presented the best antiviral activity against YFV. Structural comparisons between HCV and YFV bound to sofosbuvir showed conserved NS5-RdRp domains for the sofosbuvir interaction. Last, treatment with sofosbuvir in the two YFV-infected patients was accompanied by a reduction in viremia and an improvement in the course of the disease. These results indicate that sofosbuvir may be used as an option for treatment against yellow fever.
AbbreviationsALT alanine aminotransferase aspartate aminotransferase average infection ratio of negative control wells average infection ratio of positive control wells average infection ratio of test compound wells complete blood count concentration corresponding to 50% of cell toxicity. compound direct antiviral agents daclatasvir 4′,6-diamidino-2-phenylindole dimethyl sulfoxide Dulbecco's phosphate-buffered saline concentration corresponding to 50% viral inhibition (of activity). fetal bovineserum gammaglutamyltransferase hemoglobin/hematocrit high content analysis hepatitis A virus hepatitis B virus Hospital das Clínicas da Faculdade de Medicina da Universidade de São Paulo hepatitis C virus human immunodeficiency virus hepatocyte-derived carcinoma cell line intensive care unit interferon-α-2A international normalized ratio ledispavir multiplicity of infection not available not detected nucleotide inhibitors nonnucleotide inhibitors nonstructural phosphate-buffered saline polymerase chain reaction protein database paraformaldehyde quantitative reverse transcription polymerase chain reaction RNA-dependent-RNA-polymerase root mean square deviation of atomic positions selectivity index ratio between the CC50 value and the EC50 value sofosbuvir yellow fever yellow fever virus YFV strain (17D) expressing the reporter yellow fluorescent protein Zika virus vaccinal YFV strain
The authors declare that they do not have any conflicts of interest.
We thank Professor Amadou Alpha Sall from Institute Pasteur, Senegal, and Dr. Laura H. V. G. Gil from the Research Center Aggeu Magalhaes, FIOCRUZ, Brazil for providing the YFV used in this study. A. albopictus C6/36 cells and the human hepatoma cell line Huh7 were kindly provided by Dr. Amílcar Tanuri from Universidade Federal do Rio de Janeiro, Rio de Janeiro, Brazil. This work was supported by Fundação de Amparo à Pesquisa do Estado de São Paulo (FAPESP) Grant number 2016/20045-7. Scholarship funding was granted by FAPESP (Érica Araújo Mendes and Bruno dos Santos Pascoalino) and Conselho Nacional para o Desenvolvimento Científico e Tecnológico (CNPq) for Denise Regina Bairros de Pilger.