MicroRNAs represent a class of non-coding RNA molecules that negatively regulate gene expression either by repressing translation or by inducing degradation of messenger RNA. Studies have shown that, as regulators of gene expression, microRNAs are widely involved in various human diseases, including hepatitis B virus-related liver diseases. By modulating hepatitis B virus replication, regulating extracellular matrix formation, as well as silencing tumor suppressor genes, these small molecules are implicated in the development of chronic hepatitis, liver fibrosis/cirrhosis, and hepatocellular carcinoma caused by hepatitis B virus infection. In addition, current researches indicated a potential role of microRNA as diagnostic markers and therapeutic targets. In conclusion, microRNAs are promising tools in the diagnosis and treatment of hepatitis B virus -related liver diseases.
Almost 40 years have passed since effective anti-hepatitis B virus(HBV) therapy was firstly introduced to clinical practice.1 Despite significant progress in anti-viral treatment, HBV infection still causes more than 1 million deaths of liver failure, cirrhosis, and hepatocellular carcinoma(HCC) annually.2
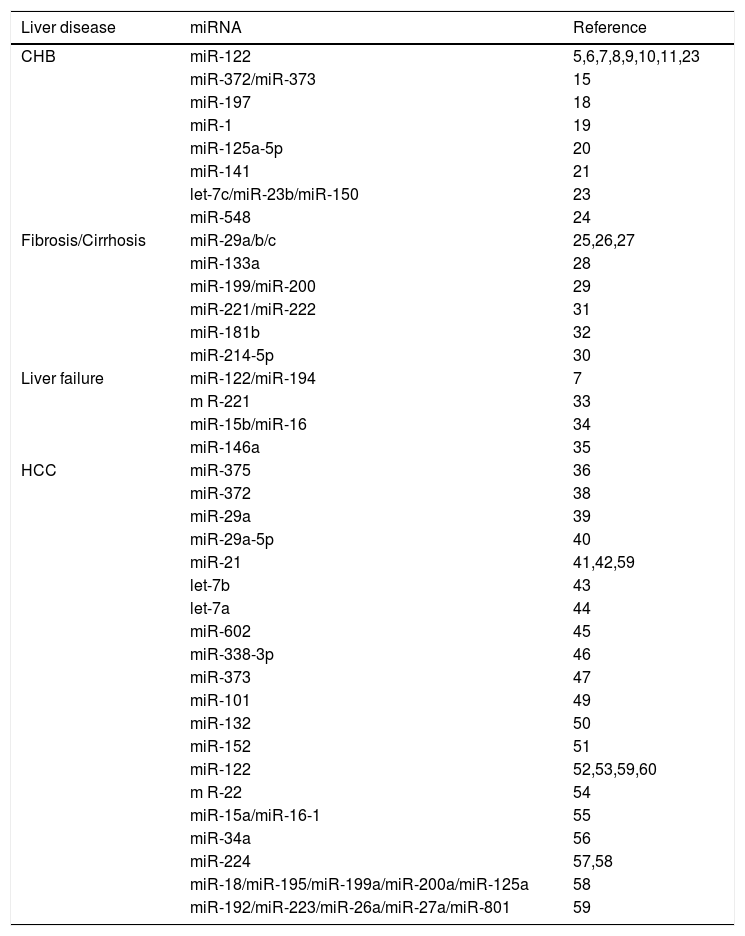
MicroRNAs(miRNA, miR) represent a class of non-coding RNA molecules that negatively regulate gene expression post-transcriptionally by interacting with the mRNA of target genes.3,4 Current studies have revealed the contribution of miRNAs to the pathogenesis of various human diseases, including HBV-related liver diseases (summarized in Table 1). A large number of studies have demonstrated altered miRNA expression profiles in chronic hepatitis B (CHB), cirrhosis, or HCC, which provide us new insight into the molecular mechanisms involved in the progression of HBV-related liver diseases. Given these findings, it is probable for aberrantly expressed miRNAs to serve as diagnostic tools, as well as promising therapeutic targets.
