Approximately 500 million people, i.e. one fifth of the world population, are chronically infected with hepatitis B virus (HBV) or hepatitis C virus (HCV).1,2 Each year, over 1.5 million people die from HBV-and/or HCV-re-lated chronic liver diseases, such as end-stage cirrhosis and hepatocellular carcinoma (HCC). HCC is one of the most common cancers worldwide and in 2002, HBV and HCV have been responsible for at least half a million of these cancers.3 Nowadays, virological tools are needed to diagnose chronic HBV and HCV infections, they may be useful to establish their prognosis, but they have found their principal application in guiding treatment decisions and assessing the virological responses to therapy.
Virological toolsViral antigen and antibody detectionThe detection (and eventually the quantification) of viral antigens and of specific antibodies in body fluids is based on the use of sandwich enzyme immunoassays (EIAs). Recombinant antigens or antibodies are used to capture circulating antibodies or antigens, respectively, onto the wells of microtiter plates, microbeads or specific holders adapted to close automated devices. The presence of antigens or antibodies is revealed by antibodies (in the case of antigen) or anti-antibodies (in the case of antibody) labeled with an enzyme that catalyzes the transformation of a substrate into a colored compound. The optical density (OD) ratio of the reaction (sample OD/internal control OD) is proportional to the amount of antigens or antibodies in the sample. EIAs are cheap, easy to use, can be fully automated and are well adapted to large volume testing.
Genome detection and quantificationViral genome detection and quantification can be achieved by using two categories of molecular biology-based techniques, including target amplification (such as polymerase chain reaction (PCR)) and signal amplification (such as hybrid capture or the branched DNA assay). Whatever technique used, international units per millili-ter must be preferred to any other quantitative unit. Conversion factors can be used to establish a relationship between the IUs and the non standardized copies.4,5 The classical techniques for viral genome detection and quantification are now progressively being replaced by real-time PCR assays in most virology laboratories.
Real-time PCR techniques have a broad dynamic range of quantification, well suited to the clinical needs. Real-time PCR is more sensitive than classical PCR, with lower limits of detection of the order of 10 to 15 IU/mL. Real-time PCR assays do not yield false-positive results due to carryover contaminations, and they can be fully automated. Real-time PCR has become the technique of choice to detect and quantify viral genomes in clinical practice.
Nucleotide sequence analysisIn practice, the identification of signature sequences in viral genomes is used: (i) to classify viral strains into phylogenetic groups of clinical interest (genotypes and subtypes); (ii) to identify natural polymorphisms with clinical significance; (iii) to identify amino acid substitutions known to confer viral resistance to specific antiviral inhibitors.
The reference method for viral genotype determination is phylogenetic analysis of sequences generated after PCR amplification of a portion of the viral genome relative to reference sequences. However, this method only identifies viral variants representing at least 20 to 25% of the circulating viral populations. Reverse hybridization of PCR amplicons to membrane-bound probes is more sensitive than direct sequence analysis to detect minor variants representing as few as 5% of the whole viral population.6 Line probe assays use a series of short immobilized oligonucleotide probes to discriminate among different PCR fragments. Direct sequence analysis and reverse hybridization methods are also used to identify amino acid substitutions known to confer resistance to antiviral drugs before the viral level increases.
Hepatitis B virus infectionCurrent HBV assaysFive serological HBV markers are used in clinical practice, including HBs antigen (HBsAg), anti-HBs antibodies, HBe antigen (HBeAg), anti-HBe antibodies, and anti-HBc antibodies (including total anti-HBc antibodies and anti-HBc IgM). They are detected by means of EIA assays. Two molecular markers are also useful in clinical practice, including the HBV DNA level and the identification of amino acid substitutions conferring HBV resistance to antiviral drugs.