MiRNAs in HBV-related liver diseases.
| Liver disease | miRNA | Reference |
|---|---|---|
| CHB | miR-122 | 5,6,7,8,9,10,11,23 |
| miR-372/miR-373 | 15 | |
| miR-197 | 18 | |
| miR-1 | 19 | |
| miR-125a-5p | 20 | |
| miR-141 | 21 | |
| let-7c/miR-23b/miR-150 | 23 | |
| miR-548 | 24 | |
| Fibrosis/Cirrhosis | miR-29a/b/c | 25,26,27 |
| miR-133a | 28 | |
| miR-199/miR-200 | 29 | |
| miR-221/miR-222 | 31 | |
| miR-181b | 32 | |
| miR-214-5p | 30 | |
| Liver failure | miR-122/miR-194 | 7 |
| m R-221 | 33 | |
| miR-15b/miR-16 | 34 | |
| miR-146a | 35 | |
| HCC | miR-375 | 36 |
| miR-372 | 38 | |
| miR-29a | 39 | |
| miR-29a-5p | 40 | |
| miR-21 | 41,42,59 | |
| let-7b | 43 | |
| let-7a | 44 | |
| miR-602 | 45 | |
| miR-338-3p | 46 | |
| miR-373 | 47 | |
| miR-101 | 49 | |
| miR-132 | 50 | |
| miR-152 | 51 | |
| miR-122 | 52,53,59,60 | |
| m R-22 | 54 | |
| miR-15a/miR-16-1 | 55 | |
| miR-34a | 56 | |
| miR-224 | 57,58 | |
| miR-18/miR-195/miR-199a/miR-200a/miR-125a | 58 | |
| miR-192/miR-223/miR-26a/miR-27a/miR-801 | 59 |
Patients who developed chronic liver diseases, such as CHB, after infection of HBV are at increased risk for developing cirrhosis and primary HCC. Therefore, the suppression of HBV replication, and ideally the eradication of HBV cccDNA are of great importance for antiviral therapies.
As liver-specific miRNA, miR-122 was found closely related to HBV replication and liver injury.5–11 One study7 showed that circulating miR-122 was significantly up-regulated in HBV-infected patients, especially in hepatitis B e antigen positive CHB individuals. Interestingly, in cell line experiments, a negative correlation was observed between the levels of miR-122 and HBV mRNA.7 Similarly, Chen and colleagues8 reported that miR-122 suppressed gene expression and replication of HBV by binding to its highly conserved RNA sequence. A research from Wang, et al.9 showed that the miR-122 levels were reversely related to intrahepatic viral load and hepatic necroinflammation, and the depletion of endogenous miR-122 led to enhanced HBV replication, whereas overexpression of miR-122 inhibited viral production. The study9 finally identified a target gene of miR-122-cyclin G(1), whose specific interact with P53 blocked the specific binding of P53 to HBV enhancer elements, which led to the inhibition of HBV transcription. Another research 10 found that treatment of IFN-α contributed to a marked decrease of miR-122 expression in hepatocytes by inducing upregulation of NT5C3 (an IFN-stimulated gene), which was identified as a target of miR-122, but could in turn suppress miR-122 by binding and sequestering its mRNA 3’-UTR, thus promoting HBV expression and replication.10 This may help to explain the poor responses to IFN-α in CHB patients, if the results could be further confirmed in clinical samples. These exciting findings indicated a promising anti-HBV treatment based on miR-122. More interestingly, a research found that serum miR-122 levels may serve as an indicator for viral translation and a potential marker for risk stratification in patients infected with HBV.11
In addition to miR-122, many other miRNAs were also correlated with HBV infection, and even the degree of liver injury.12–17 MiR-372/373 were positively associated with the hepatic HBV DNA levels in vivo.15 Nuclear factor I/B (NFIB) was identified as a direct target of miR-372/373 and cell experiment confirmed the suppression of HBV expression by NFIB.15 The study demonstrated that miR-372/373 promoted HBV expression through a pathway involving the transcription factor NFIB.15 Besides, by targeting IL-18, miR-197 might reactivate liver inflammation.18 MiR-1 was found capable of enhancing the HBV core promoter transcription activity by increasing expression of farnesoid X receptor α.19 MiR-125a-5p interacted with viral mRNA of HBV and interfered with viral translation, down-regulating the expression of surface antigen.20 MiR-141 was reported to be able to target peroxisome proliferatoractivated receptor alpha (PPARA), which regulates HBV gene expression through interaction with HBV promoter regulatory elements, thus repressing HBV replication.21 HBXIP, an inhibitor of HBV replication, was a potential target of miR-501.22 Moreover, Chen, et al.23 identified a panel of serum miRNAs (let-7c, miR-23b, miR-122, and miR-150) with extremely high sensitivity (99.9%) and specificity (99.8%) in diagnosing occult hepatitis B virus infection. Investigators also found miR-548 family members, especially miR-548c5p, which involved in type I IFN signaling, up-regulated significantly in patients with CHB compared to normal controls, which indicated a close relationship between miR-548 family and disorder of type I IFN secretion.24 On the whole, these data demonstrated the roles of miRNAs in the modulation of HBV replication, including the epigenetic regulation of HBV cccDNA function (reviewed elsewhere25). However, the application of our current knowledge to clinical practice is a huge challenge. Although trails on antiviral treatment based on miR-122 have been launched,26,27 it is still far from bench to beside.
Liver Fibrosis/CirrhosisLiver fibrosis develops through a complex network of signaling pathways regulating the deposition of extracellular matrix and collagens. The principal cells responsible for the accumulation of extracellular matrix proteins during hepatic fibrogenesis are activated hepatic stellate cells (HSC).
MiR-29 family members (miR-29a/b/c) were well documented fibrosis relevant miRNAs.28–31 It was reported that members of the miR-29 family were strikingly down-regulated in mouse models of liver fibrosis and in human fibrotic livers.28 Furthermore, the decreased expression of miR-29b in animals was significantly correlated with the degree of liver fibrosis.28 More recently, serum miR-29 levels were found negatively correlated with liver fibrotic stages and necroinflammation grades in patients chronically infected with HBV.29 Investigators confirmed that overexpression of miR-29b significantly decreased the expression of extracellular matrix genes in HSC.28 And the process might be a part of signaling network involving TGF-β and NF-κB.28 In addition, miR-29b was capable of suppressing HSC activation as well as production of type I collagen.30,31 All these data suggest that miR-29b prevent liver fibrosis. MiR-133a was another identified miRNA during fibrogenesis but down-regulated in HSC specifically.32 Researchers found that miR-133a suppressed collagen synthesis through a TGF-β-dependent way.32 In addition, overexpression of miR-199 and -200 families were tightly related to the grade of liver fibrosis.33 Other investigations have associated overexpression of several miRNA (miR-221/222, miR-181b, and miR-214-5p) with activation of HSC as well as progression of liver fibrosis.34–36
Liver FailureLiver failure is a life-threatening condition in which there is a severe deterioration of liver function, and miRNAs seem to be involved in this pathogenesis.
MiR-221 was found significantly up-regulated in liver of mice which developed FAS-induced fulminant liver failure.37 Cell line experiments indicated that miR-221 protected primary hepatocytes and hepatoma cells from apoptosis and in vivo overexpression of miR-221 postponed FAS-induced fulminant liver failure.37 The underlying mechanism is that miR-221 regulated hepatic expression of Puma (p53 up-regulated modulator of apoptosis), which functioned as a pro-apoptotic factor.37 Contrary to miR-221, up-regulation of miR-15b and miR-16 increased TNF production and hepatic apoptosis by targeting anti-apoptotic gene BCL2.38 Recent studies39 demonstrated that the single nucleotide polymorphism (SNP) of the miR-146a gene was correlated with the susceptibility of acute-on-chronic hepatitis B liver failure (ACLF-HBV). In patients with ACLF-HBV, the amount of mature miR-146a in PBMCs was significantly higher in the GG homozygote group than those in the CC and CG genotype group.39 The GG genotype was also associated with relative lower serum level of TNF-α and higher survival rate.39 Therefore, the GG genotype within the pre-miR-146a was a protective genotype for ACLF-HBV. Other studies7 showed that the expression levels of miR-122 and miR-194 were inversely correlated with the age of patients with ACLF. Clinical investigations, however, were needed to further clarify the relationship between miRNAs and HBV-related liver failure.