Current HBsAg detection assays detect at least 0.15 nanograms (ng) per milliliter (mL) of circulating HBsAg, i.e. approximately 350 international units (IU) per mL.7 In addition, the ability of HBsAg assays to detect HBV variants bearing nucleotide substitutions in the S gene leading to modifications of the tri-dimensional structure of HBsAg has been improved compared to the previous generations of tests.8–10 The specificity of current HBsAg detection assays is of more than 99.5%. False-positive results can be exceptionally observed in pregnant women, autoimmune diseases, and chronic liver diseases of other causes.10–12 They can also sometimes be observed in hep-arinized, hemolyzed (hemoglobin above 1.4 g/dL) or icteric (bilirubin above 52.8 μmol) blood specimens.11 In practice, it is recommended that the first detection of HB-sAg be confirmed by neutralization in order to eliminate a false-positive result. Under certain circumstances, HB-sAg may not be detectable during chronic hepatitis B: (i) in low-replication asymptomatic HBV carriers; (ii) in the case of HBV variants bearing nucleotide substitutions in the S gene leading to the synthesis of an HBsAg that is not recognized by commercial assays;13,14 (iii) when infection resolves spontaneously or after successful antiviral therapy in chronic HBV-infected patients who may subsequently achieve an HBs seroconversion; (iv) in hepatitis delta HBV-co-infected patients, where hepatitis delta virus most often inhibits HBV replication and ex-pression.13,15
HBsAg quantification can now be performed by means of a fully automated chemiluminescent micropar-ticle immunoassay. HBsAg quantification is easy, cheap and may provide a mean to establish the prognosis of antiviral therapy in the future. HBsAg quantification indeed appears to be a surrogate marker of the amount of covalently closed circular DNA (cccDNA), the persistent intrahepatic form of HBV DNA, in the hepatocytes and a predictor of a sustained virological response to antiviral treatment off therapy.16–18
HBV DNA assays based on real-time PCR technology are now replacing the classical techniques in most virology laboratories. Three real-time PCR platforms are cur-rently available for detection and quantification of HBV DNA: the Cobas Taqman® platform, which can be used together with automated sample preparation with the Cobas AmpliPrep® system (CAP-CTM, Roche Molecular System, Pleasanton, California), the Real-Art® HBV PCR Assay (Artus-Biotech, Qiagen, Hamburg, Germany), and the Abbott platform (Abbott Diagnostic, Chicago Illinois), which uses the m2000RT amplification platform to-gether with the m2000SP device for sample preparation. These assays have been shown to accurately quantify HBV DNA levels in clinical practice.19,20
A commercial line probe assay has been developed for HBV genotype determination (INNO-LiPA HBV Geno-typing, Innogenetics, Gent, Belgium),21 but the utility of HBV genotype determination in clinical practice is debated. The Trugene® HBV Genotyping Kit (Siemens Medical Solutions Diagnotics, Tarrytown, New York) is based on direct sequence analysis of a portion of the reverse transcriptase domain of the HBV polymerase gene and can be used to detect HBV resistance substitutions.22 A new generation of reverse hybridization assay, INNO-LiPA HBV DR v3, (innogenetics) has been developed in 2008 to detect amino acid substitutions associated with lamivudine, adefovir and entecavir resistance.23,24
Practical use of HBV assaysSerological and molecular HBV markers are used in clinical practice to diagnose chronic HBV infection, assess the prognosis of the disease, guide therapy and monitor treatment responses.
Diagnosis of chronic HBV infectionThe persistence of HBsAg for more than 6 months defines chronic HBV carriage. In a chronic HBV carrier, chronic hepatitis B is defined by an elevated serum HBV DNA level with persistent or intermittent elevation of aminotransferase levels and signs of chronic hepatitis B on liver biopsy.25–28 In a chronic HBV carrier, the HBV DNA level should be systematically measured by means of a sensitive and accurate method, ideally based on real-time PCR. The presence of HBeAg (in the absence of anti-HBe antibodies) is generally associated with high-level viral replication and high transmissibil-ity. HBV replication levels are substantially lower on average in HBeAg-negative patients. Inactive HBV carriers have a low level of viral replication (< 2 x 103 IU/ mL), normal aminotransferase levels, no HBeAg and positive anti-HBe antibodies.
Assessment of disease severity and prognosisThe HBV DNA level and aminotransferase activity provide valuable prognostic information. A high HBV DNA level is associated with a significant risk of progres-sion of chronic hepatitis B to cirrhosis and HCC, independently of the HBeAg serostatus and level of ami-notransferase activity.29–31 The risk of HCC is low in the absence of detectable HBV DNA, except in patients with cirrhosis.
Treatment of HBV infectionThe goal of chronic hepatitis B therapy is to prevent progression of liver disease to its life-threatening complications, cirrhosis and HCC. This can be achieved if HBV replication is durably abolished or significantly reduced. In HBeAg-positive patients, HBeAg clearance followed by the HBe seroconversion phase can be achieved in some cases with short-term therapy and ensures long-term control of viral replication. In HBeAg-negative patients, long-term antiviral suppression of viral replication is needed in most cases.32 The loss of HB-sAg, eventually associated with an HBs seroconversion, is the most desirable endpoint of therapy but is rarely achieved.32
Decision to treat. The decision to treat chronic hepatitis B is based on the assessment of multiple parameters including clinical, biological and histologic parameters. Antiviral treatment is currently recommended in patients with an HBV DNA titer above 2,000 IU/mL, elevated serum alanine aminotransferase activity (above the upper limit of normal), and/or evidence of chronic hepatitis with or without cirrhosis.32 Antiviral treatment should be considered in patients with a low level of viral replication who show mild to moderate necro-inflammatory activity and/or fibrosis. In HBV-infected patients with normal aminotransferase activity and an HBV DNA level below 2,000 IU/mL, repeated HBV DNA and ALT determinations are recommended every 3 to 6 months in the absence of treatment.
Selection of optimal therapy. The current treatment of chronic hepatitis B is based on the use of two categories of antiviral compounds: pegylated interferon alpha and nucleos(t)ide analogues that inhibit the reverse tran-scriptase domain of viral polymerase. Five nucleos(t)ide analogues are approved in Europe or in US for the treatment of chronic hepatitis B, including lamivudine, ade-fovir dipivoxil, entecavir, telbivudine and tenofovir dis-oproxil fumarate.
In HBeAg-positive patients, first-line treatment with pegylated interferon alpha is recommended if ami-notransferase levels are elevated and the HBV DNA level is moderate (below 2 x 106 IU/mL).32 Although HBV genotypes A and B globally appear to better respond to in-terferon-based therapy than genotypes C and D, HBV genotype determination is not yet recommended to guide the therapeutic decision in the absence of strong individual predictive value.
Most of the other patients, whatever the HBe serosta-tus, and in HBeAg-positive patients who did not achieve an HBe seroconversion during or after interferon alpha therapy, the use of nucleos(t)ide analogues is recommended. Entecavir or tenofovir must be preferred as first-line treatment because they both potently inhibit HBV replication and they have a high genetic barrier to resis-tance.32 Combinations of nucleos(t)ide analogues with no cross-resistance increases the genetic barrier to resistance and thus better prevents resistance on the long-term than mono-or sequential therapy. De novo combinations can be used as first-line treatment in patients with high HBV DNA levels who are unlikely to reach undetectable HBV DNA on monotherapy.
Treatment monitoring. HBV treatment monitoring is based on HBV DNA quantifications and ALT determinations every 3 to 6 months, whatever the HBe serostatus and antiviral treatment.33
In HBeAg-positive patients, treatment efficacy is witnessed by the loss of HBeAg which may be followed by a seroconversion to anti-HBe antibodies. It is generally associated with a profound reduction of serum HBV DNA levels and normalization of aminotransferase activity. Ideally in HBe seroconverters, HBV DNA should be undetectable with a sensitive real-time PCR assay (lower limit of detection of the order 10 to 15 IU/mL) and aminotransferase activity should be normal.34 If this is not the case, the patient may have switched from HBeAg-positive to HBeAg-negative during therapy and may need sustained therapeutic suppression of viral replication.
In HBeAg-positive patients with no HBe seroconver-sion and in HBeAg-negative patients, the goal of antiviral treatment is to achieve a profound and durable HBV DNA suppression.32 The HBV DNA level should be undetectable on treatment with a sensitive real-time PCR assay (lower limit of detection: 10 to 15 IU/mL).33 If the HBV DNA level remains detectable after 48 weeks, a second antiviral compound with no cross-resistance with the first one must be added in order to prevent subsequent resistance.
In the patients who have responded and have been compliant, resistance should be suspected if the HBV DNA level rises by more than 1 Log10 IU/mL above nadir in two consecutive samples taken one month apart.33,35 The identification of selected amino acid substitutions known to be associated with resistance to the administered drug(s) by means of molecular testing can help guide treatment adaptation. Consensus decisional algorithms will need to be established before systematic ge-notypic resistance testing can be recommended to adapt the treatment strategy to the resistance profile of the infecting viral strain.
Hepatitis C virus infectionCurrent HCV assaysThree HCV markers are useful in clinical practice, including total anti-HCV antibodies, HCV genotype and HCV RNA.