Hepatocellular CarcinomaMiRNA expression profiling studies in patients with hepatocellular carcinoma (HCC) or hepatoma cell lines have shown some unique miRNA signatures, which provided us novel insights into the pathogenesis and development of HCC.
By targeting yes-associated protein (YAP, a transcriptional co-activator involved in downstream of Hippo signaling pathway), miR-375 was found to be able to suppress proliferation and invasion of HCC cells.40 HULC (highly up-regulated in liver cancer), as a long non-coding RNA (lncRNA), plays an important role in the initiation of hepatocellular carcinoma.41 Studies42 demonstrated that cAMP-response element binding protein (CREB) up-regulated HULC expression level by interacting with miR-372. Interestingly, HULC reversely inhibited miR-372 and led to up-regulation of its target gene, PRKACB, which in turn induced phosphorylation of CREB.42 The auto-regulatory loop of HULC promoted its own expression in liver cancer. Meng, et al.43 performed a systematic miRNA expression profiling study using cell lines disposed with arsenic trioxide and found the expression of miR-29a was up-regulated. They confirmed that PPM1D is the target of miR-29a, whereby the effect of inhibiting cell growth and inducing cell apoptosis in liver cancer cells were exerted. In addition, miRNA-29a-5p was identified has the potential to serve as biomarker for early relapse of HBV-related HCC post resection.44
Tumor suppressor Sprouty1 is the target of miR-21, investigators found that in liver cancer cells, Sprouty1 expression was inversely correlated with Pokemon levels, and miR-21 was up-regulated by Pokemon.45 By overexpression and silence, investigators confirmed that Pokemon repressed Sprouty1 expression via miR-21 at both mRNA and protein levels, promoted the growth and proliferation of liver cancer cells.45 PTEN activates phosphatidylinositol 3-kinase signaling to AKT, and studies46 indicated that up-regulation of miR-21 activated PTEN, and led to progression of HCC.
The oncogenic HMGA2 (High Mobility Group AT-2 hook), a nuclear non-histone transcriptional cofactor, was found down-regulated in human hepatocellular carcinoma models by panobinostat through an anti-oncomir- hsa-let-7b-dependent way.47 In addition, down-regulation of let-7a by HBx increased STAT3 expression, promoted cell proliferation and caused tumorigenesis.48 Studies49 also revealed that miR-602 was up-regulated in HCC and targeted tumor suppressive gene RASSF1A (Ras-associated domain family member 1A). Inhibition of miR-602 promoted hepatoma cell apoptosis and inhibited cell proliferation.49 Oncogene HBV X was found to be able to promote the expression of CyclinD1 by suppressing miR-338-3p, which led to cell proliferation.50 This might contribute to the HBx-mediated hepatocarcinogenesis. Study51 also found by down-regulation of miR-373, HBx decreased E-cadherin expression, which contributed to the metastasis of HBV-related HCC.