Direct sequence analysis is the gold standard for ge-nomic sequence analysis. The Trugene® 5’NC HCV Genotyping kit (Siemens Medical Solutions Diagnostics) has been developed for HCV genotype determination by direct sequencing of a portion of the 5’ non-coding region (NCR) of the viral genome. However, it only identifies viral variants representing at least 20 to 25% of the circulating viral populations. The most recent version of the line probe assay for HCV genotype determination (VERSANT® HCV Genotype 2.0 Assay, Siemens Medical Solutions Diagnostics) bears consistently improved accuracy for HCV genotype 1 subtype and HCV genotype 6 determination compared to the previous assays.36–39 The HCV genotype can also be determined by means of a competitive ELISA assay using genotype-specific antigens.40 This assay allows to identify the 6 HCV genotypes (1 to 6) but not the subtype.
HCV RNA assays based on real time PCR are now used in clinical virology laboratories for RNA detection and quantification. Two real-time PCR platforms are cur-rently available for detection and quantification of HCV RNA: the Cobas Taqman® platform, which can be used together with automated sample preparation with the Cobas AmpliPrep® system (CAP-CTM, Roche Molecular Systems), and the Abbott platform (Abbott Diagnostic), which uses the m2000RT amplification platform together with the m2000Sp device for sample preparation. Another assay, developed by Siemens Medical Solutions Diagnostics will be available soon. The intrinsic performances of available tests differ. Indeed, approximately 15% of HCV genotype 2 and 30% of HCV genotype 4 samples are substantially underestimated in the CAP-CTM, most likely because of nucleotide mismatches, whereas this problem has not been found with the Abbott assay.41,42
Practical use of HCV assaysSerological and molecular markers are used in clinical practice to diagnose chronic hepatitis C, guide treatment decisions and monitor the antiviral efficacy of treatment.
Diagnosis of chronic HCV infectionIn patients with clinical and/or biological signs of chronic liver disease, chronic hepatitis C is diagnosed by the simultaneous presence of anti-HCV antibodies and HCV RNA. Detectable HCV replication in the absence of anti-HCV antibodies is exceptional with current anti-HCV enzyme immunoassays. It is almost exclusively observed in profoundly immunodepressed, hemodialysis or agamaglobulinemic subjects.43,44
Treatment of HCV infectionThe current standard treatment for chronic hepatitis C is the combination of pegylated interferon alpha-2a or -2b and ribavirin.45 The efficacy endpoint of chronic hepatitis C treatment is the sustained virological response (SVR), defined by an undetectable HCV RNA in serum with a sensitive assay (lower limit of detection of 10-50 IU/mL) 24 weeks after the end of treatment.