Gene inactivation caused by CpG hypermethylation is now recognized as an important mechanism in tumor initiation and progression.52 A research from Wei, et al.53 confirmed that hepatitis B virus x protein increased the expression of DNMT3A (DNA methyltransferase 3A, a target of miR-101) by down-regulating miR-101 and enhanced DNA methylation of tumor-suppressor genes. In addition, investigators revealed that proliferation and colony formation of HCC cells could be repressed by miR-132-mediated inhibition of the Akt-signaling pathway, while DNA methylation induced by HBx suppressed miR-132 expression.54 Besides, researchers found that the frequent down-regulation of miR152 deregulated DNMT1, caused subsequent aberrant DNA methylation in HBV-related HCC.55
Studies also manifested that inhibition of miR-122 by HBV mRNA up-regulated PTTG1 (pituitary tumor-transforming gene 1) binding protein, and promoted HCC tumor growth and cell invasion.56 By binding to peroxisome proliferator activated receptor-gamma (PPARy), HBx, rather than hepatitis C viral particles, suppressed the transcription of miRNA-122 in hepatoma cells.57 Interestingly, in male patients with HBV-related HCC, miR-22 over-expressed in tumor adjacent tissues, which was related to decreased expression of ERa, indicating a possible effect of weakened protective function of estrogen in HBV-related carcinogenesis in male patients.58
Other studies suggested that, in hepatocytes, HBV RNA led to HCC progression by inhibiting miR-15a/miR-16-1, the noted anti-oncomirs.59 Decreased tumor expression of miR-34a and increased tumor expression of CCL22 caused by HBV predisposed HCC patients to portal vein tumor thrombus.60 Recently, a research demonstrated that, by suppressing miR-224, autophagy inhibited carcinogenesis of HBV-related HCC.61
Researchers identified a couple of miRNAs (miR-18, precursor miR-18, miR-224, miR-199a*, miR-195, miR-199a, miR-200a, and miR-125a) that expressed higher in HCC than adjacent non-tumorous tissue, the prediction accuracy of HCC based on the miRNA panel reached as high as 97.8%.62 Besides, a plasma microRNA panel (miR-122, miR-192, miR-21, miR-223, miR-26a, miR-27a, and miR-801) was confirmed with satisfactory diagnostic accuracy of HCC regardless of the Barcelona Clinic Liver Cancer stages and was able to differentiate HCC from healthy, chronic hepatitis B, and cirrhosis.63 Still, miR-122 proved itself with the potential to serve as a biomarker for screening HCC.64
Future DirectionsProfiling studies concerning miRNAs in chronic liver diseases caused by HBV infection have demonstrated the important function of miRNAs in the progression of CHB, fibrosis/cirrhosis, liver failure, and HCC. These findings improved our understanding of the mechanisms implicated in HBV-related liver diseases.
Due to the stable presence in circulation, miR-NAs are favorable biomarkers. Further studies are needed to identify specific circulating miRNA signatures that have the potential to differentiate different liver diseases. To date, cccDNA eradication is the major challenge of anti-HBV therapies. Notably, many miRNAs can modulate HBV replication and cccDNA function directly or by epigenetic mechanisms. Elucidating the complicate nexus between miRNAs and HBV will certainly contribute to suppression and cccDNA elimination of HBV. In addition, decoding the complex molecular mechanisms of miRNAs involved in the development and metastasis of HCC is crucial for the establishment of effective therapeutic strategies. We believe that, in the next decade, application of treatments based on miRNAs will greatly benefit patients.
AcknoledgementsThe work was supported by Natural Science Foundation of China (81101240) to NL and Natural Science Foundation of China (81371821) to GS.
We thank Chong Huang (Department of Infectious Diseases, Huashan Hospital, Fudan University) for his critical comments on this manuscript.
Abbreviations- •
ACLF-HBV: acute-on-chronic hepatitis B liver failure.
- •
CHB: chronic hepatitis B.
- •
CREB: cAMP-response element binding protein.
- •
DNMT: DNA methyltransferase.
- •
HBV: hepatitis B virus.
- •
HBXIP: hepatitis B X interacting protein.
- •
HCC: hepatocellular carcinoma.
- •
HMGA2: High Mobility Group AT-2 hook.
- •
HSC: hepatic stellate cell.
- •
HULC: highly up-regulated in liver cancer.
- •
IFN: interferon.
- •
IL: interlukin.
- •
lncRNA: long non-coding RNA.
- •
miRNA: microRNA.
- •
NFIB: nuclear factor I/B.
- •
NF-κB: nuclear factor kappa B.
- •
PPARA: peroxisome proliferator-activated receptor alpha.
- •
PPARy: peroxisome proliferator activated receptor-gamma.
- •
PTEN: phosphatase and tensin homolog deleted on chromosome ten.
- •
PTTG1: pituitary tumor-transforming gene 1.
- •
RASSF1A: Ras-associated domain family member 1A.
- •
SNP: single nucleotide polymorphism.
- •
STAT3: signal transducer and activator of transcription 3.
- •
TGF-β: transforming growth factor-β.
- •
TNF: tumor necrosis factor.
- •
YAP: yes-associated protein.
Natural Science Foundation of China (81101240) to Ning Li and Natural Science Foundation of China (81371821) to Guangfeng Shi.
Conflicts of InterestThere were no financial disclosures from any authors.