Decision to treat and indication of treatment. The decision to treat chronic hepatitis C depends on multiple parameters including a precise assessment of the severity of liver disease, the presence of absolute or relative con-tra-indications to therapy, and the patient’s willingness to be treated.
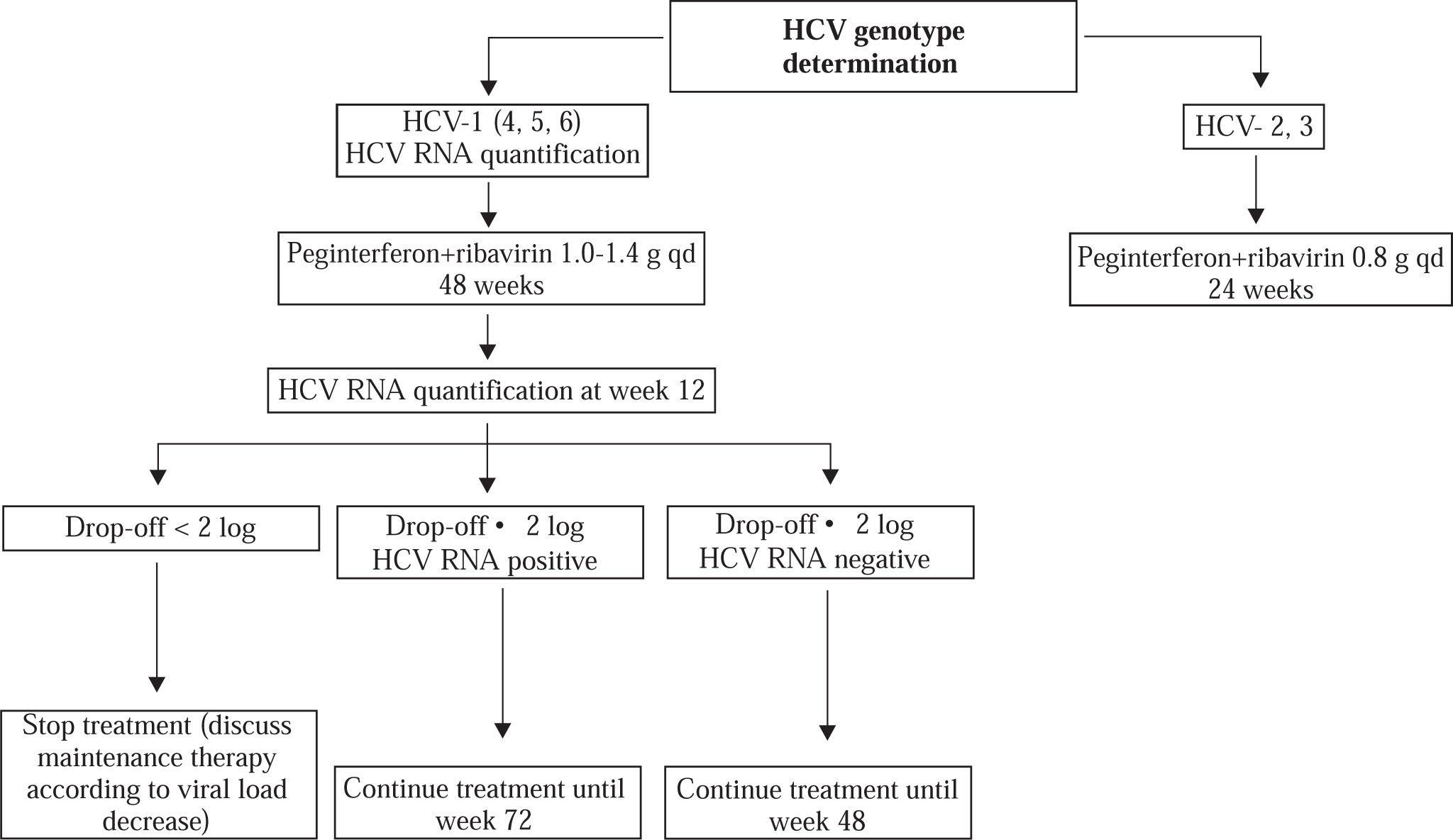
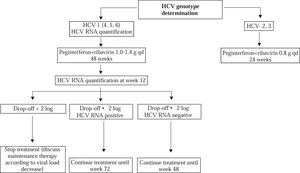
HCV genotype determination should be systematically performed before treatment, as it determines the indication, the duration of treatment, the dose of ribavirin, and the virological monitoring procedure (Figure 1).46 Genotypes 2-and 3-infected patients require 24 weeks of treatment and a low dose of ribavirin, i.e. 800 mg daily. In contrast, genotype 1-, 4-, 5-and 6-infected patients require 48 weeks of treatment and a high, body-weight based dose of ribavirin, i.e. 1,000 to 1,400 mg daily.
Treatment monitoring. Monitoring of HCV RNA levels is recommended to tailor treatment to the actual viro-logical response. A real-time PCR assay should ideally be used. In HCV genotype 1-infected patients, the HCV RNA level should be measured before therapy and 12 weeks after its initiation (Figure 1). The lack of a 12-week virological response (i.e. no change or an HCV RNA decrease of less than 2 Log10 at week 12) indicates that the patient has virtually no chance to achieve an SVR and should stop treatment. In contrast, treatment must be continued when a 2 Log10 drop in HCV RNA level has been observed at week 12, until week 48 if HCV RNA is undetectable, until week 72 if HCV RNA is still detectable at week 12.14,47
Recent studies have suggested that the patients who achive a rapid virological response, defined by an unde-tectable HCV RNA (< 50 IU/mL) at week 4 of therapy, could benefit from shorter treatment duration, i.e. 24 weeks in patients infected with HCV genotypes 1, 4, 5 or 6.48–53 These results however need confirmation and new algorithms should be drawn to tailor treatment duration to the virological response at week 4 without loosing a chance of viral eradication.
The sustained virological response corresponds to a cure of infection in more than 99% of cases. In the midterm future, triple combination therapy with pegylated interferon alpha, ribavirin and a specific HCV inhibitor will likely become the standard treatment of chronic hepatitis C. The SVR will remain the endpoint of therapy. On-treatment monitoring and the corresponding decision algorithms will need to be established.